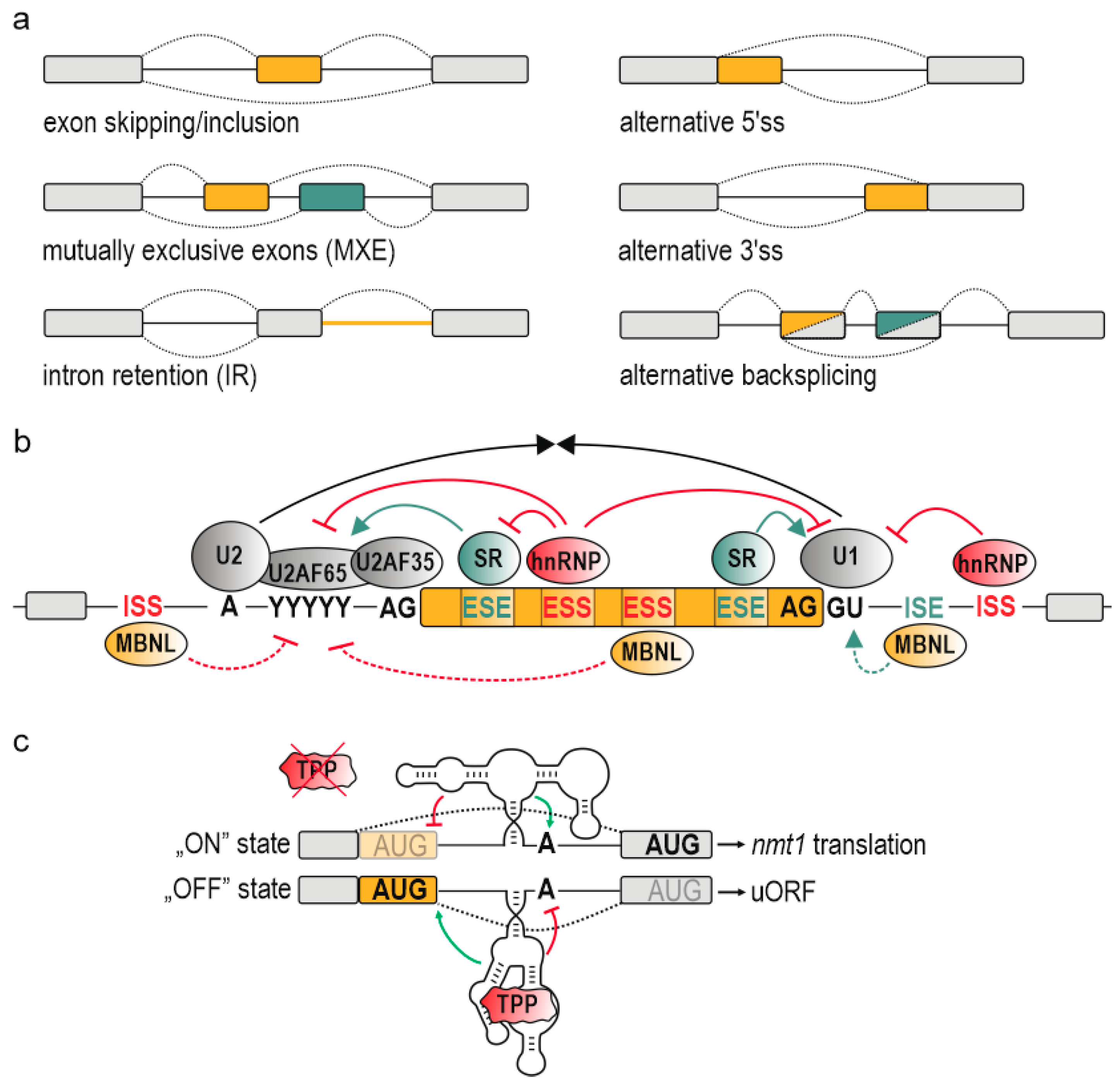
Intrinsic Regulatory Role of RNA Structural Arrangement in Alternative Splicing Control
Abstract
:1. Introduction
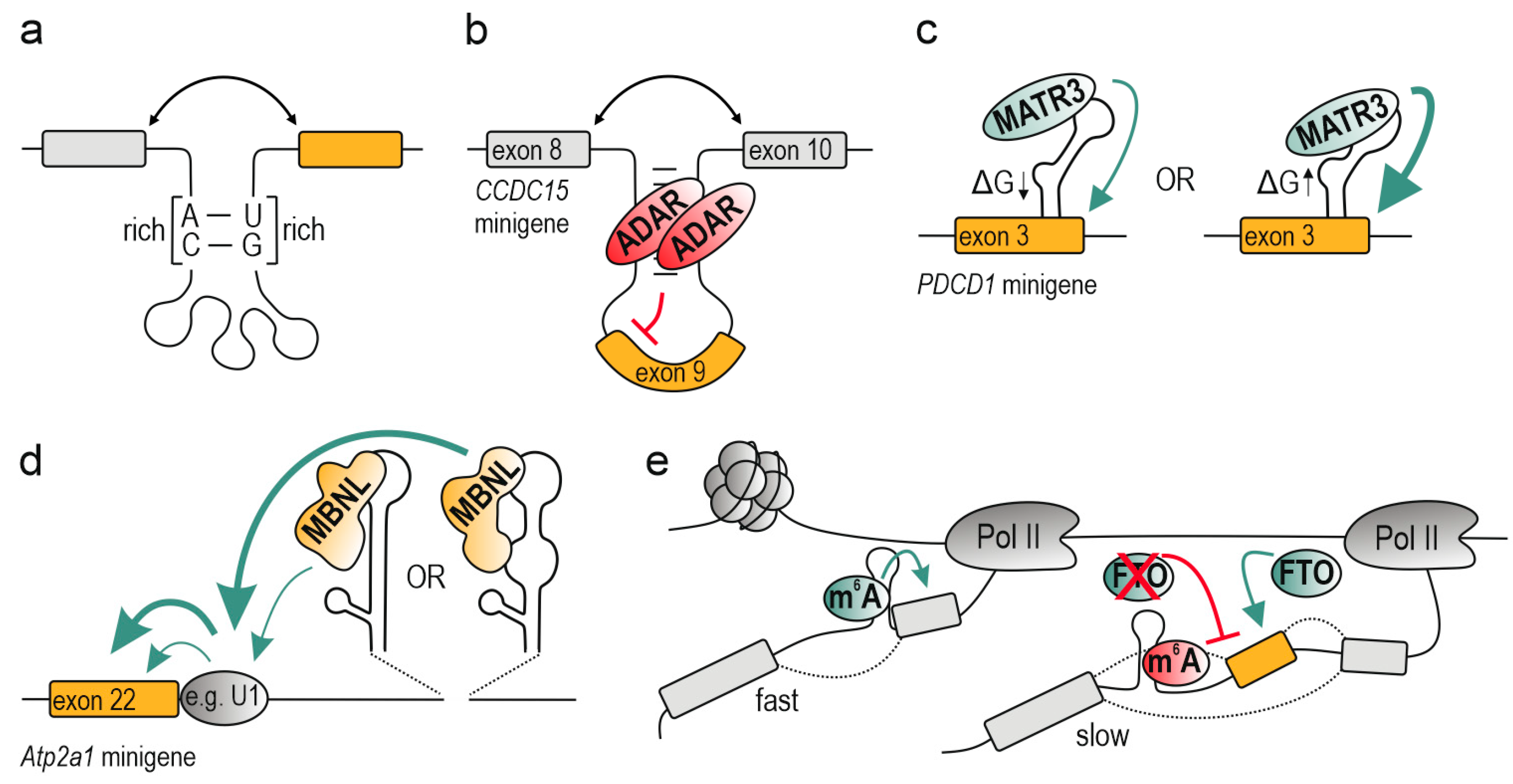
2. RNA Structural Arrangement
3. Cellular Modulators of RNA Structure
3.1. Molecular Crowding
3.2. Transcription
3.3. RNA Modifications, Editing and Sequence Composition
3.4. RNA Structural Switches
4. Mechanism of Alternative Splicing Regulation by RNA Structural Conformation
4.1. Bridging Cis-Acting Elements
4.2. Looping out Splice Sites and Entire Exons
4.3. Blocking (Steric Hindrance)/Promoting Interaction with Splicing Factors
4.4. Allosteric Activation (Enhancement)/Inhibition (Deterioration) of Splicing Factors
4.5. Modulating the Splicing Kinetics
5. Splicing-Related Diseases Mediated by RNA Structural Arrangement
5.1. Diseases Associated with Single Nucleotide Variants (SNV)
5.2. Diseases Associated with Microsatellite Mutation
5.3. Cancer
6. Conclusions and Future Directions
Funding
Acknowledgments
Conflicts of Interest
References
- Pandya-Jones, A. Pre-mRNA splicing during transcription in the mammalian system. Wiley Interdiscip. Rev. RNA 2011, 2, 700–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krchnakova, Z.; Thakur, P.K.; Krausova, M.; Bieberstein, N.; Haberman, N.; Muller-McNicoll, M.; Stanekk, D. Splicing of long non-coding RNAs primarily depends on polypyrimidine tract and 5 splice-site sequences due to weak interactions with SR proteins. Nucleic Acids Res. 2019, 47, 911–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abelson, J.; Trotta, C.R.; Li, H. tRNA splicing. J. Biol. Chem. 1998, 273, 12685–12688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konieczny, P.; Stepniak-Konieczna, E.; Sobczak, K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014, 42, 10873–10887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Jacob, A.G.; Smith, C.W.; Smith, J. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef] [Green Version]
- Kino, Y.; Washizu, C.; Oma, Y.; Onishi, H.; Nezu, Y.; Sasagawa, N.; Nukina, N.; Ishiura, S. MBNL and CELF proteins regulate alternative splicing of the skeletal muscle chloride channel CLCN1. Nucleic Acids Res. 2009, 37, 6477–6490. [Google Scholar] [CrossRef] [Green Version]
- Mankodi, A.K.; Takahashi, M.; Jiang, H.; Beck, C.L.; Bowers, W.J.; Moxley, R.T.; Cannon, S.C.; Thornton, C.A. Expanded CUG repeats trigger aberrant splicing of CLC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell 2002, 10, 35–44. [Google Scholar] [CrossRef]
- Zhang, X.-O.; Dong, R.; Zhang, Y.; Zhang, J.-L.; Luo, Z.; Zhang, J.; Chen, L.-L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Liu, S.; Zhang, L.; Issaian, A.; Hill, R.C.; Espinosa, S.; Shi, S.; Cui, Y.; Kappel, K.; Das, R.; et al. A unified mechanism for intron and exon definition and back-splicing. Nature 2019, 573, 375–380. [Google Scholar] [CrossRef]
- Xiao, M.-S.; Ai, Y.; Wilusz, J.E. Biogenesis and Functions of Circular RNAs Come into Focus. Trends Cell Biol. 2020, 30, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.K.; Black, U.L.; Zheng, S. The neurogenetics of alternative splicing. Nat. Rev. Neurosci. 2016, 17, 265–281. [Google Scholar] [CrossRef]
- Yamamoto, K.; Furukawa, M.T.; Fukumura, K.; Kawamura, A.; Yamada, T.; Suzuki, H.; Hirose, T.; Sakamoto, H.; Inoue, K. Control of the heat stress-induced alternative splicing of a subset of genes by hnRNP K. Genes Cells 2016, 21, 1006–1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, M.J.; Moreno, N.N.; Giono, L.; Botto, A.E.C.; Dujardin, G.; Bastianello, G.; Lavore, S.; Torres-Méndez, A.; Menck, C.F.M.; Blencowe, B.J.; et al. Major Roles for Pyrimidine Dimers, Nucleotide Excision Repair, and ATR in the Alternative Splicing Response to UV Irradiation. Cell Rep. 2017, 18, 2868–2879. [Google Scholar] [CrossRef] [Green Version]
- Imbriano, C.; Molinari, S. Alternative Splicing of Transcription Factors Genes in Muscle Physiology and Pathology. Genes 2018, 9, 107. [Google Scholar] [CrossRef] [Green Version]
- Rotival, M.; Quach, H.; Quintana-Murci, L. Defining the genetic and evolutionary architecture of alternative splicing in response to infection. Nat. Commun. 2019, 10, 1671. [Google Scholar] [CrossRef] [PubMed]
- Frankiw, L.; Majumdar, D.; Burns, C.; Vlach, L.; Moradian, A.; Sweredoski, M.J.; Baltimore, D. BUD13 Promotes a Type I Interferon Response by Countering Intron Retention in Irf7. Mol. Cell 2019, 73, 803–814. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.K.; Pham, M.H.C.; Ko, K.S.; Rhee, B.D.; Han, J. Alternative splicing isoforms in health and disease. Pflügers Arch. Eur. J. Physiol. 2018, 470, 995–1016. [Google Scholar] [CrossRef]
- Ule, J.; Blencowe, B.J. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol. Cell 2019, 76, 329–345. [Google Scholar] [CrossRef]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [Green Version]
- Herzel, L.; Ottoz, D.S.M.; Alpert, T.; Neugebauer, K.M. Splicing and transcription touch base: Co-transcriptional spliceosome assembly and function. Nat. Rev. Mol. Cell Biol. 2017, 18, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Shenasa, H.; Hertel, K.J. Combinatorial regulation of alternative splicing. Biochim. Biophys. Acta (BBA) Bioenerg. 2019, 1862, 194392. [Google Scholar] [CrossRef] [PubMed]
- De Conti, L.; Baralle, M.; Buratti, E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip. Rev. RNA 2012, 4, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Huang, B.; Xu, Y.-M.; Li, J.; Huang, L.-F.; Lin, J.; Zhang, J.; Min, Q.-H.; Yang, W.-M.; et al. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2014, 3, 152–158. [Google Scholar] [CrossRef] [Green Version]
- Otsuka, H.; Sasai, H.; Nakama, M.; Aoyama, Y.; Abdelkreem, E.; Ohnishi, H.; Konstantopoulou, V.; Sass, J.O.; Fukao, T. Exon 10 skipping in ACAT1 caused by a novel c.949G>A mutation located at an exonic splice enhancer site. Mol. Med. Rep. 2016, 14, 4906–4910. [Google Scholar] [CrossRef] [Green Version]
- Ramanouskaya, T.V.; Grinev, V. The determinants of alternative RNA splicing in human cells. Mol. Genet. Genom. 2017, 292, 1175–1195. [Google Scholar] [CrossRef]
- Chen, S.-Y.; Li, C.; Jia, X.; Lai, S. Sequence and Evolutionary Features for the Alternatively Spliced Exons of Eukaryotic Genes. Int. J. Mol. Sci. 2019, 20, 3834. [Google Scholar] [CrossRef] [Green Version]
- Long, J.C.; Cáceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2008, 417, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Qual. Life Res. 2016, 135, 851–867. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.-D.; Ares, M. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 689–701. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, D.; Freese, P.; Alexis, M.S.; Su, A.; Hochman, M.; Palden, T.; Bazile, C.; Lambert, N.J.; Van Nostrand, E.; Pratt, G.A.; et al. Sequence, Structure, and Context Preferences of Human RNA Binding Proteins. Mol. Cell 2018, 70, 854–867.e9. [Google Scholar] [CrossRef] [Green Version]
- Sznajder, Ł.J.; Michalak, M.; Taylor, K.; Cywoniuk, P.; Kabza, M.; Wojtkowiak-Szlachcic, A.; Matłoka, M.; Konieczny, P.; Sobczak, K. Mechanistic determinants of MBNL activity. Nucleic Acids Res. 2016, 44, 10326–10342. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Treacy, D.; Eichinger, K.; Struck, A.; Estabrook, J.; Olafson, H.; Wang, T.T.; Bhatt, K.; Westbrook, T.; Sedehizadeh, S.; et al. Transcriptome alterations in myotonic dystrophy skeletal muscle and heart. Hum. Mol. Genet. 2018, 28, 1312–1321. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Ward, A.J.; Cherone, J.M.; Giudice, J.; Wang, T.T.; Treacy, D.J.; Lambert, N.J.; Freese, P.; Saxena, T.; Cooper, T.A.; et al. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 2015, 25, 858–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.T.; Cody, N.A.L.; Jog, S.; Biancolella, M.; Wang, T.T.; Treacy, D.J.; Luo, S.; Schroth, G.P.; Housman, D.E.; Reddy, S.; et al. Transcriptome-wide Regulation of Pre-mRNA Splicing and mRNA Localization by Muscleblind Proteins. Cell 2012, 150, 710–724. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Zhang, Z.; Fregoso, O.; Krainer, A.R. Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA 2011, 18, 274–283. [Google Scholar] [CrossRef] [Green Version]
- Llorian, M.; Schwartz, S.; Clark, T.A.; Hollander, D.; Tan, L.-Y.; Spellman, R.; Gordon, A.; Schweitzer, A.C.; De La Grange, P.; Ast, G.; et al. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat. Struct. Mol. Biol. 2010, 17, 1114–1123. [Google Scholar] [CrossRef] [Green Version]
- Hiller, M.; Zhang, Z.; Backofen, R.; Stamm, S. Pre-mRNA Secondary Structures Influence Exon Recognition. PLoS Genet. 2007, 3, e204. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Kuo, C.-C.J.; Chen, L. GC content around splice sites affects splicing through pre-mRNA secondary structures. BMC Genom. 2011, 12, 90. [Google Scholar] [CrossRef] [Green Version]
- Královičová, J.; Patel, A.; Searle, M.; Vorechovsky, I. The role of short RNA loops in recognition of a single-hairpin exon derived from a mammalian-wide interspersed repeat. RNA Biol. 2015, 12, 54–69. [Google Scholar] [CrossRef]
