Effects of IGF-1 on Proliferation, Angiogenesis, Tumor Stem Cell Populations and Activation of AKT and Hedgehog Pathways in Oral Squamous Cell Carcinoma
Abstract
:1. Introduction
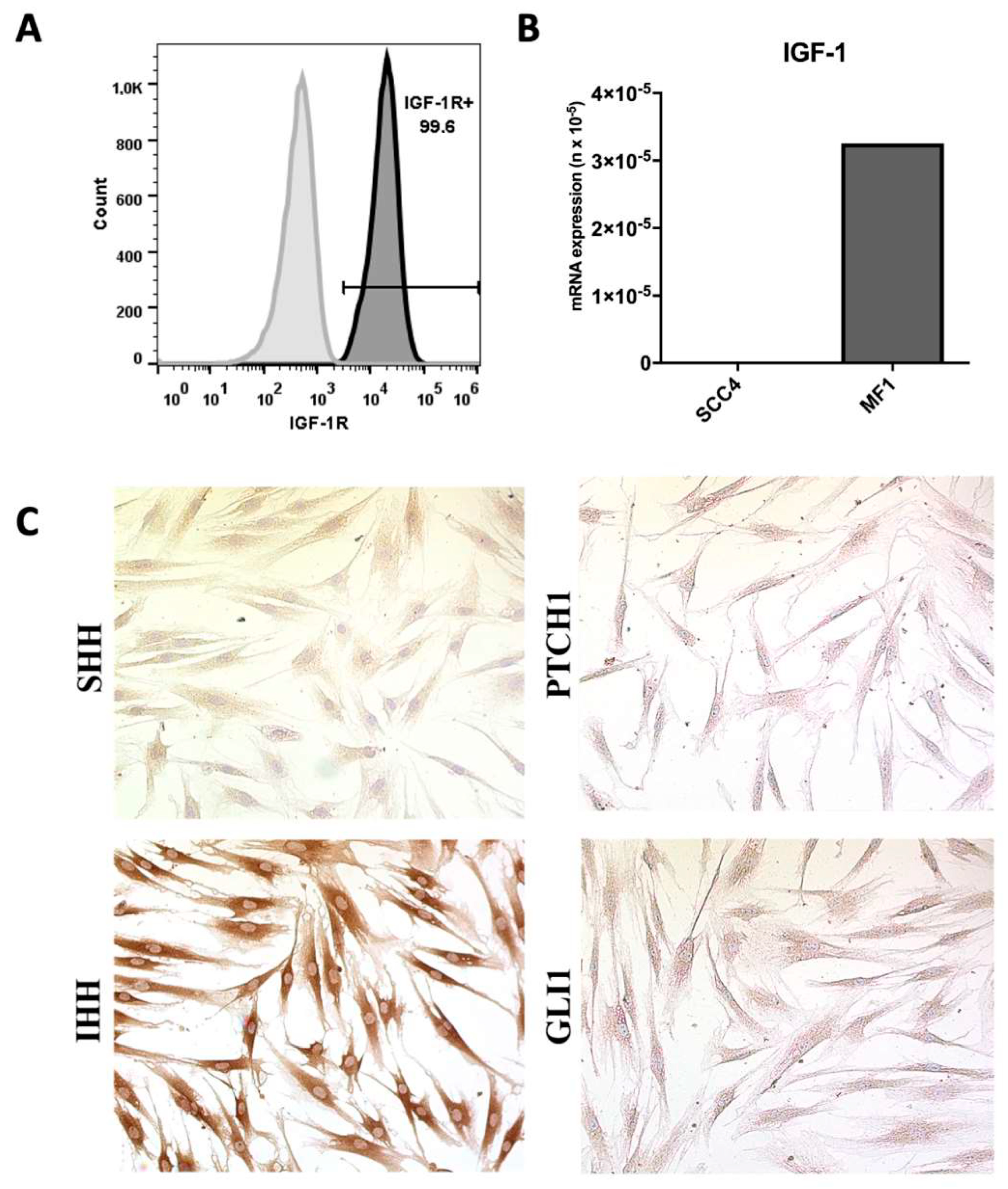
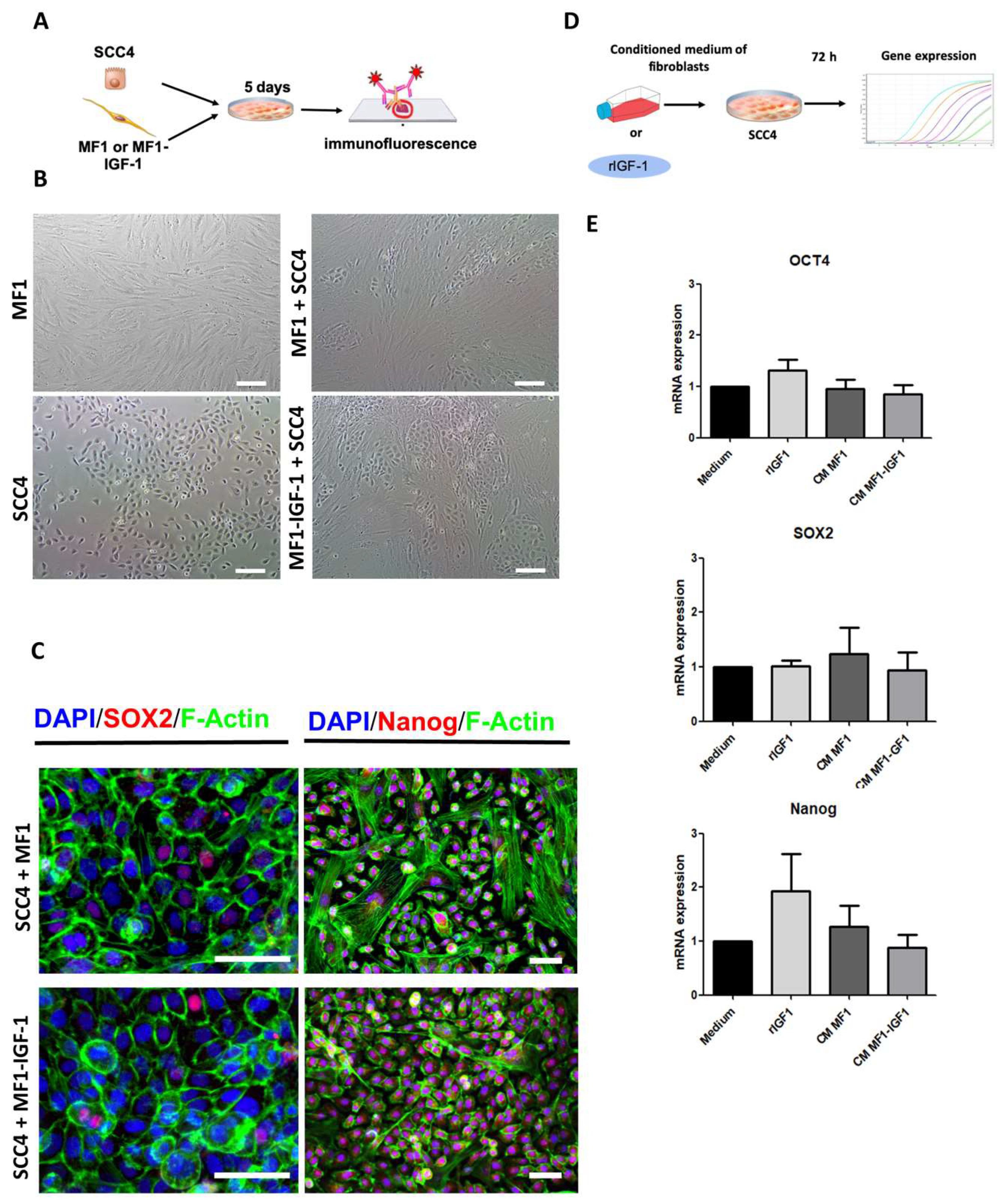
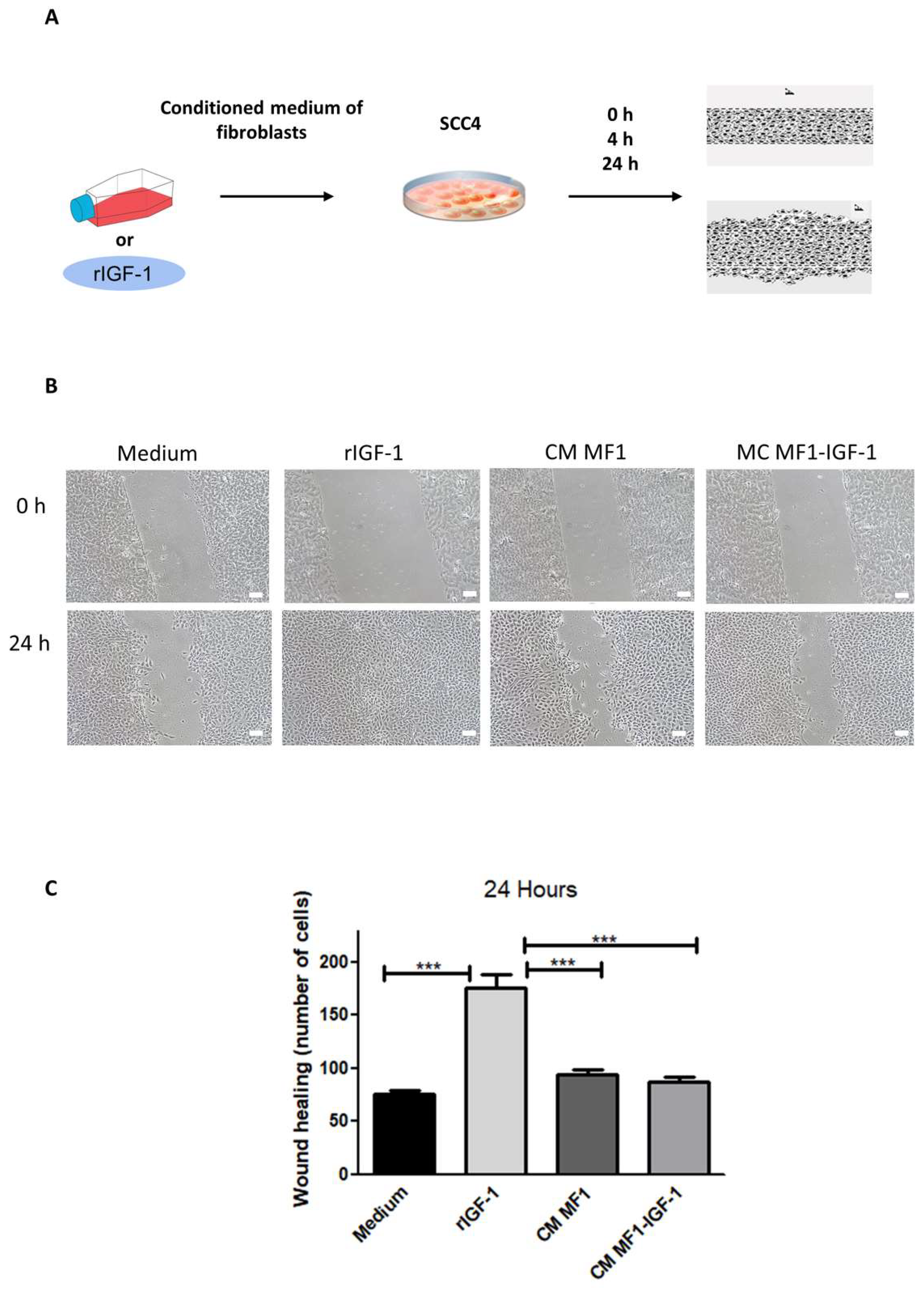
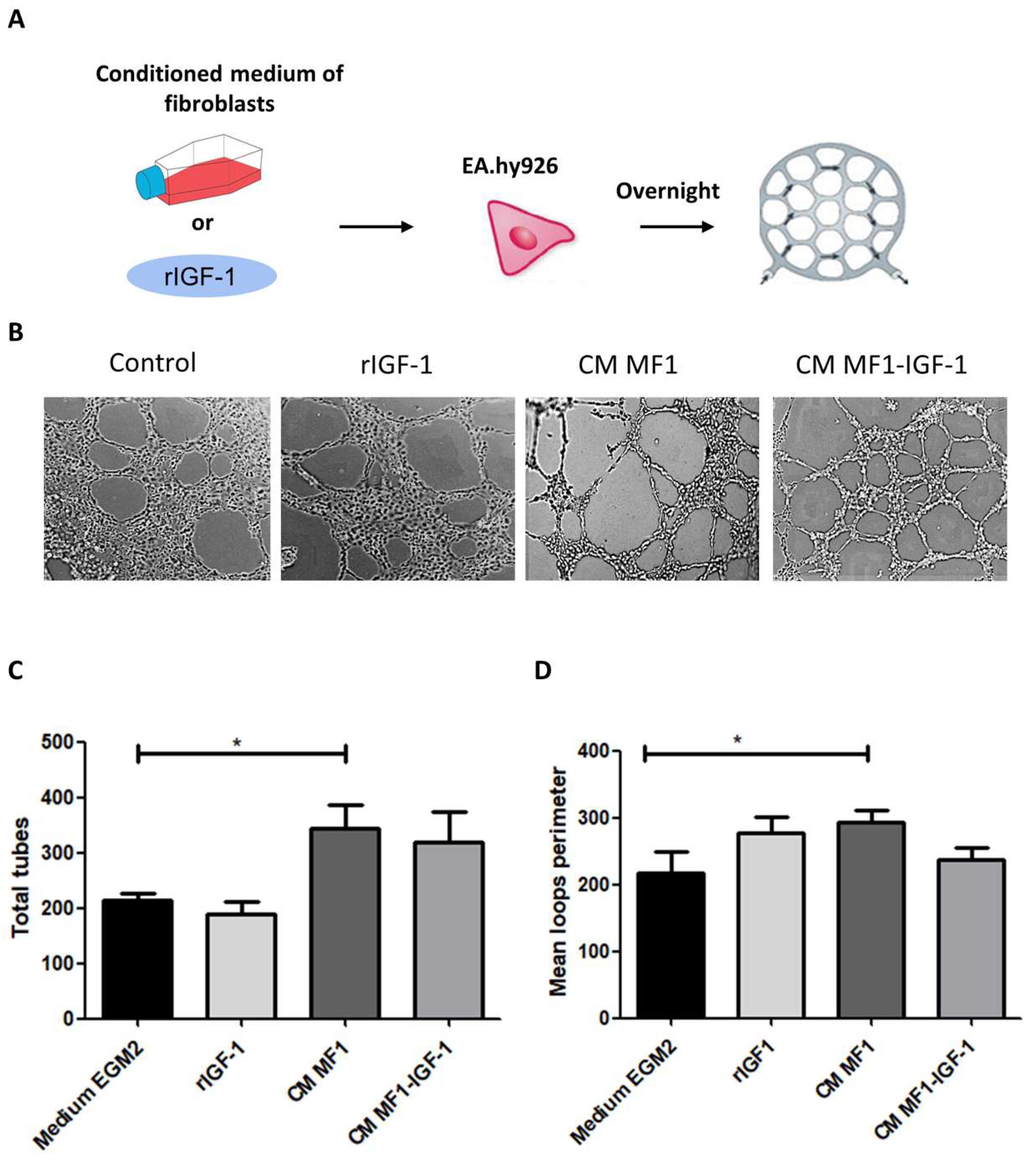
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Characterization
4.2. Genetic Modification
4.3. MF1 Conditioned Media
4.4. Gene Expression Analysis by RT-qPCR
4.5. Immunostaining
4.6. Cell Proliferation Assay
4.7. Scratch Assay
4.8. Invasion Assay
4.9. Tube Formation Assay
4.10. Tumorsphere Formation Assay
4.11. Western Blot
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| OSCC | Oral squamous cell carcinoma |
| IGF1R | Insulin-like growth factor 1 receptor |
| PI3KCA | Phosphatidylinositol 3-kinase |
| AKT-PKB | Protein kinase B |
| RAS/RAF/MAPK | Mitogen-activated protein kinases pathway |
| TCGA | The Cancer Genome Atlas |
| PRKCI | Protein kinase C iota |
| GLI | Glioma-associated oncogene |
| IGF-1 | Insulin-like growth factor 1 |
| HH | Hedgehog |
| IL | Interleukin |
| CAF | Cancer-associated fibroblasts |
| TGF-β | Transforming growth factor beta |
| MMP | Matrix metallopeptidases |
| HGF | Hepatocyte growth factor |
| EGF | Epidermal growth factor |
| CTGF | Connective tissue growth factor |
| SMO | Smoothened |
| SHH | Sonic hedgehog |
| PTCH1 | Patched 1 |
| IHH | Indian hedgehog |
| IGFBP | Insulin-like growth factor-binding protein |
| MF1 | Human dermal fibroblasts |
| MF1-IGF1 | Human dermal fibroblasts-expressing IGF-1 |
| TNF-TRAIL | Tumor necrosis factor-related apoptosis-inducing ligand |
| CRISPR/Cas9 | clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 |
| CD | Cluster differentiation |
| MOI | Multiplicity of infection |
| sgRNA | single guide RNA |
| DMEM | Dulbecco’s modified Eagle’s medium |
| CM | Conditioned medium |
References
