Hedgehog Signaling in Skeletal Development: Roles of Indian Hedgehog and the Mode of Its Action
Abstract
:1. Introduction
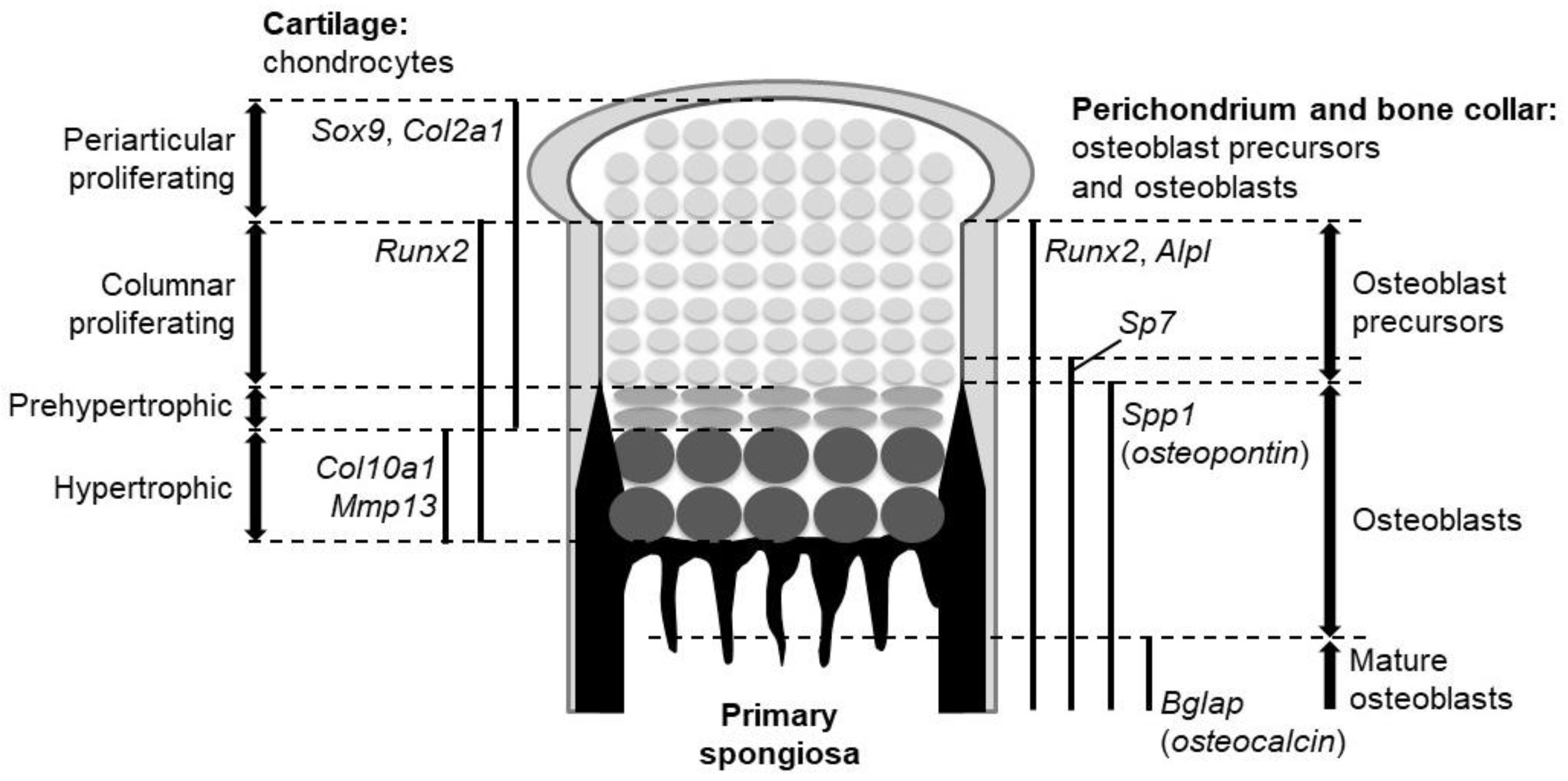
2. Two Ossification Processes in Mammals
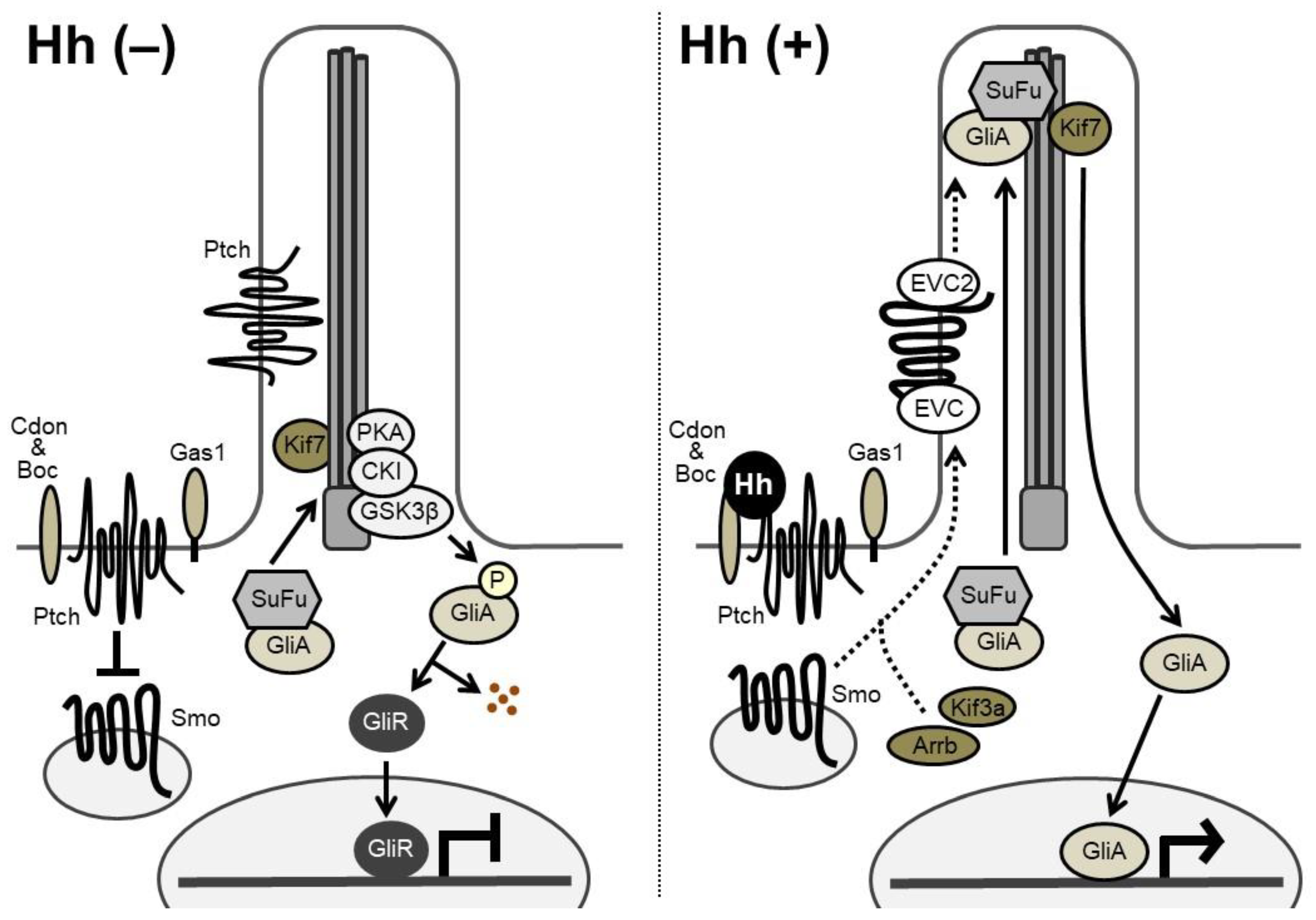
3. Hh Signaling Transduction
4. Roles of Hh Signaling in Endochondral Ossification
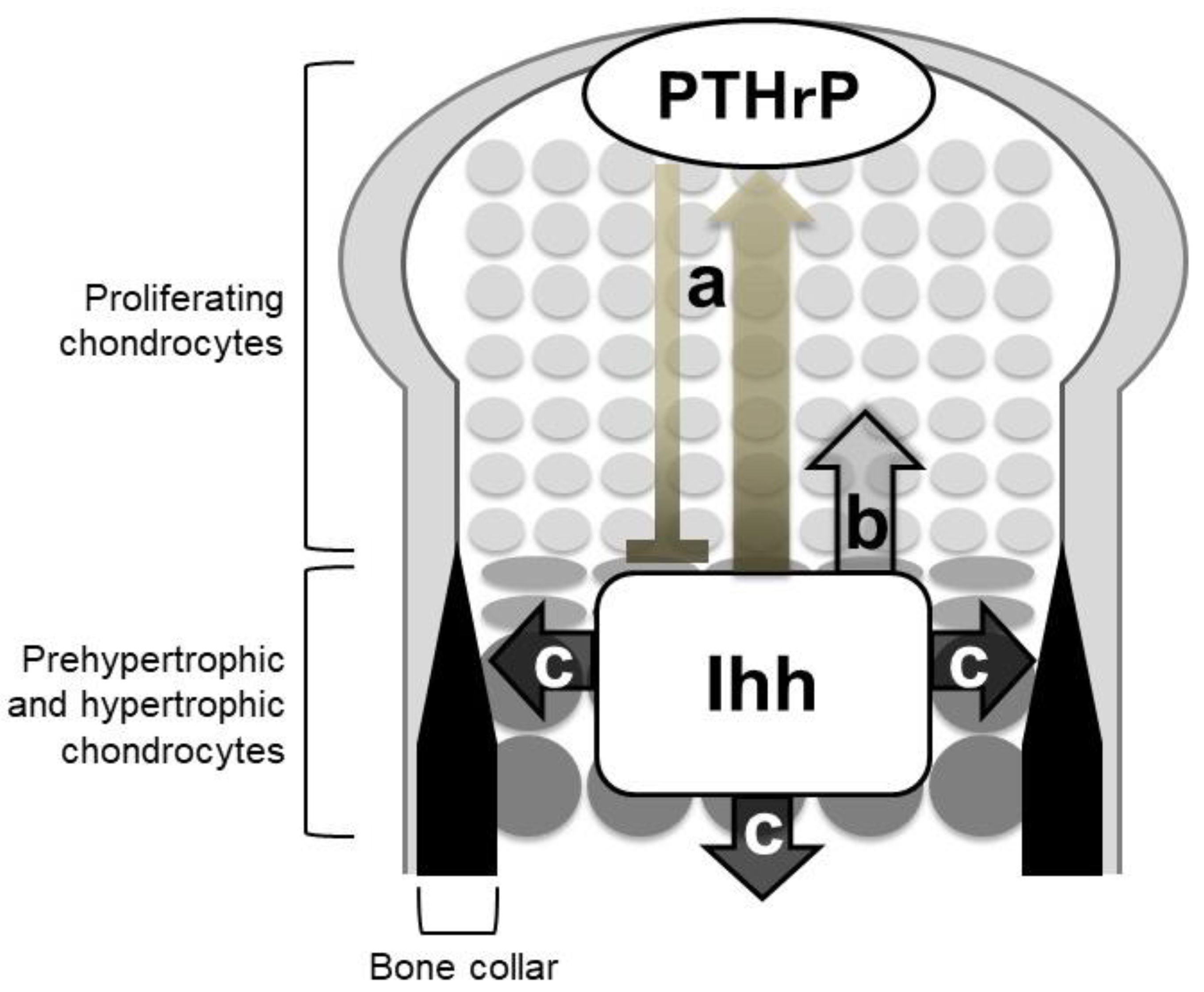
4.1. An Ihh-PTHrP Negative Feedback Loop Maintains the Growth Plate Length
4.2. Ihh Directly Regulates the Proliferation of Growth Plate Chondrocytes
4.3. Ihh Is Required for Specification of Osteoblasts
4.4. Factors Acting Downstream of Hh Signaling During Endochondral Ossification
5. Roles of Hh Signaling in Craniofacial Development
6. Postnatal Roles of Hh in Skeleton
7. Skeletal Diseases Caused by Abnormalities of Hh Signaling
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Hh | Hedgehog |
| Ihh | Indian hedgehog |
| PTHrP | parathyroid hormone-related protein |
| Shh | Sonic hedgehog |
| Dhh | Desert hedgehog |
| MMP13 | matrix metalloproteinase 13 |
| VEGF | vascular endothelial growth factor |
| Runx2 | runt-related transcription factor 2 |
| Alpl | alkaline phosphatase |
| Spp1 | secreted phosphoprotein 1 |
| Bglap | bone gla protein |
| Ski | skinny hedgehog |
| MBOAT | membrane-bound O-acyltransferase |
| Disp | Dispatched |
| Smo | Smoothened |
| Ptch | Patched |
| Cdon | cell adhesion molecule-related/downregulated by oncogenes |
| Boc | brother of Cdo |
| Gas1 | growth arrest specific 1 |
| SuFu | Suppressor of Fused |
| PKA | protein kinase A |
| CKI | casein kinase 1α |
| GSK3β | glycogen synthase kinase 3β |
| Spop | Speckle-type POZ protein |
| Gpcrk2 | G protein-coupled receptor kinase 2 |
| Arrb | β-arrestin |
| EVC | Ellis-van Creveld syndrome protein |
| PTH1R | parathyroid hormone 1 receptor |
| PPR | PTH/PTHrP receptor |
| WT | wild-type |
| BMP | bone morphogenetic protein |
| CNCC | cranial neural crest cell |
| FGF | Fibroblast growth factor |
| Fox | Forkhead box |
| IFT | intraflagellar transport proteins |
| RANKL | Receptor activator of nuclear factor kappa-B ligand |
| CREB | cAMP responsive element binding protein |
| OA | osteoarthritis |
| MMP | metaphyseal mesenchymal progenitor |
| iPSC | induced pluripotent stem cell |
| MAS | McCune Albright syndrome |
| GWAS | genome-wide association study |
| HHIP | hedgehog interacting protein |
| BDA1 | brachydactyly type AI |
| GCPS | Greig cephalopolysyndactyly syndrome |
| PHS | Pallister-Hall syndrome |
| PAPA1 | postaxial polydactyly type A1 |
| PAPB | postaxial polydactyly type B3 |
| BCNS | basal cell nevus syndrome |
| SLOS | Smith-Lemli-Opitz syndrome |
| DHCR7 | 7-dehydrocholesterol reductase |
| 7DHC | 7-dehydrocholesterol |
| POH | progressive osseous heteroplasia |
| NGS | next-generation sequencers |
References