- Lin, C.-L.; Taggart, A.J.; Lim, K.H.; Cygan, K.J.; Ferraris, L.; Creton, R.; Huang, Y.-T.; Fairbrother, W.G. RNA structure replaces the need for U2AF2 in splicing. Genome Res. 2015, 26, 12–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soemedi, R.; Cygan, K.J.; Rhine, C.; Glidden, D.T.; Taggart, A.; Lin, C.-L.; Fredericks, A.M.; Fairbrother, W.G. The effects of structure on pre-mRNA processing and stability. Methods 2017, 125, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Andrews, R.J.; Moss, W.N. Computational approaches for the discovery of splicing regulatory RNA structures. Biochim. Biophys. Acta (BBA) Bioenerg. 2019, 1862, 194380. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Breaker, R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005, 59, 487–517. [Google Scholar] [CrossRef]
- Duesterberg, V.; Fischer-Hwang, I.; Perez, C.F.; Hogan, D.; Block, S.M. Observation of long-range tertiary interactions during ligand binding by the TPP riboswitch aptamer. eLife 2015, 4, 17. [Google Scholar] [CrossRef]
- Spitale, R.C.; Flynn, R.A.; Zhang, Q.C.; Crisalli, P.; Lee, B.; Jung, J.-W.; Kuchelmeister, H.Y.; Batista, P.J.; Torre, E.A.; Kool, E.T.; et al. Erratum: Structural imprints in vivo decode RNA regulatory mechanisms. Nature 2015, 527, 264. [Google Scholar] [CrossRef]
- Lu, Z.; Chang, H.Y. The RNA Base-Pairing Problem and Base-Pairing Solutions. Cold Spring Harb. Perspect. Biol. 2018, 10, a034926. [Google Scholar] [CrossRef]
- Weng, X.; Gong, J.; Chen, Y.; Wu, T.; Wang, F.; Yang, S.; Yuan, Y.; Luo, G.; Chen, K.; Hu, L.; et al. Keth-seq for transcriptome-wide RNA structure mapping. Nat. Methods 2020, 16, 489–492. [Google Scholar] [CrossRef]
- Sun, L.; Fazal, F.M.; Li, P.; Broughton, J.P.; Lee, B.; Tang, L.; Huang, W.; Kool, E.T.; Chang, H.Y.; Zhang, Q.C. RNA structure maps across mammalian cellular compartments. Nat. Struct. Mol. Biol. 2019, 26, 322–330. [Google Scholar] [CrossRef]
- Dethoff, E.A.; Chugh, J.; Mustoe, A.M.; Al-Hashimi, H. Functional complexity and regulation through RNA dynamics. Nature 2012, 482, 322–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheah, M.T.; Wachter, A.; Sudarsan, N.; Breaker, R.R. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 2007, 447, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Breathnach, R.; Benoist, C.; O’Hare, K.; Gannon, F.; Chambon, P. Ovalbumin gene: Evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc. Natl. Acad. Sci. USA 1978, 75, 4853–4857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eperon, L.P.; Graham, I.R.; Griffiths, A.D.; Eperon, I. Effects of RNA secondary structure on alternative splicing of Pre-mRNA: Is folding limited to a region behind the transcribing RNA polymerase? Cell 1988, 54, 393–401. [Google Scholar] [CrossRef]
- Doudna, J.A.; A Doherty, E. Emerging themes in RNA folding. Fold. Des. 1997, 2, R65–R70. [Google Scholar] [CrossRef] [Green Version]
- Hendrix, D.K.; Brenner, S.E.; Holbrook, S.R. RNA structural motifs: Building blocks of a modular biomolecule. Q. Rev. Biophys. 2005, 38, 221–243. [Google Scholar] [CrossRef] [Green Version]
- Dawson, W.; Maciejczyk, M.; Jankowska, E.J.; Bujnicki, J.M. Coarse-grained modeling of RNA 3D structure. Methods 2016, 103, 138–156. [Google Scholar] [CrossRef]
- Espinosa, S.; Zhang, L.; Li, X.; Zhao, R. Understanding pre-mRNA splicing through crystallography. Methods 2017, 125, 55–62. [Google Scholar] [CrossRef]
- Schön, P. Atomic force microscopy of RNA: State of the art and recent advancements. Semin. Cell Dev. Biol. 2018, 73, 209–219. [Google Scholar] [CrossRef]
- Nowakowski, J.; Tinoco, I. RNA Structure and Stability. Semin. Virol. 1997, 8, 153–165. [Google Scholar] [CrossRef]
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwala, P.; Pandey, S.; Maiti, S. The tale of RNA G-quadruplex. Org. Biomol. Chem. 2015, 13, 5570–5585. [Google Scholar] [CrossRef]
- Kwok, C.K.; Marsico, G.; Sahakyan, A.B.; Chambers, V.; Balasubramanian, S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods 2016, 13, 841–844. [Google Scholar] [CrossRef] [PubMed]
- Pyle, A.M. Metal ions in the structure and function of RNA. JBIC J. Biol. Inorg. Chem. 2002, 7, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Theillet, F.-X.; Binolfi, A.; Frembgen-Kesner, T.; Hingorani, K.; Sarkar, M.; Kyne, C.; Li, C.; Crowley, P.B.; Gierasch, L.; Pielak, G.J.; et al. Physicochemical Properties of Cells and Their Effects on Intrinsically Disordered Proteins (IDPs). Chem. Rev. 2014, 114, 6661–6714. [Google Scholar] [CrossRef]
- Gracia, B.; Xue, Y.; Bisaria, N.; Herschlag, D.; Al-Hashimi, H.M.; Russell, R. RNA Structural Modules Control the Rate and Pathway of RNA Folding and Assembly. J. Mol. Biol. 2016, 428, 3972–3985. [Google Scholar] [CrossRef] [Green Version]
- Gracia, B.; Al-Hashimi, H.M.; Bisaria, N.; Das, R.; Herschlag, D.; Russell, R. Hidden Structural Modules in a Cooperative RNA Folding Transition. Cell Rep. 2018, 22, 3240–3250. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Gracia, B.; Herschlag, D.; Russell, R.; Al-Hashimi, H.M. Visualizing the formation of an RNA folding intermediate through a fast highly modular secondary structure switch. Nat. Commun. 2016, 7, 11. [Google Scholar] [CrossRef] [Green Version]
- Lai, W.-J.C.; Kayedkhordeh, M.; Cornell, E.V.; Farah, E.; Bellaousov, S.; Rietmeijer, R.; Salsi, E.; Mathews, D.H.; Ermolenko, D.N. mRNAs and lncRNAs intrinsically form secondary structures with short end-to-end distances. Nat. Commun. 2018, 9, 4328. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Loper, J.; Geman, S. Base-pair ambiguity and the kinetics of RNA folding. BMC Bioinform. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Spitale, R.C.; Crisalli, P.; Flynn, R.A.; Torre, E.A.; Kool, E.T.; Chang, H.Y. RNA SHAPE analysis in living cells. Nat. Methods 2012, 9, 18–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strobel, E.J.; Yu, A.; Lucks, J.B. High-throughput determination of RNA structures. Nat. Rev. Genet. 2018, 19, 615–634. [Google Scholar] [CrossRef]
- Rouskin, S.; Zubradt, M.; Washietl, S.; Kellis, M.; Weissman, J.S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 2013, 505, 701–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zubradt, M.; Gupta, P.; Persad, S.; Lambowitz, A.M.; Weissman, J.S.; Rouskin, S. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 2016, 14, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Bartel, D.P. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 2016, 353, aaf5371. [Google Scholar] [CrossRef] [Green Version]
- Schwalb, B.; Michel, M.; Zacher, B.; Frühauf, K.; Demel, C.; Tresch, A.; Gagneur, J.; Cramer, P. TT-seq maps the human transient transcriptome. Science 2016, 352, 1225–1228. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.A.; Smith, M.A.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.S.; et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016, 165, 1267–1279. [Google Scholar] [CrossRef] [Green Version]
- Aw, J.G.A.; Shen, Y.; Wilm, A.; Sun, M.; Ni Lim, X.; Boon, K.-L.; Tapsin, S.; Chan, Y.-S.; Tan, C.-P.; Sim, A.Y.; et al. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol. Cell 2016, 62, 603–617. [Google Scholar] [CrossRef] [Green Version]
- Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B.J. Global Mapping of Human RNA-RNA Interactions. Mol. Cell 2016, 62, 618–626. [Google Scholar] [CrossRef] [Green Version]
- Beaudoin, J.-D.; Novoa, E.M.; Vejnar, C.E.; Yartseva, V.; Takacs, C.M.; Kellis, M.; Giraldez, A.J. Analyses of mRNA structure dynamics identify embryonic gene regulatory programs. Nat. Struct. Mol. Biol. 2018, 25, 677–686. [Google Scholar] [CrossRef]
- De Groot, N.S.; Armaos, A.; Graña-Montes, R.; Alriquet, M.; Calloni, G.; Vabulas, R.M.; Tartaglia, G.G. RNA structure drives interaction with proteins. Nat. Commun. 2019, 10, 3246. [Google Scholar] [CrossRef]
- Sear, R.P. The cytoplasm of living cells: A functional mixture of thousands of components. J. Phys. Condens. Matter 2005, 17, S3587–S3595. [Google Scholar] [CrossRef] [Green Version]
- Benny, P.; Raghunath, M. Making microenvironments: A look into incorporating macromolecular crowding into in vitro experiments, to generate biomimetic microenvironments which are capable of directing cell function for tissue engineering applications. J. Tissue Eng. 2017, 8, 8. [Google Scholar] [CrossRef]
- Rivas, G.; Minton, A.P. Macromolecular Crowding In Vitro, In Vivo, and In Between. Trends Biochem. Sci. 2016, 41, 970–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klumpp, S.; Bode, W.; Puri, P. Life in crowded conditions. Eur. Phys. J. Speéc. Top. 2019, 227, 2315–2328. [Google Scholar] [CrossRef]
- Zhou, H.-X.; Rivas, G.; Minton, A.P. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008, 37, 375–397. [Google Scholar] [CrossRef] [Green Version]
- Paudel, B.P.; Fiorini, E.; Börner, R.; Sigel, R.K.O.; Rueda, D.S. Optimal molecular crowding accelerates group II intron folding and maximizes catalysis. Proc. Natl. Acad. Sci. USA 2018, 115, 11917–11922. [Google Scholar] [CrossRef] [Green Version]
- Dupuis, N.F.; Holmstrom, E.D.; Nesbitt, D.J. Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics. Proc. Natl. Acad. Sci. USA 2014, 111, 8464–8469. [Google Scholar] [CrossRef] [Green Version]
