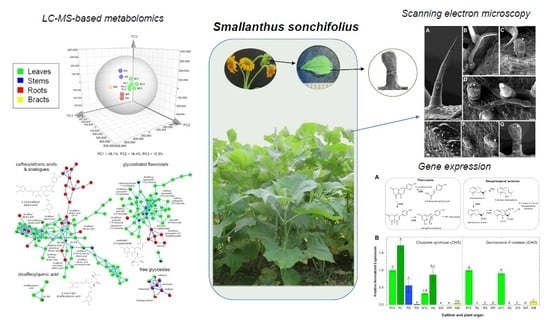
Metabolomic and Gene Expression Studies Reveal the Diversity, Distribution and Spatial Regulation of the Specialized Metabolism of Yacón (Smallanthus sonchifolius, Asteraceae)
Abstract
:1. Introduction
2. Results and Discussion
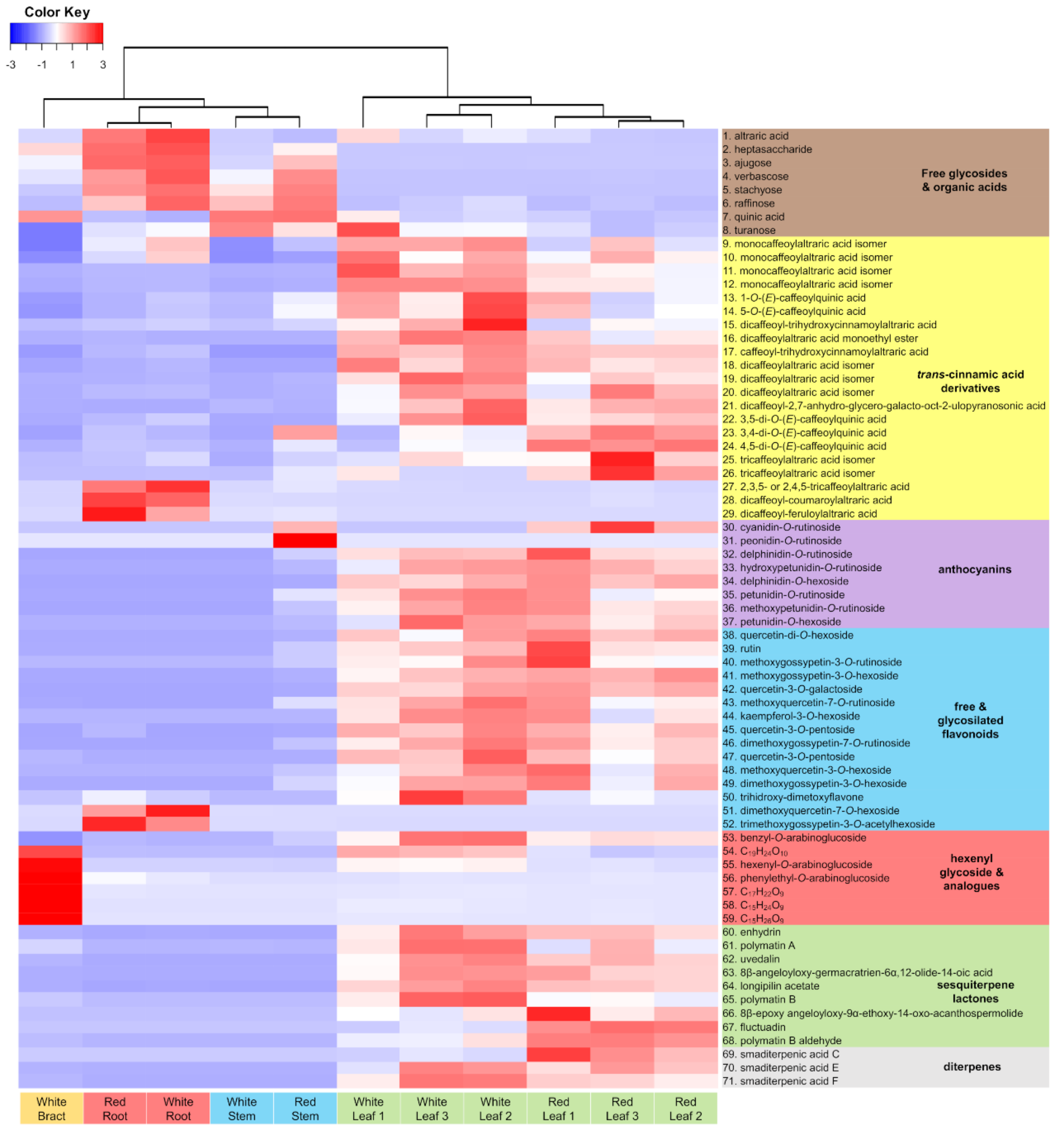
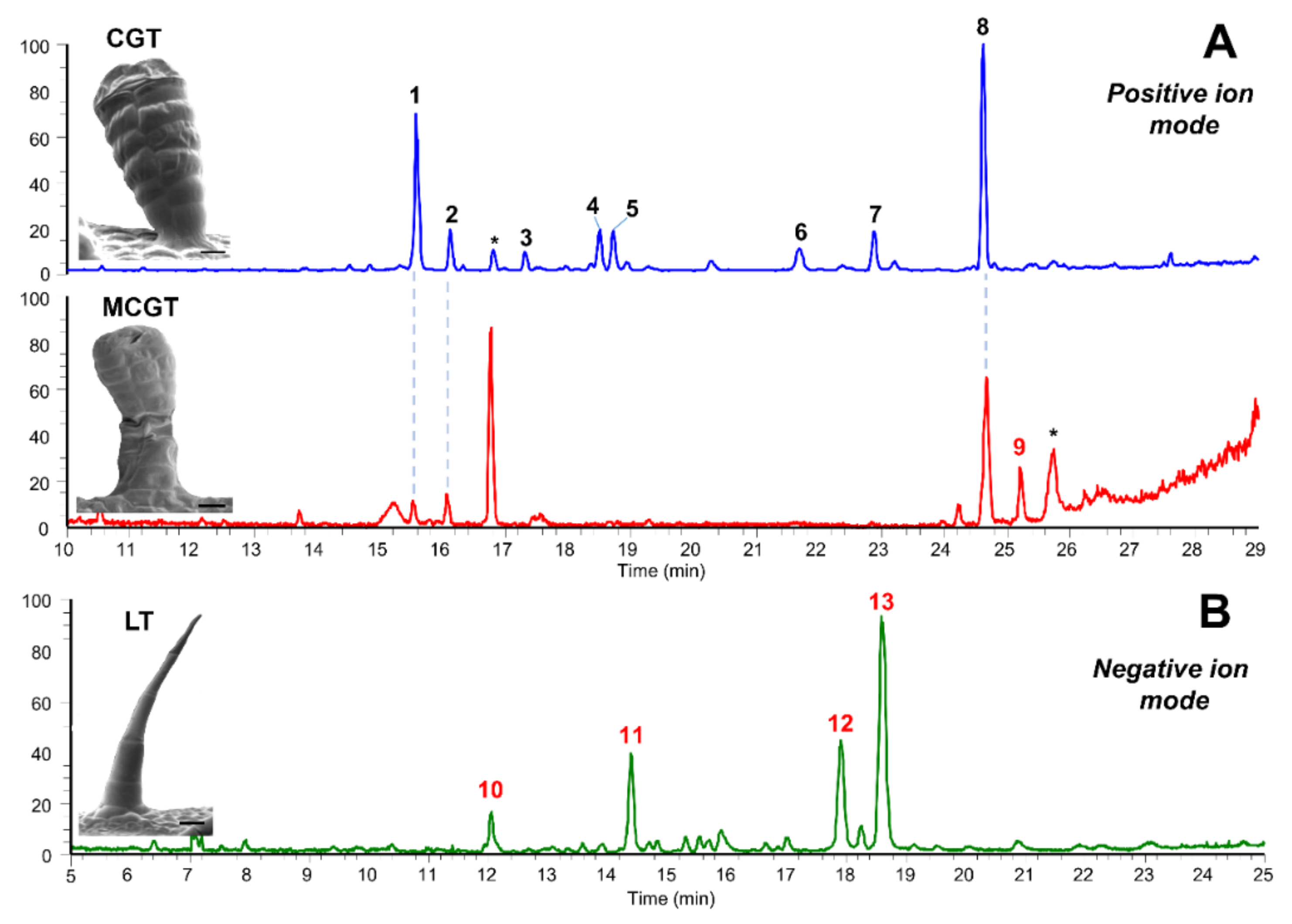
2.1. Metabolic Fingerprinting of Different Organs and Cultivars
2.2. Gene Expression Patterns in Different Organs and Cultivars
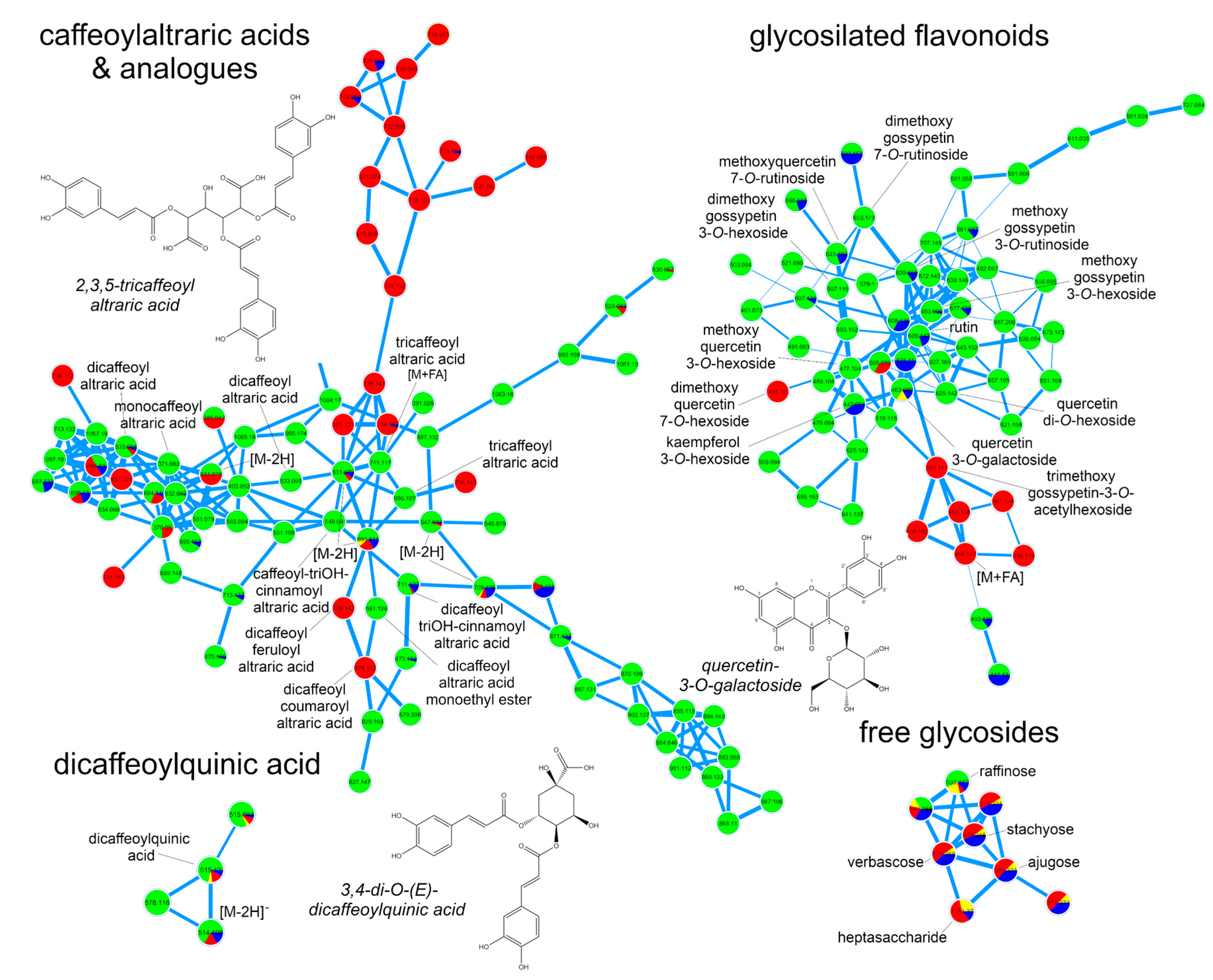
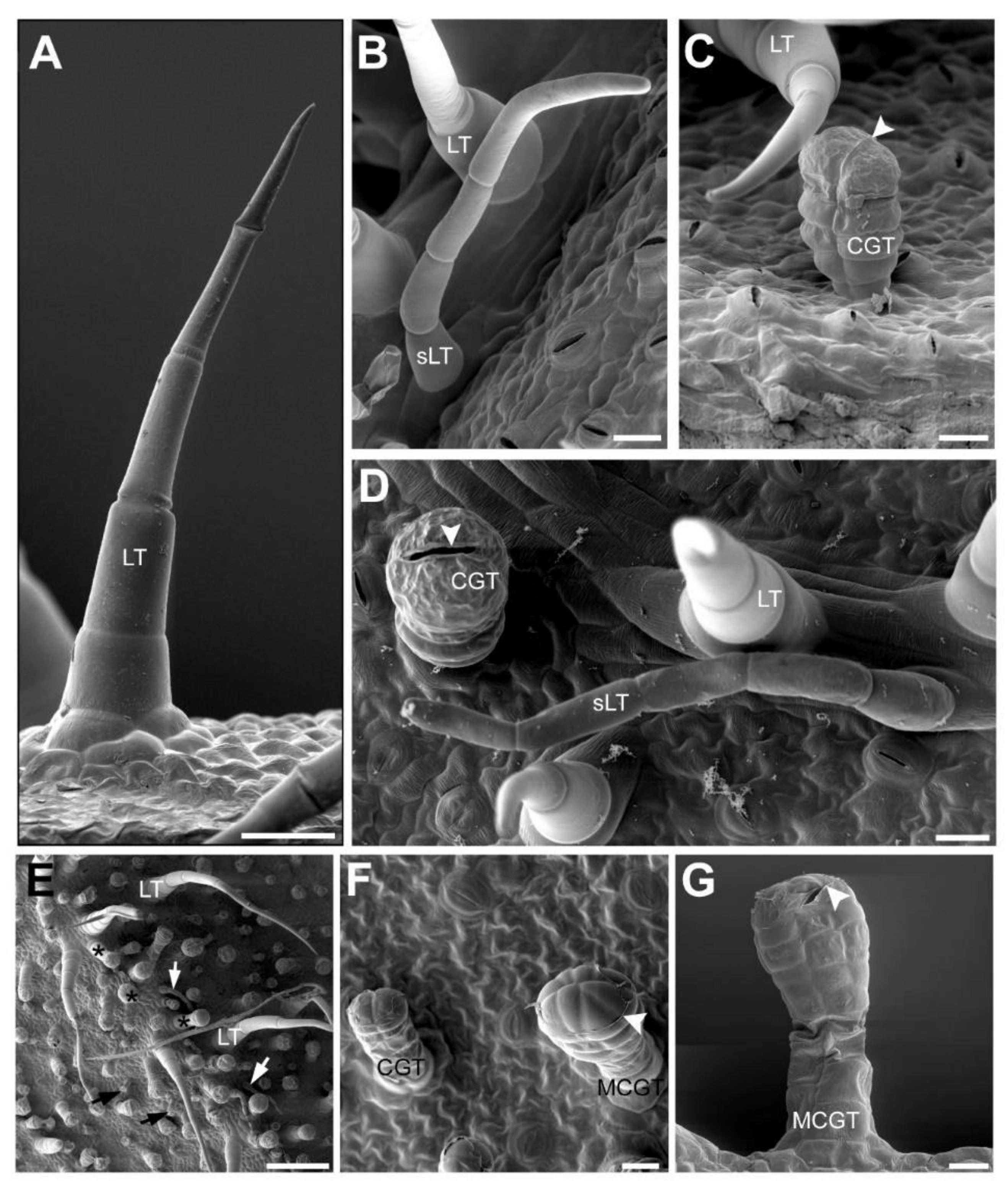
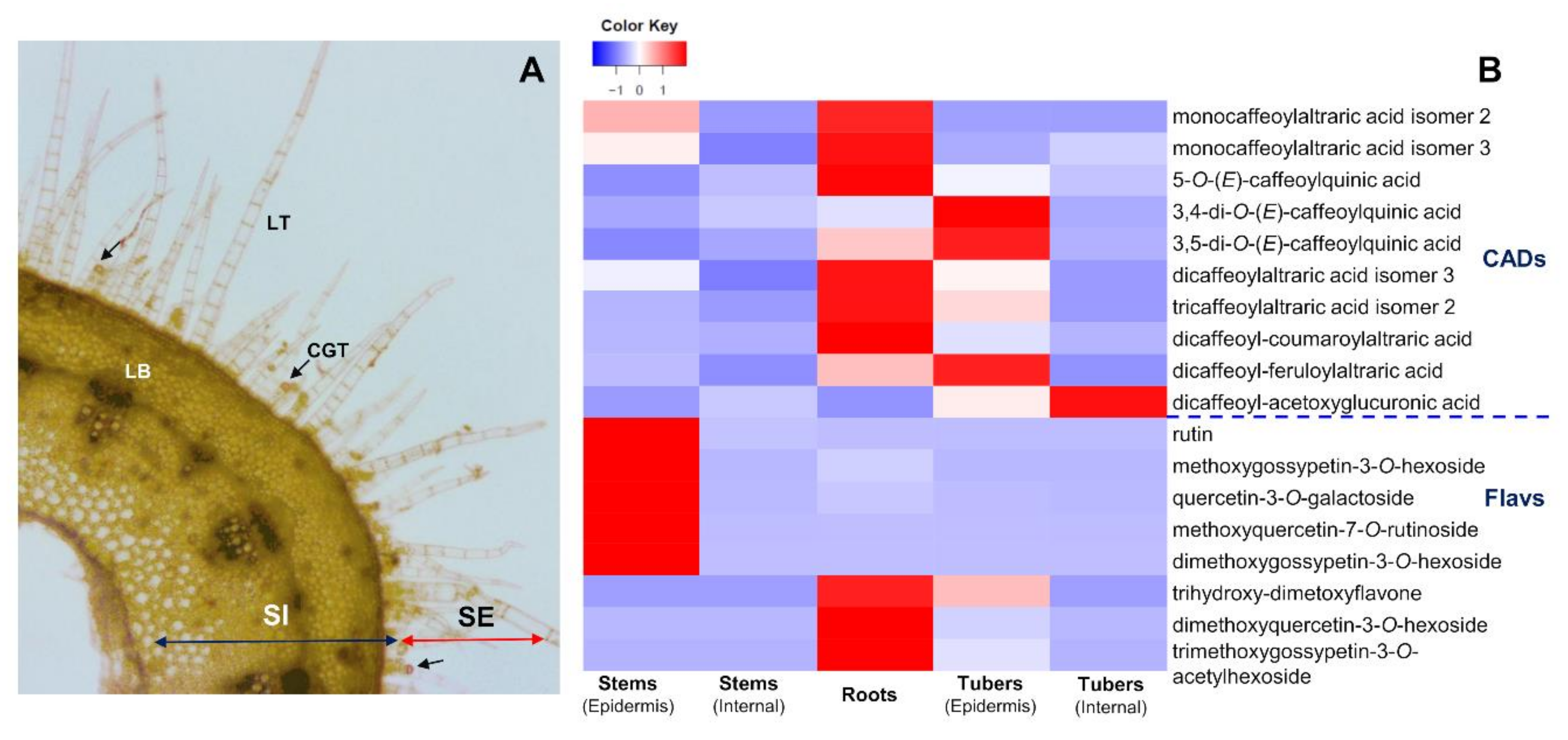
2.3. Accumulation Patterns of Specialized Metabolites in Specific Organs and Tissues
3. Materials and Methods
3.1. Plant Material
3.2. Microscopic Analyses
3.3. Metabolite Extraction and UHPLC-UV-HRMS/MS Analysis
3.4. Data Preprocessing and Multivariate Analyses
3.5. Dereplication of Plant Extracts
3.6. Isolation of Tricaffeoylaltraric Acid
3.7. RNA Extraction and RT-qPCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SM | Specialized metabolism |
| STLs | Sesquiterpene lactones |
| CADs | trans-Cinnamic acid derivatives |
| CHS | Chalcone synthase |
| GAO | Germacrene A oxidase |
| UHPLC-UV-HRMS/MS | Ultra-high-performance liquid chromatography coupled to UV detection and high-resolution tandem mass spectrometry |
| PCA | Principal component analysis |
| OPLS-DA | Orthogonal projections to latent structures discriminant analysis |
| ANS | Anthocyanidin synthase |
| HaG8H | Helianthus annuus GAA 8β-hydroxylase CYP71BL1 |
| HaES | Helianthus annuus eupatolide synthase |
| CGT | Capitate glandular trichome |
| LT | Linear trichome |
| sLT | Small linear trichome previously referred as “flexuous type” |
| MCGT | Multiseriate capitate glandular trichome |
| CPD | #critical point drying |
| PDA | Photodiode array detector |
| AGC | Automatic gain control |
| DNP | Dictionary of Natural Products |
| Ha_qACT | Helianthus annus qPCR actin primers |
| Ha_qEF | Helianthus annus qPCR elongation factor primers |
| Ha_qGAPDH | Helianthus annus qPCR glycerin aldehyde phosphate dehydrogenase primers |
References
- Fernandez, C.E.; Viehmannova, I.; Bechyně, M.; Lachman, J.; Milella, L.; Martelli, G. The cultivation and phenological growth stages of yacon [Smallanthus sonchifolius (Poepp. et Endl.) H. Robinson. Agric. Trop. Subtrop. 2007, 40, 71–77. [Google Scholar]
- De la Gonzales, M.C.; Baldeón, S.M.; Santiago, H.B.; Jullian, V.; Bourdy, G. Hot and cold: Medicinal plant uses in Quechua speaking communities in the high Andes (Callejón de Huaylas, Ancash, Perú). J. Ethnopharmacol. 2014, 155, 1093–1117. [Google Scholar] [CrossRef] [PubMed]
- Abrams, S.A.; Hawthorne, K.M.; Aliu, O.; Hicks, P.D.; Chen, Z.; Griffin, I.J. An inulin-type fructan enhances calcium absorption primarily via an effect on colonic absorption in humans. J. Nutr. 2007, 137, 2208–2212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yu, H.; Xiao, X.; Hu, L.; Xin, F.; Yu, X. Inulin-type fructan improves diabetic phenotype and gut microbiota profiles in rats. PeerJ 2018, 6, e4446. [Google Scholar] [CrossRef] [PubMed]
- Causey, J.L.; Feirtag, J.M.; Gallaher, D.D.; Tungland, B.C.; Slavin, J.L. Effects of dietary inulin on serum lipids, blood glucose and the gastrointestinal environment in hypercholesterolemic men. Nutr. Res. 2000, 20, 191–201. [Google Scholar] [CrossRef]
