Validation of Breast Cancer Margins by Tissue Spray Mass Spectrometry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue Handling
2.2. Chemicals
2.3. Tissue Spray Mass Spectrometry of Breast Samples
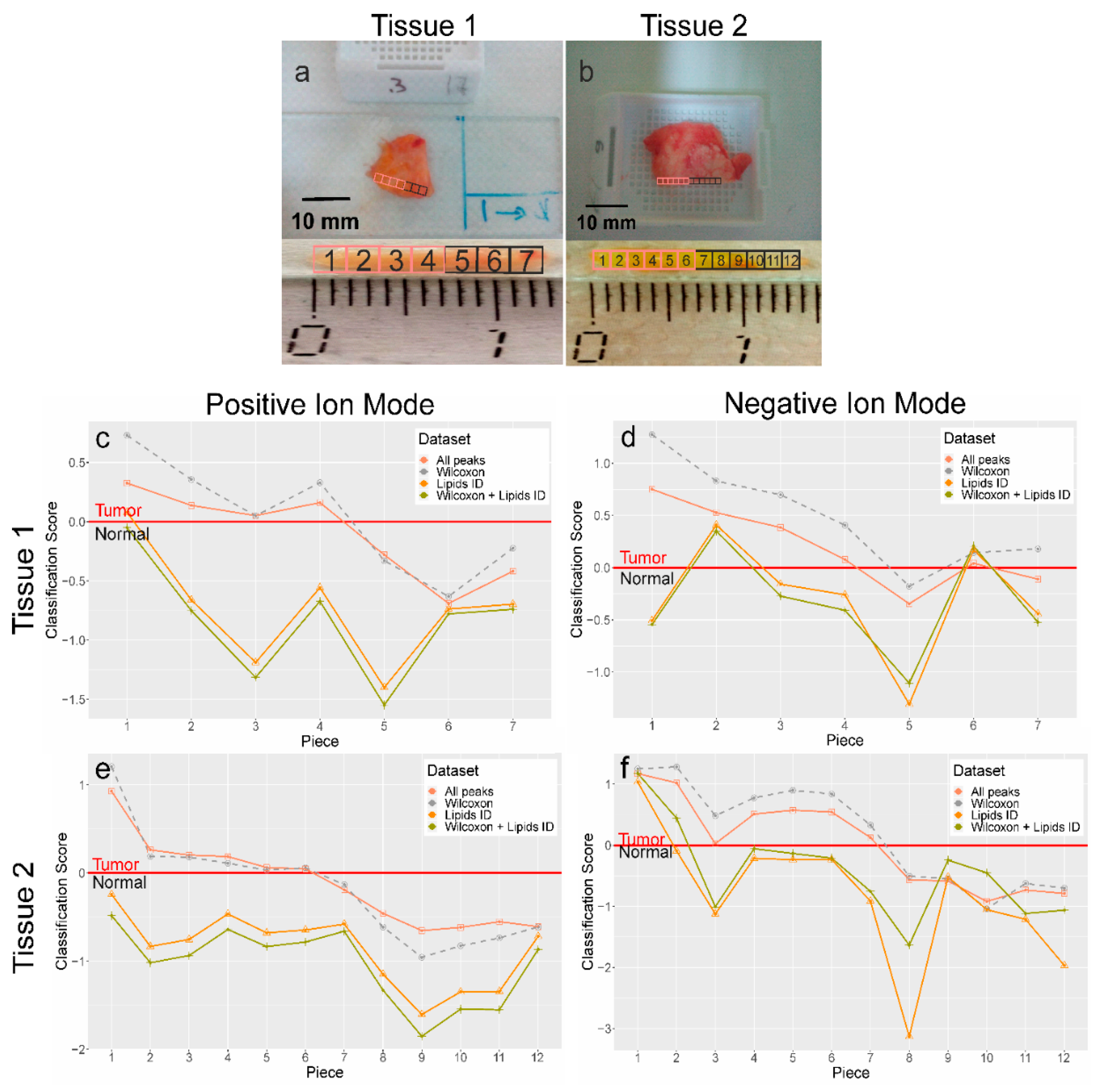
2.4. Histological and Pathological Data
2.5. Statistical Analysis of Mass Spectrometry Data and MS Peak Annotation
2.6. Results and Discussion
2.6.1. Tissue Spray MS Data
2.6.2. Classification Models Training
2.6.3. Classification Model Testing
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Miller, K.D.; Siegel, R.L.; Lin, C.C.; Mariotto, A.B.; Kramer, J.L.; Rowland, J.H.; Stein, K.D.; Alteri, R.; Jemal, A. Cancer treatment and survivorship statistics, 2016. CA. Cancer J. Clin. 2016, 66, 271–289. [Google Scholar] [CrossRef] [Green Version]
- Jeevan, R.; Cromwell, D.A.; Trivella, M.; Lawrence, G.; Kearins, O.; Pereira, J.; Sheppard, C.; Caddy, C.M.; Van Der Meulen, J.H.P. Reoperation rates after breast conserving surgery for breast cancer among women in England: Retrospective study of hospital episode statistics. BMJ 2012, 345, e4505. [Google Scholar] [CrossRef] [Green Version]
- Fisher, B.; Anderson, S.; Bryant, J.; Margolese, R.; Deutsch, M.; Fisher, E.R.; Jeong, J.-H.; Wolmark, N. Twenty-Year Follow-Up of a Randomized Trial Comparing Total for the Treatment of Invasive Breast Cancer. N. Engl. J. Med. 2002, 347, 1233–1241. [Google Scholar] [CrossRef] [Green Version]
- van Dongen, J.A. Long-term results of a randomized trial comparing breast-conserving therapy with mastectomy: European organization for research and treatment of cancer 10801 trial 4606. J. Natl. Cancer Inst. 2000, 92, 1143–1150. [Google Scholar] [CrossRef] [Green Version]
- Veronesi, U.; Cascinelli, N.; Mariani, L.; Greco, M.; Saccozzi, R.; Luini, A.; Aguilar, M.; Marubini, E. Twenty-Year Follow-up of a Randomized Study Comparing Breast-Conserving Surgery with Radical Mastectomy for Early Breast Cancer. N. Engl. J. Med. 2002, 347, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- McCahill, L.E.; Single, R.M.; Aiello Bowles, E.J.; Feigelson, H.S.; James, T.A.; Barney, T.; Engel, J.M.; Onitilo, A.A. Variability in reexcision following breast conservation surgery. JAMA J. Am. Med. Assoc. 2012, 307, 467–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waljee, J.F.; Hu, E.S.; Newman, L.A.; Alderman, A.K. Predictors of re-excision among women undergoing breast-conserving surgery for cancer. Ann. Surg. Oncol. 2008, 15, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Olson, T.P.; Harter, J.; Muñoz, A.; Mahvi, D.M.; Breslin, T.M. Frozen section analysis for intraoperative margin assessment during breast-conserving surgery results in low rates of re-excision and local recurrence. Ann. Surg. Oncol. 2007, 14, 2953–2960. [Google Scholar] [CrossRef]