- Sarradin, V.; Siegfried, A.; Uro-Coste, E.; Delord, J.P. Classification de l’OMS 2017 des tumeurs de la tête et du cou: Principales nouveautés et mise à jour des méthodes diagnostiques. Bull. Cancer 2018, 105, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Foulstone, E.; Prince, S.; Zaccheo, O.; Burns, J.; Harper, J.; Jacobs, C.; Hassan, A. Insulin-like growth factor ligands, receptors, and binding proteins in cancer. J. Pathol. 2005, 205, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Laviola, L.; Natalicchio, A.; Giorgino, F. The IGF-I Signaling Pathway. Curr. Pharm. Des. 2007, 13, 663–669. [Google Scholar] [CrossRef]
- Rabiee, A.; Krüger, M.; Ardenkjær-Larsen, J.; Kahn, C.R.; Emanuelli, B. Distinct signalling properties of insulin receptor substrate (IRS)-1 and IRS-2 in mediating insulin/IGF-1 action. Cell. Signal. 2018, 47, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Jia, L.; Wang, B.; Diao, S.; Jia, R.; Shang, J. MiR-495/IGF-1/AKT signaling as a novel axis is involved in the epithelial-mesenchymal transition of oral squamous cell carcinoma. J. Oral Maxillofac. Surg. 2019, 77, 1009–1021. [Google Scholar] [CrossRef]
- Hao, Y.; Zhang, C.; Sun, Y.; Xu, H. Licochalcone A inhibits cell proliferation, migration, and invasion through regulating the PI3K/AKT signaling pathway in oral squamous cell carcinoma. Onco Targets Ther. 2019, 12, 4427–4435. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Li, J.C.; Yang, H.; Zhang, X.; Liu, L.L.; Li, Y.; Fu, L. Expansion of cancer stem cell pool initiates lung cancer recurrence before angiogenesis. Proc. Natl. Acad. Sci. USA 2018, 115, E8948–E8957. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.J.; Ho, C.C.; Chang, Y.L.; Chen, H.Y.; Lin, C.A.; Ling, T.Y.; Pan, S.H. Cancer-associated fibroblasts regulate the plasticity of lung cancer stemness via paracrine signalling. Nat. Commun. 2014, 5, 3472. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Schultz, N. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owusu-Brackett, N.; Shariati, M.; Meric-Bernstam, F. Predictive Biomarkers in Oncology; Springer: Cham, Switzerland, 2019; p. 263. [Google Scholar]
- Lyra, S.M.D.C.; Moleri, A.B.; Dezonne, R.S.; Lourenço, S.D.Q.C.; Moura-Neto, V.; Pereira, C. Evaluation of micrornas related to the sonic hedgehog pathway in oral cancer. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2020, 129, e133. [Google Scholar] [CrossRef]