- Terada, S.; Itoh, K.; Noguchi, N.; Ishida, T. Alpha-Glucosidase Inhibitor for Blood Glucose Level Elevation and Functional Food Containing Tricaffeoylaldaric Acid and Method for Producing Tricaffeoylaldaric Acid. U.S. Patent 20090209649A1, 20 August 2009. [Google Scholar]
- Russo, D.; Valentão, P.; Andrade, P.; Fernandez, E.; Milella, L. Evaluation of antioxidant, antidiabetic and anticholinesterase activities of Smallanthus sonchifolius landraces and correlation with their phytochemical profiles. Int. J. Mol. Sci. 2015, 16, 17696–17718. [Google Scholar] [CrossRef] [Green Version]
- Honoré, S.M.; Cabrera, W.M.; Genta, S.B.; Sánchez, S.S. Protective effect of yacon leaves decoction against early nephropathy in experimental diabetic rats. Food Chem. Toxicol. 2012, 50, 1704–1715. [Google Scholar] [CrossRef]
- Park, J.-S.; Yang, J.-S.; Hwang, B.-Y.; Yoo, B.-K.; Han, K. Hypoglycemic effect of yacon tuber extract and its constituent, chlorogenic acid, in streptozotocin-induced diabetic rats. Biomol. Ther. 2009, 17, 256–262. [Google Scholar] [CrossRef] [Green Version]
- Genta, S.B.; Cabrera, W.M.; Mercado, M.I.; Grau, A.; Catalán, C.A.; Sánchez, S.S. Hypoglycemic activity of leaf organic extracts from Smallanthus sonchifolius: Constituents of the most active fractions. Chem. Biol. Interact. 2010, 185, 143–152. [Google Scholar] [CrossRef]
- Raga, D.D.; Alimboyoguen, A.B.; del Fierro, R.S.; Ragasa, C.Y. Hypoglycaemic effects of tea extracts and ent-kaurenoic acid from Smallanthus sonchifolius. Nat. Prod. Res. 2010, 24, 1771–1782. [Google Scholar] [CrossRef]
- Genta, S.; Cabrera, W.; Habib, N.; Pons, J.; Carillo, I.M.; Grau, A.; Sánchez, S. Yacon syrup: Beneficial effects on obesity and insulin resistance in humans. Clin. Nutr. 2009, 28, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.O.; Braga, C.P.; Fernandes, A.A.H. Improvement of biochemical parameters in type 1 diabetic rats after the roots aqueous extract of yacon [Smallanthus sonchifolius (Poepp.& Endl.)] treatment. Food Chem. Toxicol. 2013, 59, 256–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, R.B.; Chagas-Paula, D.A.; Secatto, A.; Gasparoto, T.H.; Faccioli, L.H.; Campanelli, A.P.; da Costa, F.B. Topical anti-inflammatory activity of yacon leaf extracts. Braz. J. Pharmacogn. 2013, 23, 497–505. [Google Scholar] [CrossRef] [Green Version]
- Lin, F.; Hasegawa, M.; Kodama, O. Purification and identification of antimicrobial sesquiterpene lactones from yacon (Smallanthus sonchifolius) leaves. Biosci. Biotechnol. Biochem. 2003, 67, 2154–2159. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.G.; Kang, O.H.; Lee, Y.S.; Oh, Y.C.; Chae, H.S.; Obiang-Obounou, B.; Park, S.C.; Shin, D.W.; Hwang, B.Y.; Kwon, D.Y. Antimicrobial activity of the constituents of Smallanthus sonchifolius leaves against methicillin-resistant Staphylococcus aureus. Eur. Rev. Med. Pharmacol. Sci. 2010, 14, 1005–1009. [Google Scholar]
- Yan, X.; Suzuki, M.; Ohnishi-Kameyama, M.; Sada, Y.; Nakanishi, T.; Nagata, T. Extraction and identification of antioxidants in the roots of yacon (Smallanthus sonchifolius). J. Agric. Food Chem. 1999, 47, 4711–4713. [Google Scholar] [CrossRef]
- Valentova, K.; Cvak, L.; Muck, A.; Ulrichova, J.; Simanek, V. Antioxidant activity of extracts from the leaves of Smallanthus sonchifolius. Eur. J. Nutr. 2003, 42, 61–66. [Google Scholar] [CrossRef]
- de Andrade, E.F.; de Souza Leone, R.; Ellendersen, L.N.; Masson, M.L. Phenolic profile and antioxidant activity of extracts of leaves and flowers of yacon (Smallanthus sonchifolius). Ind. Crops Prod. 2014, 62, 499–506. [Google Scholar] [CrossRef]