- Ingham, P.W.; McMahon, A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briscoe, J.; Therond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Andre, P.; Ye, L.; Yang, Y.Z. The Hedgehog signalling pathway in bone formation. Int. J. Oral Sci. 2015, 7, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Ohba, S. Hedgehog signaling in endochondral ossification. J. Dev. Biol. 2016, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, R.; Kitazawa, R.; Kohara, Y.; Ikedo, A.; Imai, Y.; Kitazawa, S. Recent insights into long bone development: Central role of hedgehog signaling pathway in regulating growth plate. Int. J. Mol. Sci. 2019, 20, 5840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.T.; Gao, Y.H.; Inada, M.; et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef] [Green Version]
- Pan, A.; Chang, L.; Nguyen, A.; James, A.W. A review of hedgehog signaling in cranial bone development. Front. Physiol. 2013, 4, 61. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Pan, Y.; Wang, B. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development 2010, 137, 2001–2009. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Liu, A. Spop promotes skeletal development and homeostasis by positively regulating Ihh signaling. Proc. Natl. Acad. Sci. USA 2016, 113, 14751–14756. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Deeds, J.D.; Segre, G.V. Expression of parathyroid hormone-related peptide and its receptor messenger ribonucleic acids during fetal development of rats. Endocrinology 1995, 136, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Lanske, B.; Karaplis, A.C.; Deeds, J.D.; Kohno, H.; Nissenson, R.A.; Kronenberg, H.M.; Segre, G.V. Parathyroid hormone-related peptide delays terminal differentiation of chondrocytes during endochondral bone development. Endocrinology 1996, 137, 5109–5118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amizuka, N.; Warshawsky, H.; Henderson, J.E.; Goltzman, D.; Karaplis, A.C. Parathyroid hormone-related peptide-depleted mice show abnormal epiphyseal cartilage development and altered endochondral bone formation. J. Cell Biol. 1994, 126, 1611–1623. [Google Scholar] [CrossRef] [PubMed]
- Karaplis, A.C.; Luz, A.; Glowacki, J.; Bronson, R.T.; Tybulewicz, V.L.; Kronenberg, H.M.; Mulligan, R.C. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev. 1994, 8, 277–289. [Google Scholar] [CrossRef] [Green Version]
- Lanske, B.; Karaplis, A.C.; Lee, K.; Luz, A.; Vortkamp, A.; Pirro, A.; Karperien, M.; Defize, L.H.; Ho, C.; Mulligan, R.C.; et al. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science 1996, 273, 663–666. [Google Scholar] [CrossRef]
- Miao, D.; Liu, H.; Plut, P.; Niu, M.; Huo, R.; Goltzman, D.; Henderson, J.E. Impaired endochondral bone development and osteopenia in Gli2-deficient mice. Exp. Cell Res. 2004, 294, 210–222. [Google Scholar] [CrossRef]