- Boersma, A.J.; Zuhorn, I.S.; Poolman, B. A sensor for quantification of macromolecular crowding in living cells. Nat. Methods 2015, 12, 227–229. [Google Scholar] [CrossRef]
- Murade, C.U.; Shubeita, G.T. A Molecular Sensor Reveals Differences in Macromolecular Crowding between the Cytoplasm and Nucleoplasm. ACS Sensors 2019, 4, 1835–1843. [Google Scholar] [CrossRef]
- Gomez, D.; Huber, K.; Klumpp, S. On Protein Folding in Crowded Conditions. J. Phys. Chem. Lett. 2019, 10, 7650–7656. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawyer, I.A.; Sturgill, D.; Sung, M.-H.; Hager, G.L.; Dundr, M. Cajal body function in genome organization and transcriptome diversity. BioEssays 2016, 38, 1197–1208. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.E.; Manteiga, J.C.; Henninger, J.E.; Sabari, B.R.; Dall’Agnese, A.; Hannett, N.M.; Spille, J.-H.; Afeyan, L.K.; Zamudio, A.V.; Shrinivas, K.; et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 2019, 572, 543–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galganski, L.; Urbanek, M.O.; Krzyzosiak, W.J. Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Res. 2017, 45, 10350–10368. [Google Scholar] [CrossRef] [Green Version]
- Sei-Iida, Y. Real-time monitoring of in vitro transcriptional RNA synthesis using fluorescence resonance energy transfer. Nucleic Acids Res. 2000, 28, 59. [Google Scholar] [CrossRef] [Green Version]
- Pan, T.; Sosnick, T. Rna folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 161–175. [Google Scholar] [CrossRef]
- Lai, D.; Proctor, J.R.; Meyer, I.M. On the importance of cotranscriptional RNA structure formation. RNA 2013, 19, 1461–1473. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.-R.; Hu, C.-G.; Hu, Z.C.-G. Regulatory effects of cotranscriptional RNA structure formation and transitions. Wiley Interdiscip. Rev. RNA 2016, 7, 562–574. [Google Scholar] [CrossRef]
- Jonkers, I.; Kwak, H.; Lis, J.T. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 2014, 3, e02407. [Google Scholar] [CrossRef]
- Naftelberg, S.; Schor, I.E.; Ast, G.; Kornblihtt, A. Regulation of Alternative Splicing Through Coupling with Transcription and Chromatin Structure. Annu. Rev. Biochem. 2015, 84, 165–198. [Google Scholar] [CrossRef] [PubMed]
- Wallace, E.W.J.; Beggs, J.D. Extremely fast and incredibly close: Cotranscriptional splicing in budding yeast. RNA 2017, 23, 601–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pai, A.A.; Henriques, T.; McCue, K.; Burkholder, A.; Adelman, K.; Burge, C.B. The kinetics of pre-mRNA splicing in the Drosophila genome and the influence of gene architecture. Abstract 2017, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Wachutka, L.; Caizzi, L.; Gagneur, J.; Cramer, P. Global donor and acceptor splicing site kinetics in human cells. eLife 2019, 8, 52. [Google Scholar] [CrossRef]
- Drexler, H.L.; Choquet, K.; Churchman, L.S. Splicing Kinetics and Coordination Revealed by Direct Nascent RNA Sequencing through Nanopores. Mol. Cell 2020, 77, 985–998.e8. [Google Scholar] [CrossRef]
- Fong, N.; Kim, H.; Zhou, Y.; Ji, X.; Qiu, J.; Saldi, T.; Diener, K.; Jones, K.; Fu, X.-D.; Bentley, D. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014, 28, 2663–2676. [Google Scholar] [CrossRef] [Green Version]
- Dujardin, G.; Lafaille, C.; De La Mata, M.; Marasco, L.E.; Muñoz, M.J.; Le Jossic-Corcos, C.; Corcos, L.; Kornblihtt, A. How Slow RNA Polymerase II Elongation Favors Alternative Exon Skipping. Mol. Cell 2014, 54, 683–690. [Google Scholar] [CrossRef] [Green Version]
- Aslanzadeh, V.; Huang, Y.; Sanguinetti, G.; Beggs, J.D. Corrigendum: Transcription rate strongly affects splicing fidelity and cotranscriptionality in budding yeast. Genome Res. 2018, 28, 203–213. [Google Scholar] [CrossRef] [Green Version]
- Neugebauer, K.M. Nascent RNA and the Coordination of Splicing with Transcription. Cold Spring Harb. Perspect. Biol. 2019, 11, 13. [Google Scholar] [CrossRef] [Green Version]
- Incarnato, D.; Morandi, E.; Anselmi, F.; Simon, L.M.; Basile, G.; Oliviero, S. In vivo probing of nascent RNA structures reveals principles of cotranscriptional folding. Nucleic Acids Res. 2017, 45, 9716–9725. [Google Scholar] [CrossRef] [Green Version]
- Saldi, T.; Fong, N.; Bentley, D.L. Corrigendum: Transcription elongation rate affects nascent histone pre-mRNA folding and 3′ end processing. Genes Dev. 2018, 32, 592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyons, S.M.; Cunningham, C.H.; Welch, J.D.; Groh, B.; Guo, A.Y.; Wei, B.; Whitfield, M.L.; Xiong, Y.; Marzluff, W.F. A subset of replication-dependent histone mRNAs are expressed as polyadenylated RNAs in terminally differentiated tissues. Nucleic Acids Res. 2016, 44, 9190–9205. [Google Scholar] [CrossRef] [Green Version]
- Nikolaev, Y.; Ripin, N.; Soste, M.; Picotti, P.; Iber, D.; Allain, F.H.-T. Publisher Correction: Systems NMR: Single-sample quantification of RNA, proteins and metabolites for biomolecular network analysis. Nat. Methods 2019, 16, 932. [Google Scholar] [CrossRef] [Green Version]
- Roundtree, I.A.; He, C. RNA epigenetics—chemical messages for posttranscriptional gene regulation. Curr. Opin. Chem. Biol. 2016, 30, 46–51. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kierzek, R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 2003, 31, 4472–4480. [Google Scholar] [CrossRef] [PubMed]
- Roost, C.; Lynch, S.R.; Batista, P.J.; Qu, K.; Chang, H.Y.; Kool, E.T. Correction to “Structure and Thermodynamics of N6-Methyladenosine in RNA: A Spring-Loaded Base Modification”. J. Am. Chem. Soc. 2015, 137, 8308. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, Y.; Sun, B.-F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.-J.; Ping, X.-L.; Chen, Y.-S.; Wang, W.-J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- Bartosovic, M.; Molares, H.C.; Gregorova, P.; Hrossova, D.; Kudla, G.; Vaňáčová, Š. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 2017, 45, 11356–11370. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef] [Green Version]
- Lewis, C.; Pan, T.; Kalsotra, A. RNA modifications and structures cooperate to guide RNA–protein interactions. Nat. Rev. Mol. Cell Biol. 2017, 18, 202–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, C.; Klukovich, R.; Peng, H.; Wang, Z.; Yu, T.; Zhang, Y.; Zheng, H.; Klungland, A.; Yan, W. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. USA 2017, 115, E325–E333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasowitz, S.; Ma, J.; Anderson, S.J.; Leu, N.A.; Xu, Y.; Gregory, B.D.; Schultz, R.M.; Wang, P.J. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018, 14, e1007412. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, A.L.; Hongay, C.F. The m6A Dynamics of Profilin in Neurogenesis. Front. Genet. 2019, 10, 987. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Louloupi, A.; Ntini, E.; Conrad, T.; Ørom, U.A.V. Transient N-6-Methyladenosine Transcriptome Sequencing Reveals a Regulatory Role of m6A in Splicing Efficiency. Cell Rep. 2018, 23, 3429–3437. [Google Scholar] [CrossRef]
- Schwartz, S.; Bernstein, U.A.; Mumbach, M.; Jovanovic, M.; Herbst, R.H.; León-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [Green Version]
- Boo, S.H.; Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020, 52, 400–408. [Google Scholar] [CrossRef] [Green Version]
- Carlile, T.M.; Martinez, N.M.; Schaening, C.; Su, A.; Bell, T.A.; Zinshteyn, B.; Gilbert, W.V. mRNA structure determines modification by pseudouridine synthase 1. Nat. Methods 2019, 15, 966–974. [Google Scholar] [CrossRef]
- Kierzek, E.; Malgowska, M.; Lisowiec-Wachnicka, J.; Turner, U.H.; Gdaniec, Z.; Kierzek, R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2013, 42, 3492–3501. [Google Scholar] [CrossRef]
- Hudson, G.A.; Bloomingdale, R.J.; Znosko, B.M. Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides. RNA 2013, 19, 1474–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charette, M.W.G.M.; Gray, M.W. Pseudouridine in RNA: What, Where, How, and Why. IUBMB Life 2000, 49, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dunker, W.; Yu, Y.-T.; Karijolich, J. The Role of Noncoding RNA Pseudouridylation in Nuclear Gene Expression Events. Front. Bioeng. Biotechnol. 2018, 6, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adachi, H.; De Zoysa, M.D.; Yu, Y.-T. Post-transcriptional pseudouridylation in mRNA as well as in some major types of noncoding RNAs. Biochim. Biophys. Acta (BBA) Bioenerg. 2019, 1862, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Polson, A.G.; Crain, P.F.; Pomerantz, S.C.; McCloskey, J.A.; Bass, B.L. The mechanism of adenosine to inosine conversion by the double-stranded RNA unwinding/modifying activity: A high-performance liquid chromatography-mass spectrometry analysis. Biochemistry 1991, 30, 11507–11514. [Google Scholar] [CrossRef]
- Rieder, L.E.; Staber, C.J.; Hoopengardner, B.; Reenan, R.A. Tertiary structural elements determine the extent and specificity of messenger RNA editing. Nat. Commun. 2013, 4, 2232. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, Y.-H.E.; Bahn, J.H.; Yang, Y.; Lin, X.; Tran, S.; Yang, E.-W.; Quinones-Valdez, G.; Xiao, X. RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res. 2018, 28, 812–823. [Google Scholar] [CrossRef] [Green Version]