- Bohlmann, F.; Jakupovic, J.; Zdero, C.; King, R.M.; Robinson, H. Neue melampolide und cis,cis-germacranolide aus vertretern der subtribus melampodiinae. Phytochemistry 1979, 18, 625–630. [Google Scholar] [CrossRef]
- Bohlmann, F.; Knoll, K.H.; Robinson, H.; King, R.M.R.M.; Karl-Heinz, K.; Robinson, H.; King, R.M.R.M.; Knoll, K.H.; Robinson, H.; King, R.M.R.M. Neue kauren-derivate und melampolide aus Smallanthus uvedalia. Phytochemistry 1980, 19, 107–110. [Google Scholar] [CrossRef]
- Bohlmann, F.; Ziesche, J.; King, R.M.; Robinson, H. Neue melampolide aus Smallanthus fruticosus. Phytochemistry 1980, 19, 973–974. [Google Scholar] [CrossRef]
- Simonovska, B.; Vovk, I.; Andrenšek, S.; Valentová, K.; Ulrichová, J. Investigation of phenolic acids in yacon (Smallanthus sonchifolius) leaves and tubers. J. Chromatogr. A 2003, 1016, 89–98. [Google Scholar] [CrossRef]
- Takenaka, M.; Yan, X.; Ono, H.; Yoshida, M.; Nagata, T.; Nakanishi, T. Caffeic acid derivatives in the roots of yacon (Smallanthus sonchifolius). J. Agric. Food Chem. 2003, 51, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.-J.J.; Chzn, L.; Chzn, Y.-K.K.; Liao, Z.; Liu, W.; Yang, G.-Y.Y.; Hu, Q.-F.F.; Wang, L.; Chzn, Y.-K.K.; Liao, Z.; et al. Lignans from the leaf of Smallanthus sonchifolius. Asian J. Chem. 2011, 23, 933–934. [Google Scholar]
- Mercado, M.I.; Coll Aráoz, M.V.; Manrique, I.; Grau, A.; Catalán, C.A.N. Variability in sesquiterpene lactones from the leaves of yacon (Smallanthus sonchifolius) accessions of different geographic origin. Genet. Resour. Crop Evol. 2014, 61, 1209–1217. [Google Scholar] [CrossRef]
- Coll Aráoz, M.V.; Mercado, M.I.; Grau, A.; Catalán, C.A.N. Intraspecific variation of sesquiterpene lactones associated to a latitudinal gradient in Smallanthus macroscyphus (Heliantheae: Asteraceae). Chemoecology 2016, 26, 143–151. [Google Scholar] [CrossRef]
- Padilla-González, G.F.G.F.; Frey, M.; Gómez-Zeledón, J.; Da Costa, F.B.F.B.; Spring, O. Metabolomic and gene expression approaches reveal the developmental and environmental regulation of the secondary metabolism of yacón (Smallanthus sonchifolius, Asteraceae). Sci. Rep. 2019, 9, 13178. [Google Scholar] [CrossRef]
- Campos, D.; Betalleluz-Pallardel, I.; Chirinos, R.; Aguilar-Galvez, A.; Noratto, G.; Pedreschi, R. Prebiotic effects of yacon (Smallanthus sonchifolius Poepp. & Endl), a source of fructooligosaccharides and phenolic compounds with antioxidant activity. Food Chem. 2012, 135, 1592–1599. [Google Scholar] [CrossRef]
- Inoue, A.; Tamogami, S.; Kato, H.; Nakazato, Y.; Akiyama, M.; Kodama, O.; Akatsuka, T.; Hashidoko, Y. Antifungal melampolides from leaf extracts of Smallanthus sonchifolius. Phytochemistry 1995, 39, 845–848. [Google Scholar] [CrossRef]
- Mercado, M.I.; Coll-Aráoz, M.V.; Grau, A.; Catalán, C.A.N.; Coll Aráoz, M.V.; Grau, A.; Catalán, C.A.N. New acyclic diterpenic acids from yacon (Smallanthus sonchifolius) leaves. Nat. Prod. Commun. 2010, 5, 1721–1726. [Google Scholar] [CrossRef] [Green Version]