- Weir, E.C.; Philbrick, W.M.; Amling, M.; Neff, L.A.; Baron, R.; Broadus, A.E. Targeted overexpression of parathyroid hormone-related peptide in chondrocytes causes chondrodysplasia and delayed endochondral bone formation. Proc. Natl. Acad. Sci. USA 1996, 93, 10240–10245. [Google Scholar] [CrossRef] [Green Version]
- Schipani, E.; Lanske, B.; Hunzelman, J.; Luz, A.; Kovacs, C.S.; Lee, K.; Pirro, A.; Kronenberg, H.M.; Juppner, H. Targeted expression of constitutively active receptors for parathyroid hormone and parathyroid hormone-related peptide delays endochondral bone formation and rescues mice that lack parathyroid hormone-related peptide. Proc. Natl. Acad. Sci. USA 1997, 94, 13689–13694. [Google Scholar] [CrossRef] [Green Version]
- Philbrick, W.M.; Dreyer, B.E.; Nakchbandi, I.A.; Karaplis, A.C. Parathyroid hormone-related protein is required for tooth eruption. Proc. Natl. Acad. Sci. USA 1998, 95, 11846–11851. [Google Scholar] [CrossRef] [Green Version]
- Vortkamp, A.; Lee, K.; Lanske, B.; Segre, G.V.; Kronenberg, H.M.; Tabin, C.J. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 1996, 273, 613–622. [Google Scholar] [CrossRef]
- St-Jacques, B.; Hammerschmidt, M.; McMahon, A.P. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999, 13, 2072–2086. [Google Scholar] [CrossRef] [Green Version]
- Mak, K.K.; Chen, M.H.; Day, T.F.; Chuang, P.T.; Yang, Y. Wnt/β-catenin signaling interacts differentially with Ihh signaling in controlling endochondral bone and synovial joint formation. Development 2006, 133, 3695–3707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karp, S.J.; Schipani, E.; St-Jacques, B.; Hunzelman, J.; Kronenberg, H.; McMahon, A.P. Indian hedgehog coordinates endochondral bone growth and morphogenesis via parathyroid hormone related-protein-dependent and -independent pathways. Development 2000, 127, 543–548. [Google Scholar] [PubMed]
- Chung, U.I.; Lanske, B.; Lee, K.; Li, E.; Kronenberg, H. The parathyroid hormone/parathyroid hormone-related peptide receptor coordinates endochondral bone development by directly controlling chondrocyte differentiation. Proc. Natl. Acad. Sci. USA 1998, 95, 13030–13035. [Google Scholar] [CrossRef] [Green Version]
- Chung, U.I.; Schipani, E.; McMahon, A.P.; Kronenberg, H.M. Indian hedgehog couples chondrogenesis to osteogenesis in endochondral bone development. J. Clin. Investig. 2001, 107, 295–304. [Google Scholar] [CrossRef] [Green Version]
- Hilton, M.J.; Tu, X.; Long, F. Tamoxifen-inducible gene deletion reveals a distinct cell type associated with trabecular bone, and direct regulation of PTHrP expression and chondrocyte morphology by Ihh in growth region cartilage. Dev. Biol. 2007, 308, 93–105. [Google Scholar] [CrossRef] [Green Version]