- Licht, K.; Kapoor, U.; Amman, F.; Picardi, E.; Martin, D.; Bajad, P.; Jantsch, M.F. A high resolution A-to-I editing map in the mouse identifies editing events controlled by pre-mRNA splicing. Genome Res. 2019, 29, 1453–1463. [Google Scholar] [CrossRef]
- Hong, H.; An, O.; Chan, T.H.M.; Ng, V.H.E.; Kwok, H.S.; Lin, S.J.; Qi, L.; Han, J.; Tay, T.J.D.; Tang, S.J.; et al. Bidirectional regulation of adenosine-to-inosine (A-to-I) RNA editing by DEAH box helicase 9 (DHX9) in cancer. Nucleic Acids Res. 2018, 46, 7953–7969. [Google Scholar] [CrossRef]
- Tang, S.J.; Shen, H.; An, O.; Hong, H.; Li, J.; Song, Y.; Han, J.; Tay, D.J.T.; Ng, V.H.E.; Molias, F.B.; et al. Cis- and trans-regulations of pre-mRNA splicing by RNA editing enzymes influence cancer development. Nat. Commun. 2020, 11, 799. [Google Scholar] [CrossRef] [Green Version]
- Daniel, C.; Widmark, A.; Rigardt, D.; Öhman, M. Editing inducer elements increases A-to-I editing efficiency in the mammalian transcriptome. Genome Biol. 2017, 18, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Athanasiadis, A.; Rich, A.; Maas, S. Widespread A-to-I RNA Editing of Alu-Containing mRNAs in the Human Transcriptome. PLoS Biol. 2004, 2, e391. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Yang, L. ALU ternative Regulation for Gene Expression. Trends Cell Biol. 2017, 27, 480–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Versteeg, R.; Van Schaik, B.D.; Van Batenburg, M.F.; Roos, M.; Monajemi, R.; Caron, H.; Bussemaker, H.J.; Van Kampen, A.H. The Human Transcriptome Map Reveals Extremes in Gene Density, Intron Length, GC Content, and Repeat Pattern for Domains of Highly and Weakly Expressed Genes. Genome Res. 2003, 13, 1998–2004. [Google Scholar] [CrossRef] [Green Version]
- Lev-Maor, G.; Ram, O.; Kim, O.; Sela, N.; Goren, A.; Levanon, E.Y.; Ast, G. Intronic Alus Influence Alternative Splicing. PLoS Genet. 2008, 4, e1000204. [Google Scholar] [CrossRef] [PubMed]
- Nakama, M.; Otsuka, H.; Ago, Y.; Sasai, H.; Abdelkreem, E.; Aoyama, Y.; Fukao, T. Intronic antisense Alu elements have a negative splicing effect on the inclusion of adjacent downstream exons. Gene 2018, 664, 84–89. [Google Scholar] [CrossRef]
- Payer, L.M.; Steranka, J.P.; Ardeljan, D.; Walker, J.; Fitzgerald, K.C.; Calabresi, A.P.; Cooper, A.T.; Burns, K.H. Alu insertion variants alter mRNA splicing. Nucleic Acids Res. 2018, 47, 421–431. [Google Scholar] [CrossRef] [Green Version]
- Lev-Maor, G.; Sorek, R.; Shomron, N.; Ast, G. The Birth of an Alternatively Spliced Exon: 3′ Splice-Site Selection in Alu Exons. Science 2003, 300, 1288–1291. [Google Scholar] [CrossRef]
- Miriami, E. Conserved sequence elements associated with exon skipping. Nucleic Acids Res. 2003, 31, 1974–1983. [Google Scholar] [CrossRef]
- Dardenne, E.; Espinoza, M.P.; Fattet, L.; Germann, S.; Lambert, M.-P.; Neil, H.; Zonta, E.; Mortada, H.; Gratadou, L.; Deygas, M.; et al. RNA Helicases DDX5 and DDX17 Dynamically Orchestrate Transcription, miRNA, and Splicing Programs in Cell Differentiation. Cell Rep. 2014, 7, 1900–1913. [Google Scholar] [CrossRef] [Green Version]
- Lemaire, S.; Fontrodona, N.; Aubé, F.; Claude, J.-B.; Polvèche, H.; Modolo, L.; Bourgeois, C.F.; Mortreux, F.; Auboeuf, D. Characterizing the interplay between gene nucleotide composition bias and splicing. Genome Biol. 2019, 20, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Wachter, A. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 2010, 7, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Breaker, R.R. Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing. Nucleic Acids Res. 2013, 41, 3022–3031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donovan, P.D.; Holland, L.M.; Lombardi, L.; Coughlan, A.Y.; Higgins, D.G.; Wolfe, K.H.; Butler, G. TPP riboswitch-dependent regulation of an ancient thiamin transporter in Candida. PLoS Genet. 2018, 14, e1007429. [Google Scholar] [CrossRef] [PubMed]
- Antunes, D.; Jorge, N.; Costa, M.G.D.S.; Passetti, F.; Caffarena, E. Unraveling RNA dynamical behavior of TPP riboswitches: A comparison between Escherichia coli and Arabidopsis thaliana. Sci. Rep. 2019, 9, 4197. [Google Scholar] [CrossRef]
- Wu, M.T.-P.; D’Souza, V. Alternate RNA Structures. Cold Spring Harb. Perspect. Biol. 2020, 12, a032425. [Google Scholar] [CrossRef] [Green Version]
- Rajkowitsch, L.; Chen, R.; Stampfl, S.; Semrad, K.; Waldsich, C.; Mayer, O.; Jantsch, M.F.; Konrat, R.; Bläsi, U.; Schroeder, R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007, 4, 118–130. [Google Scholar] [CrossRef] [Green Version]
- Doetsch, M.; Schroeder, R.; Fürtig, B. Transient RNA–protein interactions in RNA folding. FEBS J. 2011, 278, 1634–1642. [Google Scholar] [CrossRef] [Green Version]
- Subbaiah, K.C.V.; Hedaya, O.; Wu, J.; Jiang, F.; Yao, P. Mammalian RNA switches: Molecular rheostats in gene regulation, disease, and medicine. Comput. Struct. Biotechnol. J. 2019, 17, 1326–1338. [Google Scholar] [CrossRef]
- Bourgeois, C.; Mortreux, F.; Auboeuf, D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 426–438. [Google Scholar] [CrossRef]
- Putnam, A.; Jankowsky, E. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim. Biophys. Acta (BBA) Bioenerg. 2013, 1829, 884–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilman, B.; Tijerina, P.; Russell, R. Distinct RNA-unwinding mechanisms of DEAD-box and DEAH-box RNA helicase proteins in remodeling structured RNAs and RNPs. Biochem. Soc. Trans. 2017, 45, 1313–1321. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, P.; Huang, J.T.J.; Hiom, K. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat. Commun. 2018, 9, 4346. [Google Scholar] [CrossRef] [PubMed]
- Moy, R.H.; Cole, B.S.; Yasunaga, A.; Gold, B.; Shankarling, G.; Varble, A.; Molleston, J.M.; Tenoever, B.R.; Lynch, K.W.; Cherry, S. Stem-Loop Recognition by DDX17 Facilitates miRNA Processing and Antiviral Defense. Cell 2014, 158, 764–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McRae, E.; Booy, E.P.; Moya-Torres, A.; Ezzati, P.; Stetefeld, J.; McKenna, S.A. Human DDX21 binds and unwinds RNA guanine quadruplexes. Nucleic Acids Res. 2017, 45, 6656–6668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.J.; Wang, Q.; Rio, N.C. Coordinate regulation of alternative pre-mRNA splicing events by the human RNA chaperone proteins hnRNPA1 and DDX5. Genes Dev. 2018, 32, 1060–1074. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Bai, J.; Jiang, T.; Gao, Y.; Hua, Y. Modulation of PDCD1 exon 3 splicing. RNA Biol. 2019, 16, 1794–1805. [Google Scholar] [CrossRef]
- Ngo, T.D.; Partin, A.C.; Nam, Y. RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis. Cell Rep. 2019, 29, 4024–4035.e5. [Google Scholar] [CrossRef] [Green Version]
- Hönig, A.; Auboeuf, D.; Parker, M.M.; O’Malley, B.W.; Berget, S.M. Regulation of Alternative Splicing by the ATP-Dependent DEAD-Box RNA Helicase p72. Mol. Cell. Biol. 2002, 22, 5698–5707. [Google Scholar] [CrossRef] [Green Version]
- Damianov, A.; Ying, Y.; Lin, C.-H.; Lee, J.-A.; Tran, D.; Vashisht, A.A.; Bahrami-Samani, E.; Xing, Y.; Martin, K.C.; Wohlschlegel, J.A.; et al. Rbfox Proteins Regulate Splicing as Part of a Large Multiprotein Complex LASR. Cell 2016, 165, 606–619. [Google Scholar] [CrossRef] [Green Version]
- Nakata, D.; Nakao, S.; Nakayama, K.; Araki, S.; Nakayama, Y.; Aparicio, S.; Hara, T.; Nakanishi, A. The RNA helicase DDX39B and its paralog DDX39A regulate androgen receptor splice variant AR-V7 generation. Biochem. Biophys. Res. Commun. 2017, 483, 271–276. [Google Scholar] [CrossRef]
- Taylor, K.; Sznajder, Ł.J.; Cywoniuk, P.; Thomas, J.D.; Swanson, M.S.; Sobczak, K. MBNL splicing activity depends on RNA binding site structural context. Nucleic Acids Res. 2018, 46, 9119–9133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gates, D.P.; Coonrod, L.A.; Berglund, J.A. Autoregulated Splicing of muscleblind-like 1 (MBNL1) Pre-mRNA. J. Biol. Chem. 2011, 286, 34224–34233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falaleeva, M.; Pages, A.; Matuszek, Z.; Hidmi, S.; Agranat-Tamir, L.; Korotkov, K.V.; Nevo, Y.; Eyras, E.; Sperling, R.; Stamm, S. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2016, 113, E1625–E1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishore, S.; Stamm, S. The snoRNA HBII-52 Regulates Alternative Splicing of the Serotonin Receptor 2C. Science 2006, 311, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Kishore, S.; Khanna, A.; Zhang, Z.; Hui, J.; Balwierz, P.J.; Stefan, M.; Beach, C.; Nicholls, R.D.; Zavolan, M.; Stamm, S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 2010, 19, 1153–1164. [Google Scholar] [CrossRef] [Green Version]
- Georgomanolis, T.; Sofiadis, K.; Papantonis, A. Cutting a Long Intron Short: Recursive Splicing and Its Implications. Front. Physiol. 2016, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Howe, K.J.; Ares, M. Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc. Natl. Acad. Sci. USA 1997, 94, 12467–12472. [Google Scholar] [CrossRef] [Green Version]