- D’Halluin, F.; Tas, P.; Rouquette, S.; Bendavid, C.; Foucher, F.; Meshba, H.; Blanchot, J.; Coué, O.; Levêque, J. Intra-operative touch preparation cytology following lumpectomy for breast cancer: A series of 400 procedures. Breast 2009, 18, 248–253. [Google Scholar] [CrossRef]
- Mesurolle, B.; El-Khoury, M.; Hori, D.; Phancao, J.P.; Kary, S.; Kao, E.; Fleiszer, D. Sonography of postexcision specimens of nonpalpable breast lesions: Value, limitations, and description of a method. Am. J. Roentgenol. 2006, 186, 1014–1024. [Google Scholar] [CrossRef]
- Ciccarelli, G.; Di Virgilio, M.R.; Menna, S.; Garretti, L.; Ala, A.; Giani, R.; Bussone, R.; Canavese, G.; Berardengo, E. Radiography of the surgical specimen in early stage breast lesions: Diagnostic reliability in the analysis of the resection margins. Radiol. Med. 2007, 112, 366–376. [Google Scholar] [CrossRef]
- Balog, J.; Sasi-Szabó, L.; Kinross, J.; Lewis, M.R.; Muirhead, L.J.; Veselkov, K.; Mirnezami, R.; Dezső, B.; Damjanovich, L.; Darzi, A.; et al. Intraoperative Tissue Identification Using Rapid Evaporative Ionization Mass Spectrometry. Sci. Transl. Med. 2013, 5, ra93–ra194. [Google Scholar] [CrossRef]
- Balog, J.; Szaniszló, T.; Schaefer, K.-C.; Denes, J.; Lopata, A.; Godorhazy, L.; Szalay, D.; Balogh, L.; Sasi-Szabó, L.; Toth, M.; et al. Identification of biological tissues by rapid evaporative ionization mass spectrometry. Anal. Chem. 2010, 82, 7343–7350. [Google Scholar] [CrossRef]
- St John, E.R.; Rossi, M.; Pruski, P.; Darzi, A.; Takats, Z. Intraoperative tissue identification by mass spectrometric technologies. TrAC Trends Anal. Chem. 2016, 85, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Dill, A.L.; Ifa, D.R.; Manicke, N.E.; Ouyang, Z.; Cooks, R.G. Mass spectrometric imaging of lipids using desorption electrospray ionization. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2009, 877, 2883–2889. [Google Scholar] [CrossRef] [Green Version]
- Eberlin, L.S.; Norton, I.; Orringer, D.; Dunn, I.F.; Liu, X.; Ide, J.L.; Jarmusch, A.K.; Ligon, K.L.; Jolesz, F.A.; Golby, A.J.; et al. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc. Natl. Acad. Sci. USA 2013, 110, 1611–1616. [Google Scholar] [CrossRef] [Green Version]