- Takabatake, K.; Shimo, T.; Murakami, J.; Anqi, C.; Kawai, H.; Yoshida, S.; Nagatsuka, H. The Role of Sonic Hedgehog Signaling in the Tumor Microenvironment of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2019, 20, 5779. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582. [Google Scholar] [CrossRef] [PubMed]
- De Palma, M.; Biziato, D.; Petrova, T.V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 2017, 17, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Tan, Z.W.; Zhu, P.; Tan, N.S. Cancer-associated fibroblasts in tumor microenvironment—Accomplices in tumor malignancy. Cell. Immunol. 2018. [Google Scholar] [CrossRef]
- Javed, S.; Bhattacharyya, S.; Bagga, R.; Srinivasan, R. Insulin growth factor-1 pathway in cervical carcinoma cancer stem cells. Mol. Cell. Biochem. 2020, 1–12. [Google Scholar]
- Sun, L.; Yuan, W.; Wen, G.; Yu, B.; Xu, F.; Gan, X.; Tang, J.; Zeng, Q.; Zhu, L.; Zhang, W.; et al. Parthenolide inhibits human lung cancer cell growth by modulating the IGF-1R/PI3K/Akt signaling pathway. Oncol. Rep. 2020, 44, 1184–1193. [Google Scholar]
- Peng, Y.; Li, F.; Zhang, P.; Wang, X.; Shen, Y.; Feng, Y.; Jia, Y.; Zhang, R.; Hu, J.; He, A. IGF-1 promotes multiple myeloma progression through PI3K/Akt-mediated epithelial-mesenchymal transition. Life Sci. 2020, 249, 117503. [Google Scholar] [CrossRef]
- Wang, L.; Song, Y.; Wang, H.; Liu, K.; Shao, Z.; Shang, Z. MiR-210-3p-EphrinA3-PI3K/AKT axis regulates the progression of oral cancer. J. Cell. Mol. Med. 2020, 24, 4011–4022. [Google Scholar] [CrossRef] [Green Version]
- Chetty, R. Gene of the month: GLI-1. J. Clin. Pathol. 2020, 73, 228–230. [Google Scholar] [CrossRef] [PubMed]
- Tamayo-Orrego, L.; Gallo, D.; Racicot, F.; Bemmo, A.; Mohan, S.; Ho, B.; Salameh, S.; Hoang, T.; Jackson, A.P.; Charron, F.; et al. Sonic hedgehog accelerates DNA replication to cause replication stress promoting cancer initiation in medulloblastoma. Nat. Cancer 2020, 1–15. [Google Scholar] [CrossRef]
- Lim, K.P.; Cirillo, N.; Hassona, Y.; Wei, W.; Thurlow, J.K.; Cheong, S.C.; Prime, S.S. Fibroblast gene expression profile reflects the stage of tumour progression in oral squamous cell carcinoma. J. Pathol. 2011, 223, 459–469. [Google Scholar] [CrossRef]
