Norditerpenoids with Selective Anti-Cholinesterase Activity from the Roots of Perovskia atriplicifolia Benth.
Abstract
:1. Introduction
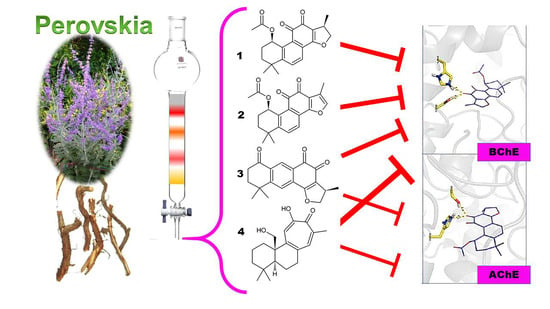
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Chemicals
3.3. Plant Material
3.4. Isolation Procedure of Compounds 1–4
3.5. Identification of Compounds 1–4
3.6. Microtiter Assays for AChE and BChE Enzyme Inhibition
3.7. Data Processing for Enzyme Inhibition Assays
3.8. Statistical Analysis of Data
3.9. Molecular Modeling
3.10. Computational Prediction of ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) Properties
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AChE | acetylcholinesterase |
| AD | Alzheimer’s disease |
| Aβ | amyloid plaques |
| BChE | butyrylcholinesterase |
| CAS | catalytic site |
| DFT | density functional theory |
| ECD | electronic circular dichroism |
| FA | Formic acid |
| GR | glucocorticoid receptor |
| HRQTOF-MS | high-resolution quadrupole time-of-flight mass spectrometer |
| MeOH-d4 | deuterated methanol |
| PAS | peripheral anionic site |
| PDB | Protein Data Bank |
References
- Erdogan Orhan, I.; Orhan, G.; Gurkas, E. An overview on natural cholinesterase inhibitors—A multi-targeted drug class—And their mass production. Mini-Rev. Med. Chem. 2011, 11, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, I.R.; Maxwell, S.P.; Reid, G.A.; Cash, M.K.; DeBay, D.R.; Darvesh, S. Quantification of butyrylcholinesterase activity as a sensitive and specific biomarker of Alzheimer’s disease. J. Alzheimer’s Dis. 2017, 58, 491–505. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Duysen, E.G.; Carlson, M.; Lockridge, O.; Rizzino, A.; McComb, R.D.; Taylor, P.; Hinrichs, S.H.; Lockridge, O. The butyrylcholinesterase knockout mouse as a model for human butyrylcholinesterase deficiency. J. Pharmacol. Exp. Ther. 2008, 324, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Makin, S. The amyloid hypothesis on trial. Nature 2018, 559, S4–S7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tundis, R.; Bonesi, M.; Menichini, F.; R Loizzo, M. Recent knowledge on medicinal plants as source of cholinesterase inhibitors for the treatment of dementia. Mini-Rev. Med. Chem. 2016, 16, 605–618. [Google Scholar] [CrossRef]
- Ashare, R.L.; Kimmey, B.A.; Rupprecht, L.E.; Bowers, M.E.; Hayes, M.R.; Schmidt, H.D. Repeated administration of an acetylcholinesterase inhibitor attenuates nicotine taking in rats and smoking behavior in human smokers. Transl. Psychiatry 2016, 6, e713. [Google Scholar] [CrossRef] [PubMed]
- Ashare, R.L.; Kimmey, B.A.; Rupprecht, L.E.; Bowers, M.E.; Hayes, M.R.; Schmidt, H.D. Repeated administration of an acetylcholinesterase inhibitor attenuates nicotine taking in rats and smoking behavior in human smokers. Transl. Psychiatry 2017, 7, e1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casida, J.E.; Durkin, K.A. Neuroactive insecticides: Targets, selectivity, resistance, and secondary effects. Annu. Rev. Entomol. 2013, 58, 99–117. [Google Scholar] [CrossRef]
- Bhavya, M.L.; Chandu, A.G.S.; Devi, S.S. Ocimum tenuiflorum oil, a potential insecticide against rice weevil with anti-acetylcholinesterase activity. Ind. Crops Prod. 2018, 126, 434–439. [Google Scholar] [CrossRef]