- Rogic, S.; Montpetit, B.; Hoos, H.H.; Mackworth, A.K.; Ouellette, B.F.F.; Hieter, P. Correlation between the secondary structure of pre-mRNA introns and the efficiency of splicing in Saccharomyces cerevisiae. BMC Genom. 2008, 9, 355. [Google Scholar] [CrossRef] [Green Version]
- Neil, C.R.; Fairbrother, W.G. Intronic RNA: Ad’junk’ mediator of post-transcriptional gene regulation. Biochim. Biophys. Acta (BBA) Bioenerg. 2019, 1862, 194439. [Google Scholar] [CrossRef]
- Meyer, M.; Plass, M.; Valle, J.P.; Eyras, E.; Vilardell, J. Deciphering 3′ss Selection in the Yeast Genome Reveals an RNA Thermosensor that Mediates Alternative Splicing. Mol. Cell 2011, 43, 1033–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovci, M.T.; Ghanem, D.; Marr, H.; Arnold, J.; Gee, S.; Parra, M.; Liang, T.Y.; Stark, T.J.; Gehman, L.T.; Hoon, S.; et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 2013, 20, 1434–1442. [Google Scholar] [CrossRef] [PubMed]
- Nasim, F.-U.H.; Hutchison, S.; Cordeau, M.; Chabot, B. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA 2002, 8, 1078–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberstrass, F.C.; Auweter, S.D.; Erat, M.; Hargous, Y.; Henning, A.; Wenter, P.; Reymond, L.; Amir-Ahmady, B.; Pitsch, S.; Black, U.L.; et al. Structure of PTB Bound to RNA: Specific Binding and Implications for Splicing Regulation. Science 2005, 309, 2054–2057. [Google Scholar] [CrossRef]
- Raker, V.A.; Mironov, A.A.; Gelfand, M.S.; Pervouchine, D.D. Modulation of alternative splicing by long-range RNA structures in Drosophila. Nucleic Acids Res. 2009, 37, 4533–4544. [Google Scholar] [CrossRef] [Green Version]
- Muh, S.J.; Hovhannisyan, R.H.; Carstens, R.P. A Non-sequence-specific Double-stranded RNA Structural Element Regulates Splicing of Two Mutually Exclusive Exons of Fibroblast Growth Factor Receptor 2 (FGFR2). J. Biol. Chem. 2002, 277, 50143–50154. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, T.M.; Pervouchine, D.D. An Evolutionary Mechanism for the Generation of Competing RNA Structures Associated with Mutually Exclusive Exons. Genes 2018, 9, 356. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Yang, Y.; Dai, L.; Cao, G.; Chen, R.; Hong, W.; Liu, B.; Shi, Y.; Meng, Y.; Shi, F.; et al. Corrigendum: Long-range RNA pairings contribute to mutually exclusive splicing. RNA 2016, 22, 1640–1642. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Shi, Y.; Wu, Y.; Meng, Y.; Jin, Y. Role of RNA secondary structures in regulating Dscam alternative splicing. Biochim. Biophys. Acta (BBA) Bioenerg. 2019, 1862, 194381. [Google Scholar] [CrossRef]
- Shi, H.; Hoffman, E.B.; Lis, J.T. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol. 1997, 17, 2649–2657. [Google Scholar] [CrossRef] [Green Version]
- Muro, A.F.; Caputi, M.; Pariyarath, R.; Pagani, F.; Buratti, E.; Baralle, F. Regulation of Fibronectin EDA Exon Alternative Splicing: Possible Role of RNA Secondary Structure for Enhancer Display. Mol. Cell. Biol. 1999, 19, 2657–2671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, M.-Y.; Underwood, J.G.; Nikolic, J.; Luu, M.H.; Black, D.L. Multisite RNA Binding and Release of Polypyrimidine Tract Binding Protein during the Regulation of c-src Neural-Specific Splicing. Mol. Cell 2000, 5, 949–957. [Google Scholar] [CrossRef]
- Buratti, E.; Muro, A.F.; Giombi, M.; Gherbassi, D.; Iaconcig, A.; Baralle, F. RNA Folding Affects the Recruitment of SR Proteins by Mouse and Human Polypurinic Enhancer Elements in the Fibronectin EDA Exon. Mol. Cell. Biol. 2004, 24, 1387–1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cywoniuk, P.; Taylor, K.; Sznajder, Ł.J.; Sobczak, K. Hybrid splicing minigene and antisense oligonucleotides as efficient tools to determine functional protein/RNA interactions. Sci. Rep. 2017, 7, 17587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Gribskov, M. The Role of RNA Sequence and Structure in RNA–Protein Interactions. J. Mol. Biol. 2011, 409, 574–587. [Google Scholar] [CrossRef]
- Iwakiri, J.; Tateishi, H.; Chakraborty, A.; Patil, P.; Kenmochi, N. Dissecting the protein–RNA interface: The role of protein surface shapes and RNA secondary structures in protein–RNA recognition. Nucleic Acids Res. 2011, 40, 3299–3306. [Google Scholar] [CrossRef] [Green Version]
- Kligun, E.; Mandel-Gutfreund, Y. The role of RNA conformation in RNA-protein recognition. RNA Biol. 2015, 12, 720–727. [Google Scholar] [CrossRef] [Green Version]
- Lambert, N.; Robertson, A.; Jangi, M.; McGeary, S.; Sharp, P.A.; Burge, C.B. RNA Bind-n-Seq: Quantitative Assessment of the Sequence and Structural Binding Specificity of RNA Binding Proteins. Mol. Cell 2014, 54, 887–900. [Google Scholar] [CrossRef] [Green Version]
- Lee, F.; Ule, J. Advances in CLIP Technologies for Studies of Protein-RNA Interactions. Mol. Cell 2018, 69, 354–369. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Miles, W.O. Beyond CLIP: Advances and opportunities to measure RBP–RNA and RNA–RNA interactions. Nucleic Acids Res. 2019, 47, 5490–5501. [Google Scholar] [CrossRef] [Green Version]
- Taliaferro, J.M.; Lambert, N.J.; Sudmant, P.H.; Dominguez, D.; Merkin, J.J.; Alexis, M.S.; Bazile, C.A.; Burge, C.B. RNA Sequence Context Effects Measured In Vitro Predict In Vivo Protein Binding and Regulation. Mol. Cell 2016, 64, 294–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Nostrand, E.; Pratt, G.A.; Shishkin, A.A.; Gelboin-Burkhart, C.; Fang, M.Y.; Sundararaman, B.; Blue, S.M.; Nguyen, T.B.; Surka, C.; Elkins, K.; et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 2016, 13, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Yang, Q.; Yang, L. RNA Structure Switches RBP Binding. Mol. Cell 2016, 64, 219–220. [Google Scholar] [CrossRef] [Green Version]
- Maticzka, D.; Lange, S.J.; Costa, F.; Backofen, R. GraphProt: Modeling binding preferences of RNA-binding proteins. Genome Biol. 2014, 15, R17. [Google Scholar] [CrossRef] [Green Version]
- Cereda, M.; Pozzoli, U.; Rot, G.; Juvan, P.; Schweitzer, A.; Clark, T.A.; Ule, J. RNAmotifs: Prediction of multivalent RNA motifs that control alternative splicing. Genome Biol. 2014, 15, R20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polishchuk, M.; Paz, I.; Kohen, R.; Mesika, R.; Yakhini, Z.; Mandel-Gutfreund, Y. A combined sequence and structure based method for discovering enriched motifs in RNA from in vivo binding data. Methods 2017, 118, 73–81. [Google Scholar] [CrossRef]
- Cook, K.B.; Vembu, S.; Ha, K.C.; Zheng, H.; Laverty, K.U.; Hughes, T.R.; Ray, D.; Morris, Q.D. RNAcompete-S: Combined RNA sequence/structure preferences for RNA binding proteins derived from a single-step in vitro selection. Methods 2017, 126, 18–28. [Google Scholar] [CrossRef]
- Ray, D.; Kazan, H.; Cook, K.; Weirauch, M.T.; Najafabadi, H.S.; Li, X.; Gueroussov, S.; Albu, M.; Zheng, H.; Yang, A.; et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 2013, 499, 172–177. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.M.; Long, J.; De Moor, C.H.; Emsley, J.; Searle, M.S. Structural insights into the targeting of mRNA GU-rich elements by the three RRMs of CELF1. Nucleic Acids Res. 2013, 41, 7153–7166. [Google Scholar] [CrossRef] [Green Version]
- Vasilyev, N.; Polonskaia, A.; Darnell, J.C.; Darnell, R.B.; Patel, D.J.; Serganov, A. Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. Proc. Natl. Acad. Sci. USA 2015, 112, E5391–E5400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoell, J.I.; Larsson, E.; Runge, S.; Nusbaum, J.D.; Duggimpudi, S.; Farazi, T.A.; Hafner, M.; Borkhardt, A.; Sander, C.; Tuschl, T. RNA targets of wild-type and mutant FET family proteins. Nat. Struct. Mol. Biol. 2011, 18, 1428–1431. [Google Scholar] [CrossRef] [PubMed]
- Lerga, A.; Hallier, M.; Delva, L.; Orvain, C.; Gallais, I.; Marie, J.; Moreau-Gachelin, F. Identification of an RNA Binding Specificity for the Potential Splicing Factor TLS. J. Biol. Chem. 2000, 276, 6807–6816. [Google Scholar] [CrossRef] [Green Version]
- Ince-Dunn, G.; Okano, H.J.; Jensen, K.B.; Park, W.-Y.; Zhong, R.; Ule, J.; Mele, A.; Fak, J.J.; Yang, C.; Zhang, C.; et al. Neuronal Elav-like (Hu) Proteins Regulate RNA Splicing and Abundance to Control Glutamate Levels and Neuronal Excitability. Neuron 2012, 75, 1067–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ule, J.; Jensen, K.B.; Mele, A.; Ruggiu, M.; Darnell, R.B. CLIP Identifies Nova-Regulated RNA Networks in the Brain. Science 2003, 302, 1212–1215. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, T.; Ozaki, H.; Terai, G.; Asai, K.; Iwasaki, W.; Kiryu, H. CapR: Revealing structural specificities of RNA-binding protein target recognition using CLIP-seq data. Genome Biol. 2014, 15, R16. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Maris, C.; Allain, F.H.-T.; Black, U.L. U1 snRNA directly interacts with polypyrimidine tract-binding protein during splicing repression. Mol. Cell 2011, 41, 579–588. [Google Scholar] [CrossRef] [Green Version]
- Královicová, J.; Ševcíková, I.; Stejskalová, E.; Obuca, M.; Hiller, M.; Staněk, D.; Vorechovsky, I. PUF60-activated exons uncover altered 3′ splice-site selection by germline missense mutations in a single RRM. Nucleic Acids Res. 2018, 46, 6166–6187. [Google Scholar] [CrossRef]