- Oh, Y.; Nagalla, S.R.; Yamanaka, Y.; Kim, H.S.; Wilson, E.; Rosenfeld, R.G. Synthesis and characterization of insulin-like growth factor-binding protein (IGFBP)-7 Recombinant human mac25 protein specifically binds IGF-I and-II. J. Biol. Chem. 1996, 271, 30322–30325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Annunziata, M.; Granata, R.; Ghigo, E. The IGF system. Acta Diabetol. 2011, 48, 1–9. [Google Scholar] [CrossRef]
- Evdokimova, V.; Tognon, C.E.; Benatar, T.; Yang, W.; Krutikov, K.; Pollak, M.; Sorensen, P.H.; Seth, A. IGFBP7 binds to the IGF-1 receptor and blocks its activation by insulin-like growth factors. Sci. Signal. 2012, 5, ra92. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, Z.; Liang, Z.; Liu, J.; Shi, W.; Bai, P.; Lin, X.; Magaye, R.; Zhao, J. Expression and clinical significance of IGF-1, IGFBP-3, and IGFBP-7 in serum and lung cancer tissues from patients with non-small cell lung cancer. Onco Targets Ther. 2013, 6, 1437. [Google Scholar]
- Bartram, I.; Erben, U.; Ortiz-Tanchez, J.; Blunert, K.; Schlee, C.; Neumann, M.; Heesch, S.; Baldus, C.D. Inhibition of IGF1-R overcomes IGFBP7-induced chemotherapy resistance in T-ALL. BMC Cancer 2015, 15, 663. [Google Scholar] [CrossRef] [Green Version]
- Baserga, R.; Peruzzi, F.; Reiss, K. The IGF-1 receptor in cancer biology. Int. J. Cancer 2003, 107, 873–877. [Google Scholar] [CrossRef]
- Xiao, J.; He, C.; Zhong, Y.; Liu, L.; Xiao, Y.; Liu, W. miR-223 regulates proliferation and apoptosis by targeting insulin-like growth factor 1 receptor (IGF1R) in osteosarcoma cells. Mater. Express 2019, 9, 459–466. [Google Scholar] [CrossRef]
- Hilmi, C.; Larribere, L.; Giuliano, S.; Bille, K.; Ortonne, J.P.; Ballotti, R.; Bertolotto, C. IGF1 Promotes Resistance to Apoptosis in Melanoma Cells through an Increased Expression of BCL2, BCL-X(L), and Survivin. J. Investig. Dermatol. 2008, 128, 1499–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muraguchi, T.; Nanba, D.; Nishimura, E.K.; Tashiro, T. IGF-1R deficiency in human keratinocytes disrupts epidermal homeostasis and stem cell maintenance. J. Dermatol. Sci. 2019, 94, 298–305. [Google Scholar] [CrossRef] [Green Version]
- Bagordakis, E.; Sawazaki-Calone, I.; Macedo, C.C.S.; Carnielli, C.M.; De Oliveira, C.E.; Rodrigues, P.C.; Coletta, R.D. Secretome profiling of oral squamous cell carcinoma-associated fibroblasts reveals organization and disassembly of extracellular matrix and collagen metabolic process signatures. Tumor Biol. 2016, 37, 9045–9057. [Google Scholar] [CrossRef] [PubMed]