- Serra-Barcellona, C.; Cabrera, W.M.; Honoré, S.M.; Mercado, M.I.; Sánchez, S.S.; Genta, S.B. Safety assessment of aqueous extract from leaf Smallanthus sonchifolius and its main active lactone, enhydrin. J. Ethnopharmacol. 2012, 144, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Žiarovská, J.; Padilla-González, G.F.F.; Viehmannová, I.; Fernández, E. Genetic and chemical diversity among yacon [Smallanthus sonchifolius (Poepp. et Endl.) H. Robinson] accessions based on iPBS markers and metabolomic fingerprinting. Plant Physiol. Biochem. 2019, 141, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Allen, F.; Pon, A.; Wilson, M.; Greiner, R.; Wishart, D. CFM-ID: A web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res. 2014, 42, W94–W99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef] [Green Version]
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; van der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.M.; et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 2012, 109, E1743–E1752. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- van der Hooft, J.J.J.; Wandy, J.; Barrett, M.P.; Burgess, K.E.V.; Rogers, S. Topic modeling for untargeted substructure exploration in metabolomics. Proc. Natl. Acad. Sci. USA 2016, 113, 13738–13743. [Google Scholar] [CrossRef] [Green Version]
- da Silva, R.R.; Wang, M.; Nothias, L.-F.; van der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef]
- Kang, K.B.; Ernst, M.; Hooft, J.J.J.; Silva, R.R.; Park, J.; Medema, M.H.; Sung, S.H.; Dorrestein, P.C. Comprehensive mass spectrometry-guided phenotyping of plant specialized metabolites reveals metabolic diversity in the cosmopolitan plant family Rhamnaceae. Plant J. 2019, 98, 1134–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.-F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Kuo, G.; De-Qiang, D.; Gui-Rong, C.; Ting-Guo, K.; Yu-Yuan, S.; Xing-Tai, L.; Feng, D. A new hexenol glycoside from leaves of Smallanthus sonchifolius. Nat. Prod. Res. 2010, 24, 1592–1597. [Google Scholar] [CrossRef] [PubMed]
- Göpfert, J.; MacNevin, G.; Ro, D.-K.; Spring, O. Identification, functional characterization and developmental regulation of sesquiterpene synthases from sunflower capitate glandular trichomes. BMC Plant Biol. 2009, 9, 86. [Google Scholar] [CrossRef] [Green Version]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Geng, P.; Sun, J.; Zhang, R.; He, J.; Abliz, Z. An investigation of the fragmentation differences of isomeric flavonol-O-glycosides under different collision-induced dissociation based mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23, 1519–1524. [Google Scholar] [CrossRef]
- Castro, V.; Jakupovic, J.; Dominguez, X.A. Melampolides from Melampodium and Smallanthus species. Phytochemistry 1989, 28, 2727–2729. [Google Scholar] [CrossRef]
- Sugimoto, K.; Matsui, K.; Takabayashi, J. Uptake and conversion of volatile compounds in plant–plant communication. In Deciphering Chemical Language of Plant Communication. Signaling and Communication in Plants; Blande, J.D., Glinwood, R., Eds.; Signaling and Communication in Plants; Springer International Publishing: Cham, Switzerland, 2016; pp. 305–316. ISBN 978-3-319-33496-7. [Google Scholar]
- Takeda, K.; Harborne, J.B.; Self, R. Identification and distribution of malonated anthocyanins in plants of the Compositae. Phytochemistry 1986, 25, 1337–1342. [Google Scholar] [CrossRef]