- Galarneau, A.; Richard, S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 2005, 12, 691–698. [Google Scholar] [CrossRef]
- Auweter, S.D.; Fasan, R.; Reymond, L.; Underwood, J.G.; Black, U.L.; Pitsch, S.; Allain, F.H.-T. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2005, 25, 163–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Zubovic, L.; Yang, F.; Godin, K.; Pavelitz, T.; Castellanos, J.; Macchi, P.; Varani, G. Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer. Nucleic Acids Res. 2016, 44, 4381–4395. [Google Scholar] [CrossRef]
- Ray, D.; Kazan, H.; Chan, E.T.; Castillo, L.P.; Chaudhry, S.; Talukder, S.; Blencowe, B.J.; Morris, Q.; Hughes, T.R. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009, 27, 667–670. [Google Scholar] [CrossRef]
- Kojima, S.; Matsumoto, K.; Hirose, M.; Shimada, M.; Nagano, M.; Shigeyoshi, Y.; Hoshino, S.-I.; Ui-Tei, K.; Saigo, K.; Green, C.B.; et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl. Acad. Sci. USA 2007, 104, 1859–1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skrisovska, L.; Bourgeois, C.F.; Stefl, R.; Grellscheid, S.-N.; Kister, L.; Wenter, P.; Elliott, D.J.; Stevenin, J.; Allain, F.H.-T. The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep. 2007, 8, 372–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, M.; Haga, I.; Li, Q.-H.; Fujisawa, I.J. Identification of cellular mRNA targets for RNA-binding protein Sam68. Nucleic Acids Res. 2002, 30, 5452–5464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legendre, J.B.; Campbell, Z.T.; Kroll-Conner, P.; Anderson, P.; Kimble, J.; Wickens, M. RNA Targets and Specificity of Staufen, a Double-stranded RNA-binding Protein in Caenorhabditis elegans. J. Biol. Chem. 2012, 288, 2532–2545. [Google Scholar] [CrossRef] [Green Version]
- Kapeli, K.; Pratt, G.A.; Vu, A.Q.; Hutt, K.R.; Martinez, F.J.; Sundararaman, B.; Batra, R.; Freese, P.; Lambert, N.J.; Huelga, S.C.; et al. Distinct and shared functions of ALS-associated proteins TDP-43, FUS and TAF15 revealed by multisystem analyses. Nat. Commun. 2016, 7, 12143. [Google Scholar] [CrossRef] [Green Version]
- Kashyap, M.; Ganguly, A.; Bhavesh, N.S. Structural delineation of stem-loop RNA binding by human TAF15 protein. Sci. Rep. 2015, 5, 17298. [Google Scholar] [CrossRef] [Green Version]
- Kuo, P.-H.; Doudeva, L.G.; Wang, Y.-T.; Shen, C.-K.J.; Yuan, H.S. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009, 37, 1799–1808. [Google Scholar] [CrossRef]
- Wang, Z.; Kayikci, M.; Briese, M.; Zarnack, K.; Luscombe, N.M.; Rot, G.; Zupan, B.; Curk, T.; Ule, J. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010, 8, e1000530. [Google Scholar] [CrossRef] [Green Version]
- Meyer, C.; Garzia, A.; Mazzola, M.; Gerstberger, S.; Molina, H.; Tuschl, T. The TIA1 RNA-Binding Protein Family Regulates EIF2AK2-Mediated Stress Response and Cell Cycle Progression. Mol. Cell 2018, 69, 622–635.e6. [Google Scholar] [CrossRef] [Green Version]
- McAlinden, A.; Liang, L.; Mukudai, Y.; Imamura, T.; Sandell, L.J. Nuclear Protein TIA-1 RegulatesCOL2A1Alternative Splicing and Interacts with Precursor mRNA and Genomic DNA. J. Biol. Chem. 2007, 282, 24444–24454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warf, M.B.; Diegel, J.V.; Von Hippel, P.H.; Berglund, J.A. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl. Acad. Sci. USA 2009, 106, 9203–9208. [Google Scholar] [CrossRef] [Green Version]
- McCullough, A.J.; Berget, S.M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 1997, 17, 4562–4571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcel, V.; Tran, P.L.; Sagne, C.; Martel-Planche, G.; Vaslin, L.; Teulade-Fichou, M.-P.; Hall, J.; Mergny, J.-L.; Hainaut, P.; Van Dyck, E. G-quadruplex structures in TP53 intron 3: Role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis 2010, 32, 271–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, S.P.; Das, P. G-quadruplex structure at intron 2 of TFE3 and its role in Xp11.2 translocation and splicing. Biochim. Biophys. Acta (BBA) Gen. Subj. 2018, 1862, 630–636. [Google Scholar] [CrossRef]
- Al-Zeer, M.A.; Kurreck, J. Deciphering the Enigmatic Biological Functions of RNA Guanine-Quadruplex Motifs in Human Cells. Biochemistry 2018, 58, 305–311. [Google Scholar] [CrossRef]
- Alkan, S.A.; Martincic, K.; Milcarek, C. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem. J. 2005, 393, 361–371. [Google Scholar] [CrossRef] [Green Version]
- Dalziel, M.; Nunes, N.M.; Furger, A. Two G-Rich Regulatory Elements Located Adjacent to and 440 Nucleotides Downstream of the Core Poly(A) Site of the Intronless Melanocortin Receptor 1 Gene Are Critical for Efficient 3′ End Processing. Mol. Cell. Biol. 2006, 27, 1568–1580. [Google Scholar] [CrossRef] [Green Version]
- Decorsière, A.; Cayrel, A.; Vagner, S.; Millevoi, S. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev. 2011, 25, 220–225. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Zhang, J.; Harvey, S.E.; Hu, X.; Cheng, C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017, 31, 2296–2309. [Google Scholar] [CrossRef]
- Dominguez, C.; Fisette, J.-F.; Chabot, B.; Allain, F.H.-T. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat. Struct. Mol. Biol. 2010, 17, 853–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samatanga, B.; Dominguez, C.; Jelesarov, I.; Allain, F.H.-T. The high kinetic stability of a G-quadruplex limits hnRNP F qRRM3 binding to G-tract RNA. Nucleic Acids Res. 2012, 41, 2505–2516. [Google Scholar] [CrossRef]
- Sauer, M.; Paeschke, K. G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 2017, 45, 1173–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kar, A.N.; Fushimi, K.; Zhou, X.; Ray, P.; Shi, C.; Chen, X.; Liu, Z.; Chen, S.; Wu, J.Y. RNA Helicase p68 (DDX5) Regulates tau Exon 10 Splicing by Modulating a Stem-Loop Structure at the 5′ Splice Site. Mol. Cell. Biol. 2011, 31, 1812–1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurent, F.-X.; Sureau, A.; Klein, A.F.; Trouslard, F.; Gasnier, E.; Furling, D.; Marie, J. New function for the RNA helicase p68/DDX5 as a modifier of MBNL1 activity on expanded CUG repeats. Nucleic Acids Res. 2011, 40, 3159–3171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obrdlik, A.; Kukalev, A.; Louvet, E.; Farrants, A.-K.O.; Caputo, L.; Percipalle, P. The Histone Acetyltransferase PCAF Associates with Actin and hnRNP U for RNA Polymerase II Transcription. Mol. Cell. Biol. 2008, 28, 6342–6357. [Google Scholar] [CrossRef] [Green Version]
- Sirand-Pugnet, P.; Durosay, P.; Brody, E.; Marie, J. An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken β-tropomyosin pre-mRNA. Nucleic Acids Res. 1995, 23, 3501–3507. [Google Scholar] [CrossRef]
- Ribeiro, M.M.; Teixeira, G.S.; Martins, L.; Marques, M.R.; De Souza, A.P.; Line, S.R.P. G-quadruplex formation enhances splicing efficiency of PAX9 intron 1. Qual. Life Res. 2014, 134, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Didiot, M.-C.; Tian, Z.; Schaeffer, C.; Subramanian, M.; Mandel, J.-L.; Moine, H. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008, 36, 4902–4912. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-T.; Chang, I.Y.-F.; Liu, H.; Ma, C.-P.; Kuo, Y.-P.; Shih, C.-T.; Shih, Y.-H.; Kang, L.; Tan, B.C. Tumor-associated intronic editing ofHNRPLLgenerates a novel splicing variant linked to cell proliferation. J. Biol. Chem. 2018, 293, 10158–10171. [Google Scholar] [CrossRef] [Green Version]
- Mazloomian, A.; Meyer, I.M. Genome-wide identification and characterization of tissue-specific RNA editing events in D. melanogaster and their potential role in regulating alternative splicing. RNA Biol. 2015, 12, 1391–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.; Gu, K.; Yu, J.; Fu, X.; Wang, X.; Guo, W.; Liao, L.; Zhu, H.; Zhang, X.; Hui, J.; et al. Opposing roles of miR-294 and MBNL 1/2 in shaping the gene regulatory network of embryonic stem cells. EMBO Rep. 2018, 19, e45657. [Google Scholar] [CrossRef]
- Singh, R.N.; Singh, N.N. Mechanism of Splicing Regulation of Spinal Muscular Atrophy Genes. Adv. Neurobiol. 2018, 20, 31–61. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.N.; Hollinger, K.; Bhattacharya, D.; Singh, R.N. An antisense microwalk reveals critical role of an intronic position linked to a unique long-distance interaction in pre-mRNA splicing. RNA 2010, 16, 1167–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.N.; Lawler, M.N.; Ottesen, E.W.; Upreti, D.; Kaczynski, J.R.; Singh, R.N. An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res. 2013, 41, 8144–8165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.N.; Singh, R.N. How RNA structure dictates the usage of a critical exon of spinal muscular atrophy gene. Biochim. Biophys. Acta (BBA) Bioenerg. 2019, 1862, 194403. [Google Scholar] [CrossRef]
- Singh, N.N.; Seo, J.; Ottesen, E.W.; Shishimorova, M.; Bhattacharya, D.; Singh, R.N. TIA1 Prevents Skipping of a Critical Exon Associated with Spinal Muscular Atrophy. Mol. Cell. Biol. 2010, 31, 935–954. [Google Scholar] [CrossRef] [Green Version]
- Ravindra, N.S. Unfolding the mystery of alternative splicing through a unique method of in vivo selection. Front. Biosci. 2007, 12, 3263. [Google Scholar] [CrossRef]