- Takats, Z.; Wiseman, J.; Gologan, B.; Cooks, R. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 2004, 306, 471–473. [Google Scholar] [CrossRef] [Green Version]
- Chughtai, K.; Heeren, R.M.A. Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 2010, 110, 3237–3277. [Google Scholar] [CrossRef] [Green Version]
- Hinsch, A.; Buchholz, M.; Odinga, S.; Borkowski, C.; Koop, C.; Izbicki, J.R.; Wurlitzer, M.; Krech, T.; Wilczak, W.; Steurer, S.; et al. MALDI imaging mass spectrometry reveals multiple clinically relevant masses in colorectal cancer using large-scale tissue microarrays. J. Mass Spectrom. 2017, 52, 165–173. [Google Scholar] [CrossRef]
- Caprioli, R.M.; Farmer, T.B.; Gile, J. Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal. Chem. 1997, 69, 4751–4760. [Google Scholar] [CrossRef]
- Boxer, S.G.; Kraft, M.L.; Weber, P.K. Advances in Imaging Secondary Ion Mass Spectrometry for Biological Samples. Annu. Rev. Biophys. 2009, 38, 53–74. [Google Scholar] [CrossRef] [Green Version]
- Bluestein, B.M.; Morrish, F.; Graham, D.J.; Guenthoer, J.; Hockenbery, D.; Porter, P.L.; Gamble, L.J. An unsupervised MVA method to compare specific regions in human breast tumor tissue samples using ToF-SIMS. Analyst 2016, 141, 1947–1957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wucher, A.; Cheng, J.; Winograd, N. Protocols for three-dimensional molecular imaging using mass spectrometry. Anal. Chem. 2007, 79, 5529–5539. [Google Scholar] [CrossRef]
- Kononikhin, A.; Zhvansky, E.; Shurkhay, V.; Popov, I.; Bormotov, D.; Kostyukevich, Y.; Karchugina, S.; Indeykina, M.; Bugrova, A.; Starodubtseva, N.; et al. A novel direct spray-from-tissue ionization method for mass spectrometric analysis of human brain tumors. Anal. Bioanal. Chem. 2015, 407, 7797–7805. [Google Scholar] [CrossRef]
- Wei, Y.; Chen, L.; Zhou, W.; Chingin, K.; Ouyang, Y.; Zhu, T.; Wen, H.; Ding, J.; Xu, J.; Chen, H. Tissue spray ionization mass spectrometry for rapid recognition of human lung squamous cell carcinoma. Sci. Rep. 2015, 5, 1–7. [Google Scholar] [CrossRef]
- Chagovets, V.V.; Wang, Z.; Kononikhin, A.S.; Starodubtseva, N.L.; Borisova, A.; Salimova, D.; Popov, I.A.; Kozachenko, A.V.; Chingin, K.; Chen, H.; et al. Endometriosis foci differentiation by rapid lipid profiling using tissue spray ionization and high resolution mass spectrometry. Sci. Rep. 2017, 7, 2546. [Google Scholar] [CrossRef] [Green Version]