- Long, F.; Linsenmayer, T.F. Regulation of growth region cartilage proliferation and differentiation by perichondrium. Development 1998, 125, 1067–1073. [Google Scholar] [PubMed]
- Long, F.; Zhang, X.M.; Karp, S.; Yang, Y.; McMahon, A.P. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 2001, 128, 5099–5108. [Google Scholar]
- Kobayashi, T.; Chung, U.I.; Schipani, E.; Starbuck, M.; Karsenty, G.; Katagiri, T.; Goad, D.L.; Lanske, B.; Kronenberg, H.M. PTHrP and Indian hedgehog control differentiation of growth plate chondrocytes at multiple steps. Development 2002, 129, 2977–2986. [Google Scholar]
- Kobayashi, T.; Soegiarto, D.W.; Yang, Y.; Lanske, B.; Schipani, E.; McMahon, A.P.; Kronenberg, H.M. Indian hedgehog stimulates periarticular chondrocyte differentiation to regulate growth plate length independently of PTHrP. J. Clin. Investig. 2005, 115, 1734–1742. [Google Scholar] [CrossRef]
- Mak, K.K.; Kronenberg, H.M.; Chuang, P.T.; Mackem, S.; Yang, Y. Indian hedgehog signals independently of PTHrP to promote chondrocyte hypertrophy. Development 2008, 135, 1947–1956. [Google Scholar] [CrossRef] [Green Version]
- Hilton, M.J.; Tu, X.; Cook, J.; Hu, H.; Long, F. Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development 2005, 132, 4339–4351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, F.; Chung, U.I.; Ohba, S.; McMahon, J.; Kronenberg, H.M.; McMahon, A.P. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development 2004, 131, 1309–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodda, S.J.; McMahon, A.P. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 2006, 133, 3231–3244. [Google Scholar] [CrossRef] [Green Version]
- Koziel, L.; Wuelling, M.; Schneider, S.; Vortkamp, A. Gli3 acts as a repressor downstream of Ihh in regulating two distinct steps of chondrocyte differentiation. Development 2005, 132, 5249–5260. [Google Scholar] [CrossRef] [Green Version]
- Park, H.L.; Bai, C.; Platt, K.A.; Matise, M.P.; Beeghly, A.; Hui, C.C.; Nakashima, M.; Joyner, A.L. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 2000, 127, 1593–1605. [Google Scholar] [PubMed]
- Hojo, H.; Ohba, S.; Yano, F.; Saito, T.; Ikeda, T.; Nakajima, K.; Komiyama, Y.; Nakagata, N.; Suzuki, K.; Takato, T.; et al. Gli1 Protein Participates in Hedgehog-mediated Specification of Osteoblast Lineage during Endochondral Ossification. J. Biol. Chem. 2012, 287, 17860–17869. [Google Scholar] [CrossRef] [Green Version]
- Hojo, H.; Ohba, S.; Taniguchi, K.; Shirai, M.; Yano, F.; Saito, T.; Ikeda, T.; Nakajima, K.; Komiyama, Y.; Nakagata, N.; et al. Hedgehog-Gli Activators Direct Osteo-chondrogenic Function of Bone Morphogenetic Protein toward Osteogenesis in the Perichondrium. J. Biol. Chem. 2013, 288, 9924–9932. [Google Scholar] [CrossRef] [Green Version]