- Howell, M.D.; Ottesen, E.W.; Singh, N.N.; Anderson, R.L.; Singh, R.N. Gender-Specific Amelioration of SMA Phenotype upon Disruption of a Deep Intronic Structure by an Oligonucleotide. Mol. Ther. 2017, 25, 1328–1341. [Google Scholar] [CrossRef] [Green Version]
- Gong, S.; Du, C.; Wang, Y. Regulation of the thiamine pyrophosphate (TPP)-sensing riboswitch in NMT1 mRNA from Neurospora crassa. FEBS Lett. 2019, 594, 625–635. [Google Scholar] [CrossRef]
- Pérez-Cano, L.; Romero-Durana, M.; Fernández-Recio, J. Structural and energy determinants in protein-RNA docking. Methods 2017, 118, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Flores, J.K.; Ataide, S.F. Structural Changes of RNA in Complex with Proteins in the SRP. Front. Mol. Biosci. 2018, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, X.; Gorin, A.; Frederick, R.; Hu, W.; Majumdar, A.; Xu, W.; McLendon, G.; Ellington, A.; Patel, D.J. RNA architecture dictates the conformations of a bound peptide. Chem. Biol. 1999, 6, 657–669. [Google Scholar] [CrossRef] [Green Version]
- Xiang, S.; Gapsys, V.; Kim, H.-Y.; Bessonov, S.; Hsiao, H.-H.; Möhlmann, S.; Klaukien, V.; Ficner, R.; Becker, S.; Urlaub, H.; et al. Phosphorylation Drives a Dynamic Switch in Serine/Arginine-Rich Proteins. Structure 2013, 21, 2162–2174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Järvelin, A.I.; Noerenberg, M.; Davis, I.; Castello, A. The new (dis)order in RNA regulation. Cell Commun. Signal. 2016, 14, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, A.; Ahmad, S.; Gromiha, M.M. Deciphering RNA-Recognition Patterns of Intrinsically Disordered Proteins. Int. J. Mol. Sci. 2018, 19, 1595. [Google Scholar] [CrossRef] [Green Version]
- Grammatikakis, I.; Goo, Y.-H.; Echeverria, G.V.; Cooper, T.A. Identification of MBNL1 and MBNL3 domains required for splicing activation and repression. Nucleic Acids Res. 2010, 39, 2769–2780. [Google Scholar] [CrossRef]
- Tran, H.; Gourrier, N.; Lemercier-Neuillet, C.; Dhaenens, C.-M.; Vautrin, A.; Fernandez-Gomez, F.J.; Arandel, L.; Carpentier, C.; Obriot, H.; Eddarkaoui, S.; et al. Analysis of Exonic Regions Involved in Nuclear Localization, Splicing Activity, and Dimerization of Muscleblind-like-1 Isoforms. J. Biol. Chem. 2011, 286, 16435–16446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamft, B.; Bintu, L.; Ishibashi, T.; Bustamante, C. Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proc. Natl. Acad. Sci. USA 2012, 109, 8948–8953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turowski, T.; Petfalski, E.; Goddard, B.D.; French, S.L.; Helwak, A.; Tollervey, D. Nascent transcript folding plays a major role in determining RNA polymerase elongation rates. Mol. Cell 2020. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.G.L.; Wood, M. RNA splicing: Disease and therapy. Briefings Funct. Genom. 2011, 10, 151–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suwinski, P.; Ong, C.; Ling, M.H.T.; Poh, Y.M.; Khan, A.M.; Ong, H.S. Advancing Personalized Medicine Through the Application of Whole Exome Sequencing and Big Data Analytics. Front. Genet. 2019, 10, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abramowicz, A.; Gos, M. Correction to: Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 2019, 60, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jian, X.; Boerwinkle, E.; Liu, X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014, 42, 13534–13544. [Google Scholar] [CrossRef] [Green Version]
- Ionita-Laza, I.; McCallum, K.; Xu, B.; Buxbaum, J.D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016, 48, 214–220. [Google Scholar] [CrossRef] [Green Version]
- Jagadeesh, K.A.; Paggi, J.M.; Ye, J.S.; Stenson, P.D.; Cooper, D.N.; Bernstein, J.A.; Bejerano, G. S-CAP extends pathogenicity prediction to genetic variants that affect RNA splicing. Nat. Genet. 2019, 51, 755–763. [Google Scholar] [CrossRef]
- Halvorsen, M.; Martin, J.S.; Broadaway, S.; Laederach, A. Disease-Associated Mutations That Alter the RNA Structural Ensemble. PLoS Genet. 2010, 6, e1001074. [Google Scholar] [CrossRef] [Green Version]
- Sterne-Weiler, T.; Howard, J.; Mort, M.; Cooper, D.N.; Sanford, J.R. Loss of exon identity is a common mechanism of human inherited disease. Genome Res. 2011, 21, 1563–1571. [Google Scholar] [CrossRef] [Green Version]
- Xiong, H.Y.; Alipanahi, B.; Lee, L.J.; Bretschneider, H.; Merico, D.; Yuen, R.K.C.; Hua, Y.; Gueroussov, S.; Najafabadi, H.S.; Hughes, T.R.; et al. The human splicing code reveals new insights into the genetic determinants of disease. Science 2014, 347, 1254806. [Google Scholar] [CrossRef] [Green Version]
- Hutton, M.; Lendon, C.L.; Rizzu, P.; Baker, M.; Froelich, S.; Houlden, H.; Pickering-Brown, S.; Chakraverty, S.; Isaacs, A.; Grover, A.; et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998, 393, 702–705. [Google Scholar] [CrossRef]
- Dujardin, S.; Bégard, S.; Caillierez, R.; Lachaud, C.; Carrier, S.; Lieger, S.; Gonzalez, J.A.; Deramecourt, V.; Déglon, N.; Maurage, C.-A.; et al. Different tau species lead to heterogeneous tau pathology propagation and misfolding. Acta Neuropathol. Commun. 2018, 6, 132. [Google Scholar] [CrossRef]
- Bourdenx, M.; Koulakiotis, N.S.; Sanoudou, D.; Bezard, E.; Dehay, B.; Tsarbopoulos, A. Protein aggregation and neurodegeneration in prototypical neurodegenerative diseases: Examples of amyloidopathies, tauopathies and synucleinopathies. Prog. Neurobiol. 2017, 155, 171–193. [Google Scholar] [CrossRef]
- McCarthy, A.; Lonergan, R.; Olszewska, D.A.; O’Dowd, S.; Cummins, G.; Magennis, B.; Fallon, E.M.; Pender, N.; Huey, E.D.; Cosentino, S.; et al. Closing the tau loop: The missing tau mutation. Brain 2015, 138, 3100–3109. [Google Scholar] [CrossRef] [Green Version]
- Qian, W.; Liu, F. Regulation of alternative splicing of tau exon 10. Neurosci. Bull. 2014, 30, 367–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.L.; Moss, W.N.; Spencer, A.; Zhang, P.; Childs-Disney, J.L.; Disney, M.D. The RNA encoding the microtubule-associated protein tau has extensive structure that affects its biology. PLoS ONE 2019, 14, e0219210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W. How do SMA-linked mutations of SMN1 lead to structural/functional deficiency of the SMA protein? PLoS ONE 2017, 12, e0178519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorson, C.L.; Hahnen, E.; Androphy, E.J.; Wirth, B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 1999, 96, 6307–6311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartegni, L.; Hastings, M.L.; Calarco, J.A.; De Stanchina, E.; Krainer, A.R. Determinants of Exon 7 Splicing in the Spinal Muscular Atrophy Genes, SMN1 and SMN2. Am. J. Hum. Genet. 2005, 78, 63–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.N.; Androphy, E.J.; Singh, R.N. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2004, 315, 381–388. [Google Scholar] [CrossRef]
- Singh, N.K.; Singh, N.N.; Androphy, E.J.; Singh, R.N. Splicing of a Critical Exon of Human Survival Motor Neuron Is Regulated by a Unique Silencer Element Located in the Last Intron. Mol. Cell. Biol. 2006, 26, 1333–1346. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.N.; Howell, M.D.; Androphy, E.J.; Singh, R.N. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther. 2017, 24, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Ottesen, E.W. ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Transl. Neurosci. 2017, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, S.M. Expandable DNA repeats and human disease. Nature 2007, 447, 932–940. [Google Scholar] [CrossRef]
- Rohilla, K.J.; Gagnon, K.T. RNA biology of disease-associated microsatellite repeat expansions. Acta Neuropathol. Commun. 2017, 5, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciesiolka, A.; Jazurek, M.; Drazkowska, K.; Krzyzosiak, W.J. Structural Characteristics of Simple RNA Repeats Associated with Disease and their Deleterious Protein Interactions. Front. Cell. Neurosci. 2017, 11, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brook, J.D.; McCurrach, M.E.; Harley, H.G.; Buckler, A.J.; Church, D.; Aburatani, H.; Hunter, K.; Stanton, V.P.; Thirion, J.-P.; Hudson, T.; et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 1992, 68, 799–808. [Google Scholar] [CrossRef]
- Miller, J.W.; Urbinati, C.R.; Teng-Umnuay, P.; Stenberg, M.G.; Byrne, B.J.; Thornton, C.A.; Swanson, M.S. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 2000, 19, 4439–4448. [Google Scholar] [CrossRef] [Green Version]
- Ranum, L.P.; Rasmussen, P.F.; Benzow, K.A.; Koob, M.D.; Day, J.W. Genetic mapping of a second myotonic dystrophy locus. Nat. Genet. 1998, 19, 196–198. [Google Scholar] [CrossRef]
- Jiang, H.; Mankodi, A.K.; Swanson, M.S.; Moxley, R.T.; Thornton, C.A. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004, 13, 3079–3088. [Google Scholar] [CrossRef] [Green Version]
- Wojciechowska, M.; Krzyzosiak, W.J. Cellular toxicity of expanded RNA repeats: Focus on RNA foci. Hum. Mol. Genet. 2011, 20, 3811–3821. [Google Scholar] [CrossRef]
- Sznajder, Ł.J.; Swanson, M.S. Short Tandem Repeat Expansions and RNA-Mediated Pathogenesis in Myotonic Dystrophy. Int. J. Mol. Sci. 2019, 20, 3365. [Google Scholar] [CrossRef] [Green Version]