- Sukhikh, G.; Chagovets, V.; Wang, X.; Rodionov, V.; Kometova, V.; Tokareva, A.; Kononikhin, A.; Starodubtseva, N.; Chingin, K.; Chen, H.; et al. Combination of low-temperature electrosurgical unit and extractive electrospray ionization mass spectrometry for molecular profiling and classification of tissues. Molecules 2019, 24, 2957. [Google Scholar] [CrossRef] [Green Version]
- Adamyan, L.V.; Starodubtseva, N.; Borisova, A.; Stepanian, A.A.; Chagovets, V.; Salimova, D.; Wang, Z.; Kononikhin, A.; Popov, I.; Bugrova, A.; et al. Direct Mass Spectrometry Differentiation of Ectopic and Eutopic Endometrium in Patients with Endometriosis. J. Minim. Invasive Gynecol. 2018, 25, 426–433. [Google Scholar] [CrossRef]
- Tokareva, A.O.; Chagovets, V.V.; Starodubtseva, N.L.; Nazarova, N.M.; Nekrasova, M.E.; Kononikhin, A.S.; Frankevich, V.E.; Nikolaev, E.N.; Sukhikh, G.T. Feature selection for OPLS discriminant analysis of cancer tissue lipidomics data. J. Mass Spectrom. 2020, 55, e4457. [Google Scholar] [CrossRef]
- Chagovets, V.; Kononikhin, A.; Tokoreva, A.; Bormotov, D.; Starodubtseva, N.; Kostyukevich, Y.; Popov, I.; Frankevich, V.; Nikolaev, E. Relative quantitation of phosphatidylcholines with interfered masses of protonated and sodiated molecules by tandem and Fourier-transform ion cyclotron resonance mass spectrometry. Eur. J. Mass Spectrom. 2019, 25, 259–264. [Google Scholar] [CrossRef]
- Chagovets, V.; Kononikhin, A.; Starodubtseva, N.; Kostyukevich, Y.; Popov, I.; Frankevich, V.; Nikolaev, E. Peculiarities of data interpretation upon direct tissue analysis by Fourier transform ion cyclotron resonance mass spectrometry. Eur. J. Mass Spectrom. 2016, 22, 123. [Google Scholar] [CrossRef] [PubMed]
- Chagovets, V.; Wang, Z.; Kononikhin, A.; Starodubtseva, N.; Borisova, A.; Salimova, D.; Popov, I.; Kozachenko, A.; Chingin, K.; Chen, H.; et al. A Comparison of Tissue Spray and Lipid Extract Direct Injection Electrospray Ionization Mass Spectrometry for the Differentiation of Eutopic and Ectopic Endometrial Tissues. J. Am. Soc. Mass Spectrom. 2017, 29, 323–330. [Google Scholar] [CrossRef]
- Eriksson, L.; Johansson, E.; Kettapeh-Wold, S.; Wold, S. Scaling: Introduction to Multi- and Megavariate Data Analysis Using Projection Methods (PCA & PLS); Umetrics: Umea, Sweden, 1999. [Google Scholar]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Koelmel, J.P.; Kroeger, N.M.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Cochran, J.A.; Beecher, C.W.W.; Garrett, T.J.; Yost, R.A. LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinformatics 2017, 18, 1–11. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, 527–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Dataset | Model Parameters | ||||