- Shimoyama, A.; Wada, M.; Ikeda, F.; Hata, K.; Matsubara, T.; Nifuji, A.; Noda, M.; Amano, K.; Yamaguchi, A.; Nishimura, R.; et al. Ihh/Gli2 signaling promotes osteoblast differentiation by regulating Runx2 expression and function. Mol. Biol. Cell. 2007, 18, 2411–2418. [Google Scholar] [CrossRef] [Green Version]
- Joeng, K.S.; Long, F.X. The Gli2 transcriptional activator is a crucial effector for Ihh signaling in osteoblast development and cartilage vascularization. Development 2009, 136, 4177–4185. [Google Scholar] [CrossRef] [Green Version]
- Ohba, S.; Kawaguchi, H.; Kugimiya, F.; Ogasawara, T.; Kawamura, N.; Saito, T.; Ikeda, T.; Fujii, K.; Miyajima, T.; Kuramochi, A.; et al. Patched1 haploinsufficiency increases adult bone mass and modulates Gli3 repressor activity. Dev. Cell. 2008, 14, 689–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, X.; Joeng, K.S.; Long, F. Indian hedgehog requires additional effectors besides Runx2 to induce osteoblast differentiation. Dev. Biol. 2012, 362, 76–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milet, C.; Monsoro-Burq, A.H. Neural crest induction at the neural plate border in vertebrates. Dev. Biol. 2012, 366, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Mao, J.; Tenzen, T.; Kottmann, A.H.; McMahon, A.P. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004, 18, 937–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Balczerski, B.; Ciozda, A.; Louie, K.; Oralova, V.; Huysseune, A.; Crump, J.G. Fox proteins are modular competency factors for facial cartilage and tooth specification. Development 2018, 145, dev165498. [Google Scholar] [CrossRef] [Green Version]
- Lenton, K.; James, A.W.; Manu, A.; Brugmann, S.A.; Birker, D.; Nelson, E.R.; Leucht, P.; Helms, J.A.; Longaker, M.T. Indian hedgehog positively regulates calvarial ossification and modulates bone morphogenetic protein signaling. Genesis 2011, 49, 784–796. [Google Scholar] [CrossRef]
- Abzhanov, A.; Rodda, S.J.; McMahon, A.P.; Tabin, C.J. Regulation of skeletogenic differentiation in cranial dermal bone. Development 2007, 134, 3133–3144. [Google Scholar] [CrossRef] [Green Version]
- Amano, K.; Okuzaki, D.; Aikawa, T.; Kogo, M. Indian hedgehog in craniofacial neural crest cells links to skeletal malocclusion by regulating associated cartilage formation and gene expression. FASEB J. 2020, 34, 6791–6807. [Google Scholar] [CrossRef] [Green Version]
- Ashe, A.; Butterfield, N.C.; Town, L.; Courtney, A.D.; Cooper, A.N.; Ferguson, C.; Barry, R.; Olsson, F.; Liem, K.F., Jr.; Parton, R.G.; et al. Mutations in mouse Ift144 model the craniofacial, limb and rib defects in skeletal ciliopathies. Hum. Mol. Genet. 2012, 21, 1808–1823. [Google Scholar] [CrossRef] [Green Version]
- Koyama, E.; Young, B.; Nagayama, M.; Shibukawa, Y.; Enomoto-Iwamoto, M.; Iwamoto, M.; Maeda, Y.; Lanske, B.; Song, B.; Serra, R.; et al. Conditional Kif3a ablation causes abnormal hedgehog signaling topography, growth plate dysfunction, and excessive bone and cartilage formation during mouse skeletogenesis. Development 2007, 134, 2159–2169. [Google Scholar] [CrossRef] [Green Version]