- Politeo, O.; Bektašević, M.; Carev, I.; Jurin, M.; Roje, M. Phytochemical composition, antioxidant potential and cholinesterase inhibition potential of extracts from Mentha pulegium L. Chem. Biodivers. 2018, 15, e1800374. [Google Scholar] [CrossRef]
- Chen, X.; Guo, J.; Bao, J.; Lu, J.; Wang, Y. The anticancer properties of Salvia miltiorrhiza Bunge (Danshen): A systematic review. Med. Res. Rev. 2014, 34, 768–794. [Google Scholar] [CrossRef]
- Wu, Z.; Song, L.; Liu, S.Q.; Huang, D. Tanshinones extend chronological lifespan in budding yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2014, 98, 8617–8628. [Google Scholar] [CrossRef]
- Buenafe, O.E.; Orellana-Paucar, A.; Maes, J.; Huang, H.; Ying, X.; De Borggraeve, W.; Crawford, A.D.; Luyten, W.; Esguerra, C.V.; de Witte, P. Tanshinone IIA exhibits anticonvulsant activity in zebrafish and mouse seizure models. ACS Chem. Neurosci. 2013, 4, 1479–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.-H.; Li, Y.-R.; Jiao, P.; Zhao, Y.; Hu, H.-X.; Lou, H.-X.; Shen, T. Therapeutic potential of Salviae miltiorrhizae radix et rhizoma against human diseases based on activation of Nrf2-mediated antioxidant defense system: Bioactive constituents and mechanism of action. Oxid. Med. Cell. Longev. 2018, 2018, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.; Wang, H.; Li, S.; Xu, H. The effect of sodium tanshinone IIA sulfate and simvastatin on elevated serum levels of inflammatory markers in patients with coronary heart disease: A study protocol for a randomized controlled trial. Evid.-Based Complement. Alternat. Med. 2013, 2013, 756519. [Google Scholar] [CrossRef]
- Li, S.; Jiao, Y.; Wang, H.; Shang, Q.; Lu, F.; Huang, L.; Liu, J.; Xu, H.; Chen, K. Sodium tanshinone IIA sulfate adjunct therapy reduces high-sensitivity C-reactive protein level in coronary artery disease patients: A randomized controlled trial. Sci. Rep. 2017, 7, 17451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drew, B.; González-Gallegos, J.G.; Xiang, C.-L.; Kriebel, R.; Drummond, C.; Walker, J.; Sytsma, K. Salvia united: The greatest good for the greatest number. Taxon 2017, 66, 133–145. [Google Scholar] [CrossRef] [Green Version]
- Slusarczyk, S.; Topolski, J.; Domaradzki, K.; Adams, M.; Hamburger, M.; Matkowski, A. Isolation and fast selective determination of nor-abietanoid diterpenoids from Perovskia atriplicifolia roots using LC-ESI-MS/MS with multiple reaction monitoring. Nat. Prod. Commun. 2015, 10, 1149–1152. [Google Scholar] [CrossRef]
- Ren, Y.; Houghton, P.J.; Hider, R.C.; Howes, M.-J.R. Novel diterpenoid acetylcholinesterase inhibitors from Salvia miltiorrhiza. Planta Med. 2004, 70, 201–204. [Google Scholar]
- Senol, F.S.; Ślusarczyk, S.; Matkowski, A.; Pérez-Garrido, A.; Girón-Rodríguez, F.; Cerón-Carrasco, J.P.; den-Haan, H.; Peña-García, J.; Pérez-Sánchez, H.; Domaradzki, K.; et al. Selective in vitro and in silico butyrylcholinesterase inhibitory activity of diterpenes and rosmarinic acid isolated from Perovskia atriplicifolia Benth. and Salvia glutinosa L. Phytochemistry 2017, 133, 33–44. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, W.; Xu, L.; Chen, L. In Salvia miltiorrhiza, phenolic acids possess protective properties against amyloid β-induced cytotoxicity, and tanshinones act as acetylcholinesterase inhibitors. Environ. Toxicol. Pharmacol. 2011, 31, 443–452. [Google Scholar] [CrossRef]