- Giusti, M.M.; Polit, M.F.; Ayvaz, H.; Tay, D.; Manrique, I. Characterization and quantitation of anthocyanins and other phenolics in native Andean potatoes. J. Agric. Food Chem. 2014, 62, 4408–4416. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- He, F.; Mu, L.; Yan, G.-L.; Liang, N.-N.; Pan, Q.-H.; Wang, J.; Reeves, M.J.; Duan, C.-Q. Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules 2010, 15, 9057–9091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, M.; Shitan, N.; Sawada, K.; Van Montagu, M.C.E.; Inze, D.; Rischer, H.; Goossens, A.; Oksman-Caldentey, K.-M.; Moriyama, Y.; Yazaki, K. Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc. Natl. Acad. Sci. USA 2009, 106, 2447–2452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shitan, N. Secondary metabolites in plants: Transport and self-tolerance mechanisms. Biosci. Biotechnol. Biochem. 2016, 80, 1283–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant flavonoids—Biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef] [PubMed]
- Buer, C.S.; Muday, G.K.; Djordjevic, M.A. Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol. 2007, 145, 478–490. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, E.; García-Villaraco, A.; Lucas, J.A.; Gradillas, A.; Gutierrez-Mañero, F.J.; Ramos-Solano, B. Transcriptomics, targeted metabolomics and gene expression of blackberry leaves and fruits indicate flavonoid metabolic flux from leaf to red fruit. Front. Plant Sci. 2017, 8, 472. [Google Scholar] [CrossRef] [Green Version]
- Braidot, E.; Zancani, M.; Petrussa, E.; Peresson, C.; Bertolini, A.; Patui, S.; Macrì, F.; Vianello, A. Transport and accumulation of flavonoids in grapevine (Vitis vinifera L.). Plant Signal. Behav. 2008, 3, 626–632. [Google Scholar] [CrossRef] [Green Version]
- Göpfert, J.C.; Heil, N.; Conrad, J.; Spring, O. Cytological development and sesquiterpene lactone secretion in capitate glandular trichomes of sunflower. Plant Biol. 2005, 7, 148–155. [Google Scholar] [CrossRef]
- Frey, M.; Klaiber, I.; Conrad, J.; Bersch, A.; Pateraki, I.; Ro, D.-K.; Spring, O. Characterization of CYP71AX36 from Sunflower (Helianthus annuus L., Asteraceae). Sci. Rep. 2019, 9, 14295. [Google Scholar] [CrossRef]
- Nguyen, T.-D.; Kwon, M.; Kim, S.-U.; Fischer, C.; Ro, D.-K. Catalytic plasticity of germacrene A oxidase underlies sesquiterpene lactone diversification. Plant Physiol. 2019, 181, 945–960. [Google Scholar] [CrossRef] [Green Version]
- Mercado, M.I.; Ponessa, G.I.; Grau, A. Morfología y anatomía foliar de “Yacón”, Smallanthus sonchifolius (Asteraceae), con fines de control de calidad. Acta Farm. Bonaer. 2006, 25, 526–532. [Google Scholar]
- do Duarte, M.R.; Wolf, S.; de Paula, B.G. Smallanthus sonchifolius (Poepp.) H. Rob. (yacón): Identificação microscópica de folha e caule para o controle de qualidade farmacognóstico. Rev. Bras. Ciências Farm. 2008, 44, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Ibañez, M.S.; Mercado, M.I.; Coll Aráoz, M.V.; Zannier, M.L.; Grau, A.; Ponessa, G.I. Flower structure and developmental stages of the capitulum of Smallanthus sonchifolius (Asteraceae): Reproductive implications. J. Plant Res. 2017, 130, 327–337. [Google Scholar] [CrossRef]
- Schorr, K.; Merfort, I.; Da Costa, F.B. A novel dimeric melampolide and further terpenoids from Smallanthus sonchifolius (Asteraceae) and the inhibition of the transcription factor NF-kB. Nat. Prod. Commun. 2007, 2, 367–374. [Google Scholar]
- Schorr, K.; Costa, F.B. Da Quantitative determination of enhydrin in leaf rinse extracts and in glandular trichomes of Smallanthus sonchifolius (Asteraceae) by reversed-phase high-performance liquid chromatography. Phytochem. Anal. 2005, 16, 161–165. [Google Scholar] [CrossRef]