- Brugmann, S.A.; Allen, N.C.; James, A.W.; Mekonnen, Z.; Madan, E.; Helms, J.A. A primary cilia-dependent etiology for midline facial disorders. Hum. Mol. Genet. 2010, 19, 1577–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, Y.; Nakamura, E.; Nguyen, M.T.; Suva, L.J.; Swain, F.L.; Razzaque, M.S.; Mackem, S.; Lanske, B. Indian Hedgehog produced by postnatal chondrocytes is essential for maintaining a growth plate and trabecular bone. Proc. Natl. Acad. Sci. USA 2007, 104, 6382–6387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amano, K.; Densmore, M.J.; Lanske, B. Conditional deletion of Indian Hedgehog in limb mesenchyme results in complete loss of growth plate formation but allows mature osteoblast differentiation. J. Bone Miner. Res. 2015, 30, 2262–2272. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Schipani, E.; Densmore, M.J.; Lanske, B. Partial rescue of postnatal growth plate abnormalities in Ihh mutants by expression of a constitutively active PTH/PTHrP receptor. Bone 2010, 46, 472–478. [Google Scholar] [CrossRef] [Green Version]
- Hirai, T.; Chagin, A.S.; Kobayashi, T.; Mackem, S.; Kronenberg, H.M. Parathyroid hormone/parathyroid hormone-related protein receptor signaling is required for maintenance of the growth plate in postnatal life. Proc. Natl. Acad. Sci. USA 2011, 108, 191–196. [Google Scholar] [CrossRef] [Green Version]
- Mak, K.K.; Bi, Y.; Wan, C.; Chuang, P.T.; Clemens, T.; Young, M.; Yang, Y. Hedgehog signaling in mature osteoblasts regulates bone formation and resorption by controlling PTHrP and RANKL expression. Dev. Cell. 2008, 14, 674–688. [Google Scholar] [CrossRef] [Green Version]
- Kitaura, Y.; Hojo, H.; Komiyama, Y.; Takato, T.; Chung, U.I.; Ohba, S. Gli1 haploinsufficiency leads to decreased bone mass with an uncoupling of bone metabolism in adult mice. PLoS ONE 2014, 9, e109597. [Google Scholar] [CrossRef]
- Kohara, Y.; Haraguchi, R.; Kitazawa, R.; Imai, Y.; Kitazawa, S. Hedgehog Inhibitors suppress osteoclastogenesis in in vitro cultures, and deletion of smo in macrophage/osteoclast lineage prevents age-related bone loss. Int. J. Mol. Sci. 2020, 21, 2745. [Google Scholar] [CrossRef]
- Wei, F.; Zhou, J.; Wei, X.; Zhang, J.; Fleming, B.C.; Terek, R.; Pei, M.; Chen, Q.; Liu, T.; Wei, L.; et al. Activation of Indian hedgehog promotes chondrocyte hypertrophy and upregulation of MMP-13 in human osteoarthritic cartilage. Osteoarthr. Cartil. 2012, 20, 755–763. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Chen, Q.; Lanske, B.; Fleming, B.C.; Terek, R.; Wei, X.; Zhang, G.; Wang, S.; Li, K.; Wei, L.; et al. Disrupting the Indian hedgehog signaling pathway in vivo attenuates surgically induced osteoarthritis progression in Col2a1-CreERT2; Ihhfl/fl mice. Arthritis Res. Ther. 2014, 16, R11. [Google Scholar] [CrossRef] [Green Version]