- Jiang, H.-L.; Wang, X.-Z.; Xiao, J.; Luo, X.-H.; Yao, X.-J.; Zhao, Y.-Y.; Chen, Y.-J.; Crews, P.; Wu, Q.-X. New abietane diterpenoids from the roots of Salvia przewalskii. Tetrahedron 2013, 69, 6687–6692. [Google Scholar] [CrossRef]
- Kang, J.; Li, L.; Wang, D.; Wang, H.; Liu, C.; Li, B.; Yan, Y.; Fang, L.; Du, G.; Chen, R. Isolation and bioactivity of diterpenoids from the roots of Salvia grandifolia. Phytochemistry 2015, 116, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Sairafianpour, M.; Christensen, J.; Stærk, D.; Budnik, B.A.; Kharazmi, A.; Bagherzadeh, K.; Jaroszewski, J.W. Leishmanicidal, antiplasmodial, and cytotoxic activity of novel diterpenoid 1,2-quinones from Perovskia abrotanoides: New source of tanshinones. J. Nat. Prod. 2001, 64, 1398–1403. [Google Scholar] [CrossRef]
- Michavila, A.; De La Torre, M.C.; Rodríguez, B. 20-Nor-abietane and rearranged abietane diterpenoids from the root of Salvia argentea. Phytochemistry 1986, 25, 1935–1937. [Google Scholar] [CrossRef]
- Sabri, N.N.; Abou-Donia, A.A.; Ghazy, N.M.; Assad, A.M.; El-Lakany, A.M.; Sanson, D.R.; Gracz, H.; Barnes, C.L.; Schlemper, E.O.; Tempesta, M.S. Two new rearranged abietane diterpene quinones from Salvia aegyptiaca L. J. Org. Chem. 1989, 54, 4097–4099. [Google Scholar] [CrossRef]
- Danheiser, R.L.; Casebier, D.S.; Huboux, A.H. Total synthesis of aegyptinones A and B. J. Org. Chem. 1994, 59, 4844–4848. [Google Scholar] [CrossRef]
- Tabefam, M.; Farimani, M.; Danton, O.; Ramseyer, J.; Kaiser, M.; Ebrahimi, S.; Salehi, P.; Batooli, H.; Potterat, O.; Hamburger, M. Antiprotozoal diterpenes from Perovskia abrotanoides. Planta Med. 2018, 84, 913–919. [Google Scholar] [CrossRef]
- Eghbaliferiz, S.; Emami, S.A.; Tayarani-Najaran, Z.; Iranshahi, M.; Shakeri, A.; Hohmann, J.; Asili, J. Cytotoxic diterpene quinones from Salvia tebesana Bunge. Fitoterapia 2018, 128, 97–101. [Google Scholar] [CrossRef]
- Ahmed, F.; Ghalib, R.M.; Sasikala, P.; Ahmed, K.K.M. Cholinesterase inhibitors from botanicals. Pharmacogn. Rev. 2013, 7, 121–130. [Google Scholar] [CrossRef] [Green Version]
- Guzior, N.; Więckowska, A.; Panek, D.; Malawska, B. Recent development of multifunctional agents as potential drug candidates for the treatment of Alzheimer’s disease. Curr. Med. Chem. 2014, 22, 373–404. [Google Scholar] [CrossRef] [PubMed]
- Nachon, F.; Asojo, O.A.; Borgstahl, G.E.O.; Masson, P.; Lockridge, O. Role of water in aging of human butyrylcholinesterase inhibited by echothiophate: The crystal structure suggests two alternative mechanisms of aging. Biochemistry 2005, 44, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Skalicka-Wozniak, K.; Orhan, I.E.; Deniz, F.S.; Trædal-Henden, S.; Cerón-Carrasco, J.; den-Haan, H.; Peña-García, J.; Pérez-Sánchez, H.; Emerce, E. Profiling auspicious butyrylcholinesterase inhibitory activity of two herbal molecules: Hyperforin and hyuganin C. Chem. Biodivers. 2019, 16, e1900017. [Google Scholar]
- Orhan, I.E.; Jedrejek, D.; Sezer Senol, F.; Ekhteiari Salmas, R.; Durdagi, S.; Kowalska, I.; Pecio, L.; Oleszek, W. Molecular modeling and in vitro approaches towards cholinesterase inhibitory effect of some natural xanthohumol, naringenin, and acyl phloroglucinol derivatives. Phytomedicine 2018, 42, 25–33. [Google Scholar] [CrossRef]