- Majdi, M.; Liu, Q.; Karimzadeh, G.; Malboobi, M.A.; Beekwilder, J.; Cankar, K.; de Vos, R.; Todorović, S.; Simonović, A.; Bouwmeester, H. Biosynthesis and localization of parthenolide in glandular trichomes of feverfew (Tanacetum parthenium L. Schulz Bip.). Phytochemistry 2011, 72, 1739–1750. [Google Scholar] [CrossRef] [PubMed]
- Bombo, A.B.; Appezzato-da-Glória, B.; Aschenbrenner, A.-K.A.-K.A.-K.; Spring, O. Capitate glandular trichomes in Aldama discolor (Heliantheae - Asteraceae): Morphology, metabolite profile and sesquiterpene biosynthesis. Plant Biol. 2016, 18, 455–462. [Google Scholar] [CrossRef]
- Ambrósio, S.R.; Oki, Y.; Heleno, V.C.G.; Chaves, J.S.; Nascimento, P.G.B.D.; Lichston, J.E.; Constantino, M.G.; Varanda, E.M.; Da Costa, F.B. Constituents of glandular trichomes of Tithonia diversifolia: Relationships to herbivory and antifeedant activity. Phytochemistry 2008, 69, 2052–2060. [Google Scholar] [CrossRef]
- Wollenweber, E.; Dietz, V.H. Ocurrence and distribution of free flavonoid aglycones in plants. Phytochemistry 1981, 20, 869–932. [Google Scholar] [CrossRef]
- Silva, D.B.; Turatti, I.C.C.; Gouveia, D.R.; Ernst, M.; Teixeira, S.P.; Lopes, N.P. Mass spectrometry of flavonoid vicenin-2, based sunlight barriers in Lychnophora species. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [Green Version]
- Hassan, S.; Mathesius, U. The role of flavonoids in root-rhizosphere signalling: Opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 2012, 63, 3429–3444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khajehei, F.; Hartung, J.; Graeff-Hönninger, S. Total phenolic content and antioxidant activity of yacon (Smallanthus sonchifolius Poepp. and Endl.) chips: Effect of cultivar, pre-treatment and drying. Agriculture 2018, 8, 183. [Google Scholar] [CrossRef] [Green Version]
- Yokotani-Tomita, K.; Kato, J.; Kosemura, S.; Yamamura, S.; Kushima, M.; Kakuta, H.; Hasegawa, K. Light-induced auxin-inhibiting substance from sunflower seedlings. Phytochemistry 1997, 46, 503–506. [Google Scholar] [CrossRef]
- Yokotani-Tomita, K.; Kato, J.; Yamada, K.; Kosemura, S.; Yamamura, S.; Bruinsma, J.; Hasegawa, K. 8-Epixanthatin, a light-induced growth inhibitor, mediates the phototropic curvature in sunflower (Helianthus annuus) hypocotyls. Physiol. Plant. 1999, 106, 326–330. [Google Scholar] [CrossRef]
- Raupp, F.M.; Spring, O. New sesquiterpene lactones from sunflower root exudate as germination stimulants for Orobanche cumana. J. Agric. Food Chem. 2013, 61, 10481–10487. [Google Scholar] [CrossRef]
- Padilla-Gonzalez, G.F.G.F.; dos Santos, F.A.F.A.; Da Costa, F.B.F.B. Sesquiterpene lactones: More than protective plant compounds with high toxicity. CRC Crit. Rev. Plant Sci. 2016, 35, 18–37. [Google Scholar] [CrossRef]
- Mercado, M.I.; Coll Aráoz, M.V.; Brandán de Weth, C.I.; Ponessa, G.I.; Grau, A. Arbuscular mycorrhizal associations and dark septate endophytes in Yacon (Smallanthus sonchifolius) and a wild relative (Smallanthus macroscyphus). Boletín la Soc. Argentina Botánica 2013, 48, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef]
- Akiyama, K.; Ogasawara, S.; Ito, S.; Hayashi, H. Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 2010, 51, 1104–1117. [Google Scholar] [CrossRef]
- Spring, O. Microsampling: An alternative approach using sesquiterpene lactones for systematics. Biochem. Syst. Ecol. 1989, 17, 509–517. [Google Scholar] [CrossRef]
- Spring, O.; Bienert, U.; Klemt, V. Sesquiterpene lactones in glandular trichomes of sunflower leaves. J. Plant Physiol. 1987, 130, 433–439. [Google Scholar] [CrossRef]