- Lin, A.C.; Seeto, B.L.; Bartoszko, J.M.; Khoury, M.A.; Whetstone, H.; Ho, L.; Hsu, C.; Ali, S.A.; Alman, B.A. Modulating hedgehog signaling can attenuate the severity of osteoarthritis. Nat. Med. 2009, 15, 1421–1425. [Google Scholar] [CrossRef]
- Ransom, R.C.; Carter, A.C.; Salhotra, A.; Leavitt, T.; Marecic, O.; Murphy, M.P.; Lopez, M.L.; Wei, Y.; Marshall, C.D.; Shen, E.Z.; et al. Mechanoresponsive stem cells acquire neural crest fate in jaw regeneration. Nature 2018, 563, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.K.F.; Gulati, G.S.; Sinha, R.; Tompkins, J.V.; Lopez, M.; Carter, A.C.; Ransom, R.C.; Reinisch, A.; Wearda, T.; Murphy, M.; et al. Identification of the human skeletal stem cell. Cell 2018, 175, 43–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, C.K.; Seo, E.Y.; Chen, J.Y.; Lo, D.; McArdle, A.; Sinha, R.; Tevlin, R.; Seita, J.; Vincent-Tompkins, J.; Wearda, T.; et al. Identification and specification of the mouse skeletal stem cell. Cell 2015, 160, 285–298. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; He, G.; Lee, W.C.; McKenzie, J.A.; Silva, M.J.; Long, F. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun. 2017, 8, 2043. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, R.; Kitazawa, R.; Imai, Y.; Kitazawa, S. Growth plate-derived hedgehog-signal-responsive cells provide skeletal tissue components in growing bone. Histochem. Cell Biol. 2018, 149, 365–373. [Google Scholar] [CrossRef]
- Onodera, S.; Saito, A.; Hojo, H.; Nakamura, T.; Zujur, D.; Watanabe, K.; Morita, N.; Hasegawa, D.; Masaki, H.; Nakauchi, H.; et al. Hedgehog activation regulates human osteoblastogenesis. Stem. Cell Rep. 2020, 15, 125–139. [Google Scholar] [CrossRef]
- Lettre, G.; Jackson, A.U.; Gieger, C.; Schumacher, F.R.; Berndt, S.I.; Sanna, S.; Eyheramendy, S.; Voight, B.F.; Butler, J.L.; Guiducci, C.; et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008, 40, 584–591. [Google Scholar] [CrossRef]
- Weedon, M.N.; Lango, H.; Lindgren, C.M.; Wallace, C.; Evans, D.M.; Mangino, M.; Freathy, R.M.; Perry, J.R.; Stevens, S.; Hall, A.S.; et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008, 40, 575–583. [Google Scholar] [CrossRef] [Green Version]
- Gao, B.; Guo, J.; She, C.; Shu, A.; Yang, M.; Tan, Z.; Yang, X.; Guo, S.; Feng, G.; He, L. Mutations in IHH, encoding Indian hedgehog, cause brachydactyly type A-1. Nat. Genet. 2001, 28, 386–388. [Google Scholar] [CrossRef]
- Gao, B.; He, L. Answering a century old riddle: Brachydactyly type A1. Cell Res. 2004, 14, 179–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, B.; Hu, J.; Stricker, S.; Cheung, M.; Ma, G.; Law, K.F.; Witte, F.; Briscoe, J.; Mundlos, S.; He, L.; et al. A mutation in Ihh that causes digit abnormalities alters its signalling capacity and range. Nature 2009, 458, 1196–1200. [Google Scholar] [CrossRef] [PubMed]