- Graizel, D.; Zlotogorski-Hurvitz, A.; Tsesis, I.; Rosen, E.; Kedem, R.; Vered, M. Oral cancer-associated fibroblasts predict poor survival: Systematic review and meta-analysis. Oral Dis. 2020, 26, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Kellermann, M.G.; Sobral, L.M.; Silva, S.D.; Zecchin, K.G.; Graner, E.; Lopes, M.A.; Nishimoto, I.; Kowalski, L.P.; Coletta, R.D. Myofibroblasts in the stroma of oral squamous cell carcinoma are associated with poor prognosis. Histopathology 2007, 51, 849–853. [Google Scholar] [CrossRef]
- Kashima, H.; Noma, K.; Ohara, T.; Kato, T.; Katsura, Y.; Komoto, S.; Fujiwara, T. Cancer-associated fibroblasts (CAFs) promote the lymph node metastasis of esophageal squamous cell carcinoma. Int. J. Cancer 2019, 144, 828–840. [Google Scholar] [CrossRef] [Green Version]
- Buim, M.E.C.; Gurgel, C.A.S.; Gonçalves Ramos, E.A.; Lourenço, S.V.; Soares, F.A. Activation of sonic hedgehog signaling in oral squamous cell carcinomas: A preliminary study. Hum. Pathol. 2011, 42, 1484–1490. [Google Scholar] [CrossRef]
- De Faro Valverde, L.; De Almeida Pereira, T.; Dias, R.B.; Guimarães, V.S.N.; Ramos, E.A.G.; Santos, J.N.; Rocha, C.A.G. Macrophages and endothelial cells orchestrate tumor-associated angiogenesis in oral cancer via hedgehog pathway activation. Tumor Biol. 2016, 37, 9233–9241. [Google Scholar] [CrossRef]
- Fernandez, C.; Tatard, V.M.; Bertrand, N.; Dahmane, N. Differential Modulation of Sonic-Hedgehog-Induced Cerebellar Granule Cell Precursor Proliferation by the IGF Signaling Network. Dev. Neurosci. 2010, 32, 59–70. [Google Scholar] [CrossRef]
- Hsieh, A.; Ellsworth, R.; Hsieh, D. Hedgehog/GLI1 regulates IGF dependent malignant behaviors in glioma stem cells. J. Cell. Physiol. 2011, 226, 1118–1127. [Google Scholar] [CrossRef]