- Amrehn, E.; Heller, A.; Spring, O. Capitate glandular trichomes of Helianthus annuus (Asteraceae): Ultrastructure and cytological development. Protoplasma 2014, 251, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Amrehn, E.; Aschenbrenner, A.-K.; Heller, A.; Spring, O. Localization of sesquiterpene lactone biosynthesis in cells of capitate glandular trichomes of Helianthus annuus (Asteraceae). Protoplasma 2016, 253, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Aschenbrenner, A.-K.; Amrehn, E.; Bechtel, L.; Spring, O. Trichome differentiation on leaf primordia of Helianthus annuus (Asteraceae): Morphology, gene expression and metabolite profile. Planta 2015, 241, 837–846. [Google Scholar] [CrossRef]
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.B.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Frey, M.; Schmauder, K.; Pateraki, I.; Spring, O. Biosynthesis of Eupatolide—A metabolic route for sesquiterpene lactone formation involving the P450 enzyme CYP71DD6. ACS Chem. Biol. 2018, 13, 1536–1543. [Google Scholar] [CrossRef]
- Bao, W.; Qu, Y.; Shan, X.; Wan, Y. Screening and validation of housekeeping genes of the root and cotyledon of Cunninghamia lanceolata under abiotic stresses by using quantitative real-time PCR. Int. J. Mol. Sci. 2016, 17, 1198. [Google Scholar] [CrossRef] [Green Version]



| Rt | m/z | [pseudo-Molecular Ion] → MS2 Ions | Discriminant Substance (Comments) | Confidence * |
|---|---|---|---|---|
| Leaves | ||||
| 24.55 | 315.232 | [M+H−H2O]+ 375.253→330.894, 315.233bp, 269.223 | smaditerpenic acid F (in-source fragment) | 2 |
| 8.37 | 303.050 | [M+H]+ 465.103→303.049bp, 285.038, 257.045, 229.050 | quercetin-3-O-galactoside (in-source fragment) | 1 |
| 8.26 | 163.039 | [M+H]+ 535.108→163.039bp | dicaffeoylaltraric acid (in-source fragment) | 3 |
| 8.43 | 333.060 | [M+H]+ 495.113→333.060bp 318.037 | methoxygossypetin-3-O-hexoside (in-source fragment) | 2 |
| 8.43 | 495.113 | [M+H]+ 495.113→333.060bp 318.037 | methoxygossypetin-3-O-hexoside | 2 |
| 8.37 | 465.103 | [M+H]+ 465.103→303.049bp, 285.038, 257.045, 229.050 | quercetin-3-O-galactoside | 1 |
| 15.40 | 482.202 | [M+H]+ 465.175→405.154, 349.128, 289.107bp, 229.086 | enhydrin (NH4+ adduct) | 1 |
| Stems | ||||
| 1.26 | 381.079 | [M+Na]+ 527.258→365.106bp, 347.095, 203.053, 185.042 | raffinose (in-source fragment) | 3 |
| 1.26 | 527.158 | [M+Na]+ 527.258→365.106bp, 347.095, 203.053, 185.042 | raffinose (Na+ adduct) | 3 |
| 1.26 | 543.132 | [M+Na]+ 527.258→365.106bp, 347.095, 203.053, 185.042 | raffinose (K+ adduct) | 3 |
| Roots | ||||
| 1.31 | 504.192 | [M+NH4]+ 504.1192→163.059, 145.049bp, 127.039, 97.029, 85.029 | trisaccharide (NH4+ adduct) | 3 |
| 16.05 | 441.048 | [M+H]+ 441.048→361.092bp, 346.068, 329.066, 301.071, 167.034 | C21H12O11 | 4 |
| 11.28 | 163.039 | [M+H]+ 697.139→163.039bp | 2,3,5/2,4,5-tricaffeoylaltraric acid (in-source fragment) | 1 |
| Bracts | ||||
| 8.55 | 412.217 | [M+ NH4]+ 412.217→145.050, 127.039, 115.039, 97.029, 91.058bp, 85.029, 73.029 | hexenyl-O-arabinoglucoside (NH4+ adduct) | 2 |
| 10.86 | 366.175 | [M+ NH4]+ 366.175→145.049, 127.039, 105.019, 97.029, 85.029bp | C15H24O9 (NH4+ adduct) | 4 |
| 12.57 | 430.171 | [M+ NH4]+ 430.170→145.049, 127.039, 109.029, 105.019, 97.029, 85.029bp | C19H24O10 (NH4+ adduct) | 4 |
| 10.64 | 388.158 | [M+ NH4]+ 388.158→145.049, 127.039, 105.019, 97.029, 85.029bp | C17H22O9 (NH4+ adduct) | 4 |