- Vortkamp, A.; Gessler, M.; Grzeschik, K.H. GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families. Nature 1991, 352, 539–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, S.; Graham, J.M., Jr.; Olney, A.H.; Biesecker, L.G. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat. Genet. 1997, 15, 266–268. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishna, U.; Wild, A.; Grzeschik, K.H.; Antonarakis, S.E. Mutation in GLI3 in postaxial polydactyly type A. Nat. Genet. 1997, 17, 269–271. [Google Scholar] [CrossRef]
- Gorlin, R.J. Nevoid basal-cell carcinoma syndrome. Medicine 1987, 66, 98–113. [Google Scholar] [CrossRef]
- Shefer, S.; Salen, G.; Batta, A.K.; Honda, A.; Tint, G.S.; Irons, M.; Elias, E.R.; Chen, T.C.; Holick, M.F. Markedly inhibited 7-dehydrocholesterol-delta 7-reductase activity in liver microsomes from Smith-Lemli-Opitz homozygotes. J. Clin. Investig. 1995, 96, 1779–1785. [Google Scholar] [CrossRef]
- Blassberg, R.; Macrae, J.I.; Briscoe, J.; Jacob, J. Reduced cholesterol levels impair Smoothened activation in Smith-Lemli-Opitz syndrome. Hum. Mol. Genet. 2016, 25, 693–705. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, F.S.; Hahn, G.V.; Zasloff, M.A. Heterotopic ossification: Two rare forms and what they can teach us. J. Am. Acad. Orthop. Surg. 1994, 2, 288–296. [Google Scholar] [CrossRef]
- Regard, J.B.; Malhotra, D.; Gvozdenovic-Jeremic, J.; Josey, M.; Chen, M.; Weinstein, L.S.; Lu, J.; Shore, E.M.; Kaplan, F.S.; Yang, Y.; et al. Activation of Hedgehog signaling by loss of GNAS causes heterotopic ossification. Nat. Med. 2013, 19, 1505–1512. [Google Scholar] [CrossRef] [Green Version]
- Will, A.J.; Cova, G.; Osterwalder, M.; Chan, W.L.; Wittler, L.; Brieske, N.; Heinrich, V.; de Villartay, J.P.; Vingron, M.; Klopocki, E.; et al. Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian hedgehog). Nat. Genet. 2017, 49, 1539–1545. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohba, S. Hedgehog Signaling in Skeletal Development: Roles of Indian Hedgehog and the Mode of Its Action. Int. J. Mol. Sci. 2020, 21, 6665. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21186665
Ohba S. Hedgehog Signaling in Skeletal Development: Roles of Indian Hedgehog and the Mode of Its Action. International Journal of Molecular Sciences. 2020; 21(18):6665. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21186665
Chicago/Turabian StyleOhba, Shinsuke. 2020. "Hedgehog Signaling in Skeletal Development: Roles of Indian Hedgehog and the Mode of Its Action" International Journal of Molecular Sciences 21, no. 18: 6665. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21186665