- Canet, G.; Chevallier, N.; Zussy, C.; Desrumaux, C.; Givalois, L. Central role of glucocorticoid receptors in Alzheimer’s disease and Depression. Front. Neurosci. 2018, 12, 739. [Google Scholar] [CrossRef] [Green Version]
- Ellman, G.L.; Courtney, K.D.; Andres, V.; Featherstone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Senol, F.S.; Skalicka Woźniak, K.; Khan, M.T.H.; Erdogan Orhan, I.; Sener, B.; Głowniak, K. An in vitro and in silico approach to cholinesterase inhibitory and antioxidant effects of the methanol extract, furanocoumarin fraction, and major coumarins of Angelica officinalis, L. fruits. Phytochem. Lett. 2011, 4, 462–467. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [Green Version]
- Singh, U.C.; Kollman, P.A. An approach to computing electrostatic charges for molecules. J. Comput. Chem. 1984, 5, 129–145. [Google Scholar] [CrossRef]
- Besler, B.H.; Merz, K.M.; Kollman, P.A. Atomic charges derived from semiempirical methods. J. Comput. Chem. 1990, 11, 431–439. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision, B.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stierand, K.; Rarey, M. PoseView—Molecular interaction patterns at a glance. J. Cheminf. 2010, 2, P50. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, 257–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drwal, M.N.; Banerjee, P.; Dunkel, M.; Wettig, M.R.; Preissner, R. ProTox: A web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014, 42, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. AdmetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inform. Mod. 2012, 52, 3099–3105. [Google Scholar] [CrossRef]
- Preissner, S.; Kroll, K.; Dunkel, M.; Senger, C.; Goldsobel, G.; Kuzman, D.; Guenther, S.; Winnenburg, R.; Schroeder, M.; Preissner, R. SuperCYP: A comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucleic Acids Res. 2010, 38, 237–243. [Google Scholar] [CrossRef] [PubMed]





| Position | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δC type | δH (J in Hz) | δC type | δH (J in Hz) | δC type | δH (J in Hz) | |
| 1 | 68.7, CH | 6.40, t (3.4) | 69.0, CH | 6.41, t (3.4) | 201.8, C | |
| 2 | 25.5, CH2 | 2.13–2.19, m 2.02, tdd (14.0, 3.5, 2.5) | 25.6, CH2 | 2.13–2.19, m 2.01, tt (14.1, 2.6) | 37.3, CH2 | 2.90, t (7.2) |
| 3 | 33.1, CH2 | 1.93, td (13.3, 2.6) 1.59, ddd (13.3, 5.5, 2.5) | 33.2, CH2 | 1.93, td (13.1, 2.4) 1.58, ddd (12.9, 5.6, 2.8) | 37.5, CH2 | 2.10, t (7.2) |
| 4 | 35.9, C | 35.8, C | 36.4, C | |||
| 5 | 154.2, C | 151.9, C | 138.7, C | |||
| 6 | 134.9, CH | 7.84, d (7.7) | 135.7, CH | 7.89, d (8.3) | 157.9, C | |
| 7 | 126.4, CH | 7.67, d (7.8) | 124.2, CH | 7.80, d (8.3) | 131.7, CH | 7.78 |
| 8 | 128.3, C | 129.6, C | 128.6, C | |||
| 9 | 130.1, C | 128.2, C | 134.2, C | |||
| 10 | 138.1, C | 138.7, C | 128.2, CH | 7.78 | ||
| 11 | 184.5, C | 183.8, C | 184.9, C | |||
| 12 | 176.3, C | 176.3, C | 178.9, C | |||
| 13 | 119.6, C | 121.1, C | 120.4, C | |||
| 14 | 172.7, C | 162.7, C | 172.0, C | |||
| 15 | 35.7, CH | 3.48–3.58, m | 122.3, C | 35.8, CH | 3.54–3.64, m | |
| 16 | 83.2, CH2 | α 4.96, t (9.4) β 4.42, dd (9.3, 6.2) | 143.6, CH | 7.48, q (1.3) | 83.6, CH2 | α 5.01, t (9.6) β 4.48, dd (9.5, 6.2) |
| 17 | 18.7, CH3 | 1.32, d (6.8) | 8.8, CH3 | 2.23, d (1.2) | 18.8, CH3 | 1.347, d (6.9) |
| 18 | 31.3, CH3 | 1.28, s | 31.2, CH3 | 1.31 s | 28.9, CH3 | 1.352, s |
| 19 | 31.9, CH3 | 1.39, s | 31.9, CH3 | 1.41, s | 28.9, CH3 | 1.350, s |