| 12.12 | 368.191 | [M+ NH4]+ 368.191→145.049, 127.039, 105.019, 97.029, 85.029bp | C15H26O9 (NH4+ adduct) | 4 |
| Rt | m/z | [pseudo-Molecular Ion] → MS2 Ions | Discriminant Substance (Comments) | Confidence * |
|---|---|---|---|---|
| White cultivar | ||||
| 5.96 | 163.039 | [M+H]+ 355.102→163.039bp | 5-O-(E)-caffeoylquinic acid (in-source fragment) | 1 |
| 1.22 | 286.092 | [M+H]+ 286.092→124.039bp, 85.029 | C12H15O7N | 4 |
| 8.26 | 163.039 | [M+H]+ 535.108→163.039bp | dicaffeoylaltraric acid isomer (in-source fragment) | 3 |
| 24.61 | 315.233 | [M+H−H2O]+ 375.253→330.894, 315.233bp, 269.223 | smaditerpenic acid F (in-source fragment) | 2 |
| 11.12 | 430.171 | [M+ NH4]+ 430.171→145.049, 127.039bp, 105.019, 97.029, 85.029 | C19H24O10 (NH4+ adduct) | 4 |
| 5.96 | 377.084 | [M+H]+ 355.102→163.039bp | 5-O-(E)-caffeoylquinic acid (Na+ adduct) | 1 |
| 5.96 | 355.102 | [M+H]+ 355.102→163.039bp | 5-O-(E)-caffeoylquinic acid | 1 |
| Red cultivar | ||||
| 19.17 | 315.232 | [M+H−H2O]+ 333.242→315.232bp, 297.22098, 269.22659 | smaditerpenic acid C (in-source fragment) | 2 |
| 8.43 | 495.113 | [M+H]+ 495.113→333.060bp 318.037 | methoxygossypetin-3-O-hexoside | 2 |
| 4.29 | 449.108 | [M]+ 595.166→449.108, 287.055bp | cyanidin-O-rutinoside (in-source fragment) | 2 |
| 10.98 | 478.265 | [M+H]+ 461.238→443.228, 417.208, 213.185, 127.039bp | C22H36O10 (NH4+ adduct) | 4 |
| 19.17 | 373.235 | [M+H−H2O]+ 333.242→315.232bp, 297.22098, 269.22659 | smaditerpenic acid C (Na+ adduct) | 2 |
| 8.43 | 333.060 | [M+H]+ 495.113→333.060bp 318.037 | methoxygossypetin-3-O-hexoside (in-source fragment) | 2 |
| Code | Primer (5′ - 3′) | Size (bp) | Ta (°C) | Efficiency | R2 |
|---|---|---|---|---|---|
| Ss_qGAO | F: CGAAAACGGCAACACCACCATT | 162 | 60 | 92.3 | 0.998 |
| R: GCTCGCACCATTGGGAAGTTTC | |||||
| Ss_qCHS | F: GCCGACTACCAGCTCACCAAACTC | 193 | 60 | 87.2 | 0.999 |
| R: CCTCATTAGGGCCACGGAACG | |||||
| Ha_qACT | F: GCCGTGCTTTCTCTTTATGCCAGCGACC | 137 | 60 | 95.0 | 0.998 |
| R: AGCGAGATCAAGACGAAG | |||||
| Ha_qEF | F: ACCAAATCAATGAGCCCAAGAGACCCA | 131 | 60 | 99.6 | 0.987 |
| R: TACCGGGCTTGATCACACCAG | |||||
| Ha_qGAPDH | F: GCAAGGACTGGAGAGGTGGAAGAG | 140 | 60 | 92.6 | 0.998 |
| R: ATCAACGGTAGGGACACGGAATG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padilla-González, G.F.; Amrehn, E.; Frey, M.; Gómez-Zeledón, J.; Kaa, A.; Da Costa, F.B.; Spring, O. Metabolomic and Gene Expression Studies Reveal the Diversity, Distribution and Spatial Regulation of the Specialized Metabolism of Yacón (Smallanthus sonchifolius, Asteraceae). Int. J. Mol. Sci. 2020, 21, 4555. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124555
Padilla-González GF, Amrehn E, Frey M, Gómez-Zeledón J, Kaa A, Da Costa FB, Spring O. Metabolomic and Gene Expression Studies Reveal the Diversity, Distribution and Spatial Regulation of the Specialized Metabolism of Yacón (Smallanthus sonchifolius, Asteraceae). International Journal of Molecular Sciences. 2020; 21(12):4555. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124555
Chicago/Turabian StylePadilla-González, Guillermo F., Evelyn Amrehn, Maximilian Frey, Javier Gómez-Zeledón, Alevtina Kaa, Fernando B. Da Costa, and Otmar Spring. 2020. "Metabolomic and Gene Expression Studies Reveal the Diversity, Distribution and Spatial Regulation of the Specialized Metabolism of Yacón (Smallanthus sonchifolius, Asteraceae)" International Journal of Molecular Sciences 21, no. 12: 4555. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124555