|---|---|---|---|---|---|
| Name | Number of Variables | Number of Features with VIP > 1 | R2X | R2Y | Q2 |
| All peaks | 541 | 102 | 0.438 | 0.868 | 0.829 |
| Significantly different peaks | 231 | 52 | 0.503 | 0.888 | 0.850 |
| Identified lipids | 106 | 22 | 0.512 | 0.845 | 0.784 |
| Significantly different lipids | 60 | 14 | 0.649 | 0.826 | 0.785 |
| Dataset | Model Parameters | ||||
|---|---|---|---|---|---|
| Name | Number of Variables | Number of Features with VIP > 1 | R2X | R2Y | Q2 |
| All peaks | 514 | 79 | 0.420 | 0.734 | 0.643 |
| Significantly different peaks | 190 | 36 | 0.490 | 0.753 | 0.579 |
| Identified lipids | 118 | 16 | 0.510 | 0.504 | 0.311 |
| Significantly different lipids | 40 | 7 | 0.706 | 0.479 | 0.381 |
| Tissue Sample | Dataset | Positive Polarity | Negative Polarity | ||
|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | ||
| 1 | All peaks | 1.00 | 1.00 | 1.00 | 0.67 |
| Identified lipids | 0.25 | 1.00 | 0.25 | 0.67 | |
| Significantly different peaks | 1.00 | 1.00 | 1.00 | 0.33 | |
| Significantly different lipids | 0.00 | 1.00 | 0.25 | 0.67 | |
| 2 | All peaks | 1.00 | 1.00 | 1.00 | 0.83 |
| Identified lipid | 0.00 | 1.00 | 0.17 | 1.00 | |
| Significantly different peaks | 1.00 | 1.00 | 1.00 | 0.83 | |
| Significantly different lipids | 0.00 | 1.00 | 0.33 | 1.00 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chagovets, V.V.; Starodubtseva, N.L.; Tokareva, A.O.; Frankevich, V.E.; Rodionov, V.V.; Kometova, V.V.; Chingin, K.; Kukaev, E.N.; Chen, H.; Sukhikh, G.T. Validation of Breast Cancer Margins by Tissue Spray Mass Spectrometry. Int. J. Mol. Sci. 2020, 21, 4568. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124568
Chagovets VV, Starodubtseva NL, Tokareva AO, Frankevich VE, Rodionov VV, Kometova VV, Chingin K, Kukaev EN, Chen H, Sukhikh GT. Validation of Breast Cancer Margins by Tissue Spray Mass Spectrometry. International Journal of Molecular Sciences. 2020; 21(12):4568. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124568
Chicago/Turabian StyleChagovets, Vitaliy V., Natalia L. Starodubtseva, Alisa O. Tokareva, Vladimir E. Frankevich, Valerii V. Rodionov, Vlada V. Kometova, Konstantin Chingin, Eugene N. Kukaev, Huanwen Chen, and Gennady T. Sukhikh. 2020. "Validation of Breast Cancer Margins by Tissue Spray Mass Spectrometry" International Journal of Molecular Sciences 21, no. 12: 4568. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124568