- Nakamura, K.; Sasajima, J.; Mizukami, Y.; Sugiyama, Y.; Yamazaki, M.; Fujii, R.; Kohgo, Y. Hedgehog Promotes Neovascularization in Pancreatic Cancers by Regulating Ang-1 and IGF-1 Expression in Bone-Marrow Derived Pro-Angiogenic Cells. PLoS ONE 2010, 5, e8824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Li, J.; Cheng, J.; Lubahn, D.B. Genes targeted by the Hedgehog-signaling pathway can be regulated by Estrogen related receptor β. BMC Mol. Biol. 2015, 16, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Zhu, G.; Huang, J.; Li, L.; Du, Y.; Gao, Y.; Wu, K. Non-canonical GLI1/2 activation by PI3K/AKT signaling in renal cell carcinoma: A novel potential therapeutic target. Cancer Lett. 2016, 370, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Riobó, N.A.; Lu, K.; Ai, X.; Haines, G.M.; Emerson, C.P. Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 4505–4510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, G.; Pedone, C.A.; Valle, L.D.; Reiss, K.; Holland, E.C.; Fults, D.W. Sonic hedgehog and insulin-like growth factor signaling synergize to induce medulloblastoma formation from nestin-expressing neural progenitors in mice. Oncogene 2004, 23, 6156–6162. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.K.; Dhadve, A.; Sakpal, A.; De, A.; Ray, P. An active IGF-1R-AKT signaling imparts functional heterogeneity in ovarian CSC population. Sci. Rep. 2016, 6, 36612. [Google Scholar] [CrossRef] [Green Version]
- Mohiuddin, I.S.; Wei, S.J.; Kang, M.H. Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2019. [Google Scholar] [CrossRef]
- Lee, J.J.; Loh, K.; Yap, Y.S. PI3K/Akt/mTOR inhibitors in breast cancer. Cancer Biol. Med. 2015, 12, 342. [Google Scholar]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Shah, A.; Patel, S.; Pathak, J.; Swain, N.; Kumar, S. The Evolving Concepts of Cancer Stem Cells in Head and Neck Squamous Cell Carcinoma. Sci. World J. 2014, 2014, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Gupta, M.K.; De Jesus, D.F.; Kahraman, S.; Valdez, I.A.; Shamsi, F.; Yi, L.; Kulkarni, R.N. Insulin receptor-mediated signaling regulates pluripotency markers and lineage differentiation. Mol. Metab. 2018, 18, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Jacobo, S.M.P.; Kazlauskas, A. Insulin-like Growth Factor 1 (IGF-1) Stabilizes Nascent Blood Vessels. J. Biol. Chem. 2015, 290, 6349–6360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, A.C.; Nakatsu, M.N.; Chou, W.; Gershon, P.D.; Hughes, C.C.W. The requirement for fibroblasts in angiogenesis: Fibroblast-derived matrix proteins are essential for endothelial cell lumen formation. Mol. Biol. Cell 2011, 22, 3791–3800. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.T.; Podar, K.; Catley, L.; Tseng, Y.H.; Shringarpure, R.; Burger, R.; Richardson, P. Insulin-like growth factor-1 induces adhesion and migration in human multiple myeloma cells via activation of b1-integrin and phosphatidylinositol 3-kinase/AKT signaling beta. Blood 2003, 102, 111A. [Google Scholar]
- Shi, X.; Teng, F. Down-regulated miR-28-5p in human hepatocellular carcinoma correlated with tumor proliferation and migration by targeting insulin-like growth factor-1 (IGF-1). Mol. Cell. Biochem. 2015, 408, 283–293. [Google Scholar] [CrossRef]
- Murekatete, B.; Shokoohmand, A.; McGovern, J.; Mohanty, L.; Meinert, C.; Hollier, B.G.; Kashyap, A.S. Targeting Insulin-Like Growth Factor-I and Extracellular Matrix Interactions in Melanoma Progression. Sci. Rep. 2018, 8, 583. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Yan, M.; Guan, Z.; Pan, C.; Xia, L.; Li, C.; Liu, Q. Aurora-A down-regulates IkappaBα via Akt activation and interacts with insulin-like growth factor-1 induced phosphatidylinositol 3-kinase pathway for cancer cell survival. Mol. Cancer 2009, 8, 95. [Google Scholar] [CrossRef] [Green Version]
- Yoeli-Lerner, M.; Toker, A. Akt/PKB signaling in cancer: A function in cell motility and invasion. Cell Cycle 2006, 5, 603–605. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Sawai, H.; Matsuo, Y.; Ochi, N.; Yasuda, A.; Takahashi, H.; Takeyama, H. IGF-1 Mediates PTEN Suppression and Enhances Cell Invasion and Proliferation via Activation of the IGF-1/PI3K/Akt Signaling Pathway in Pancreatic Cancer Cells. J. Surg. Res. 2010, 2160, 90–101. [Google Scholar] [CrossRef]
- Chung, T.W.; Lee, Y.C.; Kim, C.H. Hepatitis B viral HBx induces matrix metalloproteinase-9 gene expression through activation of ERK and PI-3K/AKT pathways: Involvement of invasive potential. FASEB J. 2004, 18, 1123–1125. [Google Scholar] [CrossRef]