- Paul, S.; Dansithong, W.; Jog, S.P.; Holt, I.; Mittal, S.; Brook, J.D.; Morris, G.E.; Comai, L.; Reddy, S. Expanded CUG Repeats Dysregulate RNA Splicing by Altering the Stoichiometry of the Muscleblind 1 Complex. J. Boil. Chem. 2011, 286, 38427–38438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Cruchten, R.T.; Wieringa, B.; Wansink, D.G. Expanded CUG repeats in DMPK transcripts adopt diverse hairpin conformations without influencing the structure of the flanking sequences. RNA 2019, 25, 481–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thornton, C.A.; Wang, E.; Carrell, E.M. Myotonic dystrophy: Approach to therapy. Curr. Opin. Genet. Dev. 2017, 44, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Overby, S.J.; Cerro-Herreros, E.; Llamusi, B.; Artero, R. RNA-mediated therapies in myotonic dystrophy. Drug Discov. Today 2018, 23, 2013–2022. [Google Scholar] [CrossRef] [PubMed]
- Wojtkowiak-Szlachcic, A.; Taylor, K.; Stepniak-Konieczna, E.; Sznajder, L.J.; Mykowska, A.; Sroka, J.; Thornton, C.A.; Sobczak, K. Short antisense-locked nucleic acids (all-LNAs) correct alternative splicing abnormalities in myotonic dystrophy. Nucleic Acids Res. 2015, 43, 3318–3331. [Google Scholar] [CrossRef] [Green Version]
- Nakamori, M.; Taylor, K.; Mochizuki, H.; Sobczak, K.; Takahashi, M.P. Oral administration of erythromycin decreases RNA toxicity in myotonic dystrophy. Ann. Clin. Transl. Neurol. 2015, 3, 42–54. [Google Scholar] [CrossRef]
- Sardone, V.; Zhou, H.; Muntoni, F.; Ferlini, A.; Falzarano, M.S. Antisense Oligonucleotide-Based Therapy for Neuromuscular Disease. Molecules 2017, 22, 563. [Google Scholar] [CrossRef] [Green Version]
- Reddy, K.; Jenquin, J.R.; McConnell, O.L.; Cleary, J.D.; Richardson, J.I.; Pinto, B.S.; Haerle, M.C.; Delgado, E.; Planco, L.; Nakamori, M.; et al. A CTG repeat-selective chemical screen identifies microtubule inhibitors as selective modulators of toxic CUG RNA levels. Proc. Natl. Acad. Sci. USA 2019, 116, 20991–21000. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, T.M.; Leger, A.J.; Pandey, S.K.; MacLeod, A.R.; Nakamori, M.; Cheng, S.H.; Wentworth, B.M.; Bennett, C.F.; Thornton, C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 2012, 488, 111–115. [Google Scholar] [CrossRef] [Green Version]
- AngelBello, A.J.; Rzuczek, S.G.; McKee, K.K.; Chen, J.L.; Olafson, H.; Cameron, M.D.; Moss, W.N.; Wang, E.T.; Disney, M.D. Precise small-molecule cleavage of an r(CUG) repeat expansion in a myotonic dystrophy mouse model. Proc. Natl. Acad. Sci. USA 2019, 116, 7799–7804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castel, A.L.; Overby, S.J.; Artero, R. MicroRNA-Based Therapeutic Perspectives in Myotonic Dystrophy. Int. J. Mol. Sci. 2019, 20, 5600. [Google Scholar] [CrossRef]
- Bargiela, A.; Sabater-Arcis, M.; Espinosa-Espinosa, J.; Zulaica, M.; De Munain, A.L.; Artero, R. Increased Muscleblind levels by chloroquine treatment improve myotonic dystrophy type 1 phenotypes in in vitro and in vivo models. Proc. Natl. Acad. Sci. USA 2019, 116, 25203–25213. [Google Scholar] [CrossRef]
- Sznajder, Ł.J.; Thomas, J.D.; Carrell, E.M.; Reid, T.; McFarland, K.N.; Cleary, J.D.; Oliveira, R.; Nutter, C.A.; Bhatt, K.; Sobczak, K.; et al. Intron retention induced by microsatellite expansions as a disease biomarker. Proc. Natl. Acad. Sci. USA 2018, 115, 4234–4239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sellier, C.; Cerro-Herreros, E.; Blatter, M.; Freyermuth, F.; Gaucherot, A.; Ruffenach, F.; Sarkar, P.; Puymirat, J.; Udd, P.; Day, J.W.; et al. rbFOX1/MBNL1 competition for CCUG RNA repeats binding contributes to myotonic dystrophy type 1/type 2 differences. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gourdon, G.; Meola, G. Myotonic Dystrophies: State of the Art of New Therapeutic Developments for the CNS. Front. Cell. Neurosci. 2017, 11, 451. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Manley, J.L. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013, 3, 1228–1237. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.-C. Alternative RNA Structure-Coupled Gene Regulations in Tumorigenesis. Int. J. Mol. Sci. 2014, 16, 452–475. [Google Scholar] [CrossRef]
- Tabaglio, T.; Low, D.H.P.; Teo, W.K.L.; Goy, P.-A.; Cywoniuk, P.; Wollmann, H.; Ho, J.; Tan, D.; Aw, J.; Pavesi, A.; et al. MBNL1 alternative splicing isoforms play opposing roles in cancer. Life Sci. Alliance 2018, 1, e201800157. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Chen, R. Understanding aberrant RNA splicing to facilitate cancer diagnosis and therapy. Oncogene 2019, 39, 2231–2242. [Google Scholar] [CrossRef]
- Fischer, S.; Di Liddo, A.; Taylor, K.; Gerhardus, J.S.; Sobczak, K.; Zarnack, K.; Weigand, E.J. Muscleblind-like 2 controls the hypoxia response of cancer cells. RNA 2020, 26, 648–663. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wei, R.; Zhou, Z.; Huang, L.; Wang, Y.; Tang, J.; Zou, Y.; Shi, L.; Gu, X.; Davis, M.J.; et al. Integrative Analysis of Somatic Mutations in Non-coding Regions Altering RNA Secondary Structures in Cancer Genomes. Sci. Rep. 2019, 9, 8205. [Google Scholar] [CrossRef] [PubMed]
- David, C.J.; Manley, J.L. Alternative pre-mRNA splicing regulation in cancer: Pathways and programs unhinged. Genes Dev. 2010, 24, 2343–2364. [Google Scholar] [CrossRef] [Green Version]
- Perron, G.; Jandaghi, P.; Solanki, S.; Safisamghabadi, M.; Storoz, C.; Karimzadeh, M.; Papadakis, A.I.; Arseneault, M.; Scelo, G.; Banks, R.E.; et al. A General Framework for Interrogation of mRNA Stability Programs Identifies RNA-Binding Proteins that Govern Cancer Transcriptomes. Cell Rep. 2018, 23, 1639–1650. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.W.; Yang, Q.B.; Du, X.Y.; Chen, Y.; Zhang, T. Targeted regulation of Rell2 by microRNA-18a is implicated in the anti-metastatic effect of polyphyllin VI in breast cancer cells. Eur. J. Pharmacol. 2019, 851, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.L.; Singh, K.; Zhong, Y.; Drewe, P.; Rajasekhar, V.K.; Sanghvi, V.R.; Mavrakis, K.J.; Jiang, M.; Roderick, J.E.; Van Der Meulen, J.; et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 2014, 513, 65–70. [Google Scholar] [CrossRef] [Green Version]
- Ou, T.M.; Lu, Y.J.; Tan, J.H.; Huang, Z.S.; Wong, K.Y.; Gu, L.Q. G-quadruplexes: Targets in anticancer drug design. Chemmedchem 2008, 3, 690–713. [Google Scholar] [CrossRef] [PubMed]

| RBPs Regulating AS | Linear Sequence Motif | RNA Structural Preferences | |
|---|---|---|---|
| 1 | CELF/BRUNOL (CUG-binding protein Elav-like) | UGUGUGU [208] | ssRNA [209] |
| 2 | FMRP (Fragile X mental retardation protein) | G-rich elements [210] | dsRNA-rG4 [210] |
| 3 | FUS (Fused in sarcoma) | AU-rich element [211] GUGGU in a G-rich context [212] | ssRNA, stem-loop [211] |
| 4 | Hu/ELAV-like (Embryonic lethal/abnormal vision-like protein) | YUUR 1 interrupted by G [205] GU-rich, secondary motif AU-rich [213] | ssRNA [213] |
| 5 | MATR3 (Matrin-3) | CAUCUU, AAUCUU [208] | ssRNA [167] |
| 6 | MBNL (Muscleblind-like protein) | YGCY 2 | ssRNA, semi-stable RNA structures [172] |
| 7 | NOVA (RNA-binding protein Nova-1) | YCAY in a Y-rich context 2 [214] | ssRNA [214,215] |
| 8 | PTBP1 (Polypyrimidine tract-binding protein 1) | YUCY [205], YCTY, YGCY 2 [38] | ssRNA, Internal loop [216] |
| 9 | PUF60 (Poly-U-binding factor 60 kDa) | U-rich elements [217] | ssRNA [217] |
| 10 | QKI (Quaking STAR protein) | ACUAAC, NACUAAY-N1-20-UAAY 2,3 [218] | ssRNA, hairpin loop [215] |
| 11 | RBFOX (RNA binding protein fox) | UGCAUG [208] | ssRNA [219], stem-loop [220] |
| 12 | RBM4 (RNA binding protein 4) | CGGG [221] | ssRNA, stem-loop structure [222] |
| 13 | RBMY (RNA-binding motif protein, Y chromosome) | CA/UCAA [223] | ssRNA, stem-loop [223] |
| 14 | SAM68 (Src-Associated substrate in Mitosis of 68 kDa) | UAAA, UUAA, U-rich [224] | ssRNA, internal/hairpin loop [224] |
| 15 | STAU1 (Double-stranded RNA-binding protein Staufen homolog 1) | none | dsRNA [225] |
| 16 | TAF15 (TATA-box binding protein Associated Factor 15) | GGUAAGU [226], GGUG [227] | ssRNA, hairpin loop [227] |
| 17 | TDP-43 (TAR DNA-binding protein 43) | RUGY 1,2 [205] | ssRNA [228] |
| 18 | TIA1 (T-cell intracellular antigen 1) | AU-rich elements [229], TTTA [205], UUUUUUC/A [206] | ssRNA [230], stem-loop [231] |
| 19 | TIAL1 (T-cell intracellular antigen 1-like 1) | AU-rich elements [229], Poly(U) [230] | ssRNA [230] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, K.; Sobczak, K. Intrinsic Regulatory Role of RNA Structural Arrangement in Alternative Splicing Control. Int. J. Mol. Sci. 2020, 21, 5161. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21145161
Taylor K, Sobczak K. Intrinsic Regulatory Role of RNA Structural Arrangement in Alternative Splicing Control. International Journal of Molecular Sciences. 2020; 21(14):5161. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21145161
Chicago/Turabian StyleTaylor, Katarzyna, and Krzysztof Sobczak. 2020. "Intrinsic Regulatory Role of RNA Structural Arrangement in Alternative Splicing Control" International Journal of Molecular Sciences 21, no. 14: 5161. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21145161