| 1-O-CO-Me | 171.8 | 172.2 | ||||
| 1-O-CO-Me | 21.0 | 1.99, s | 21.0 | 2.01, s | ||
| Compound Number | Compound | % Inhibition ± S.D. at 10.0 µg·mL−1 | BChE Inhibition IC50 | Ki app * | Content in Dried Roots | ||
|---|---|---|---|---|---|---|---|
| AChE | BChE | µg·mL−1 | µM | µM | mg (100 g)−1 | ||
| 1 | (1R,15R)-1-Acetoxycryptotanshinone | 22.8 ± 2.4 | 95.9 ± 0.0 | 0.84 ± 0.09 | 2.37 | 1.34 | 28.5 ± 2.5 |
| 2 | (1R)-1-Acetoxytanshinone IIA | 28.0 ± 0.9 | 85.3 ± 4.3 | 2.77 ± 0.48 | 7.86 | 4.59 | 8.1 ± 0.4 |
| 3 | (15R)-1-oxoaegyptinone A | 49.6 ± 1.8 | 87.3 ± 1.0 | 15.75 ± 1.12 | 50.80 | 30.0 | 21.3 ± 0.7 |
| 4 | Isograndifoliol | 50.0 ± 1.8 | 98.6 ± 0.0 | 0.27 ± 0.02 | 0.89 | 0.47 | 302.0 ± 9.1 |
| REF ** | Galanthamine hydrobromide | 97.2 ± 2.9 | 86.8 ± 2.9 | 28.16 ± 1.51 | 76.4 | --- | -------------- |
| Compound Name | Predicted LD50 Value and Tox class | Prediction Accuracy (%) | Toxicity Targets | Avg Similarity to Known Ligands (%) | Toxicity Endpoints | Prediction Probability | Cytochrome Inhibition Prediction |
|---|---|---|---|---|---|---|---|
| 1. Acetoxycrypto-tanshinone | 260 (mg/kg), Tox class: 3 | 54.26 | - | - | Immunotoxicity | 0.99 | - |
| 2. Acetoxytanshinone IIA | 1230 (mg/kg), Tox class: 4 | 54.26 | - | - | Immunotoxicity | 0.83 | CYP2C9 (62%) |
| 3. 1-oxoaegyptinone A | 260 (mg/kg), Tox class: 3 | 54.26 | - | - | Immunotoxicity | 0.96 | - |
| 4. Isograndifoliol | 2000 (mg/kg) Tox class: 4 | 68.27 | Glucocorticoid Receptor | 73.25 | - | - | CYP3A4 (78%) |
| Progesterone Receptor | 72.37 | CYP2C9 (63%) |
| Compounds | Blood-Brain Barrier | Probability | Water Solubility (logS) | Lipophilicity (AlogP) | Plasma Protein Binding (100 %) | Human Intestinal Absorption | Probability |
|---|---|---|---|---|---|---|---|
| 1. Acetoxycryptotanshinone | positive | 0.97 | −4.43 | 3.51 | 0.89 | positive | 0.99 |
| 2. Acetoxytanshinone IIA | positive | 0.93 | −4.42 | 4.31 | 1.06 | positive | 0.98 |
| 3. oxoaegyptinone A | positive | 0.96 | −3.78 | 3.08 | 0.95 | positive | 0.99 |
| 4. Isograndifoliol | positive | 0.93 | −4.28 | 3.06 | 0.82 | positive | 0.99 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ślusarczyk, S.; Senol Deniz, F.S.; Abel, R.; Pecio, Ł.; Pérez-Sánchez, H.; Cerón-Carrasco, J.P.; den-Haan, H.; Banerjee, P.; Preissner, R.; Krzyżak, E.; et al. Norditerpenoids with Selective Anti-Cholinesterase Activity from the Roots of Perovskia atriplicifolia Benth. Int. J. Mol. Sci. 2020, 21, 4475. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124475
Ślusarczyk S, Senol Deniz FS, Abel R, Pecio Ł, Pérez-Sánchez H, Cerón-Carrasco JP, den-Haan H, Banerjee P, Preissner R, Krzyżak E, et al. Norditerpenoids with Selective Anti-Cholinesterase Activity from the Roots of Perovskia atriplicifolia Benth. International Journal of Molecular Sciences. 2020; 21(12):4475. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124475
Chicago/Turabian StyleŚlusarczyk, Sylwester, F. Sezer Senol Deniz, Renata Abel, Łukasz Pecio, Horacio Pérez-Sánchez, José P. Cerón-Carrasco, Helena den-Haan, Priyanka Banerjee, Robert Preissner, Edward Krzyżak, and et al. 2020. "Norditerpenoids with Selective Anti-Cholinesterase Activity from the Roots of Perovskia atriplicifolia Benth." International Journal of Molecular Sciences 21, no. 12: 4475. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124475