- Nanta, R.; Shrivastava, A.; Sharma, J.; Shankar, S.; Srivastava, R.K. Inhibition of sonic hedgehog and PI3K/Akt/mTOR pathways cooperate in suppressing survival, self-renewal and tumorigenic potential of glioblastoma-initiating cells. Mol. Cell. Biochem. 2018, 454, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Martins, G.L.S.; Paredes, B.D.; Sampaio, G.L.A.; Nonaka, C.K.V.; Da Silva, K.N.; Allahdadi, K.J.; Souza, B.S.F. Generation of human iPS cell line CBTCi001-A from dermal fibroblasts obtained from a healthy donor. Stem Cell Res. 2019, 41, 101630. [Google Scholar] [CrossRef] [PubMed]
- Brady, G.; Crean, S.J.; Naik, P.; Kapas, S. Upregulation of IGF-2 and IGF-1 receptor expression in oral cancer cell lines. Int. J. Oncol. 2007, 31, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, G.V.M.; Silva, D.N.; Carvalho, R.H.; Souza, B.S.F.; Da Silva, K.N.; Vasconcelos, J.F.; Soares, M.B.P. Generation and characterization of transgenic mouse mesenchymal stem cell lines expressing hIGF-1 or hG-CSF. Cytotechnology 2017, 70, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferreira Mendes, J.M.; de Faro Valverde, L.; Torres Andion Vidal, M.; Paredes, B.D.; Coelho, P.; Allahdadi, K.J.; Coletta, R.D.; Souza, B.S.d.F.; Rocha, C.A.G. Effects of IGF-1 on Proliferation, Angiogenesis, Tumor Stem Cell Populations and Activation of AKT and Hedgehog Pathways in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2020, 21, 6487. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21186487
Ferreira Mendes JM, de Faro Valverde L, Torres Andion Vidal M, Paredes BD, Coelho P, Allahdadi KJ, Coletta RD, Souza BSdF, Rocha CAG. Effects of IGF-1 on Proliferation, Angiogenesis, Tumor Stem Cell Populations and Activation of AKT and Hedgehog Pathways in Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2020; 21(18):6487. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21186487
Chicago/Turabian StyleFerreira Mendes, Jéssica Mariane, Ludmila de Faro Valverde, Manuela Torres Andion Vidal, Bruno Diaz Paredes, Paulo Coelho, Kyan James Allahdadi, Ricardo Della Coletta, Bruno Solano de Freitas Souza, and Clarissa Araújo Gurgel Rocha. 2020. "Effects of IGF-1 on Proliferation, Angiogenesis, Tumor Stem Cell Populations and Activation of AKT and Hedgehog Pathways in Oral Squamous Cell Carcinoma" International Journal of Molecular Sciences 21, no. 18: 6487. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21186487



