P. gingivalis Lipopolysaccharide Stimulates the Upregulated Expression of the Pancreatic Cancer-Related Genes Regenerating Islet-Derived 3 A/G in Mouse Pancreas
Abstract
:1. Introduction
2. Results
2.1. Microarray
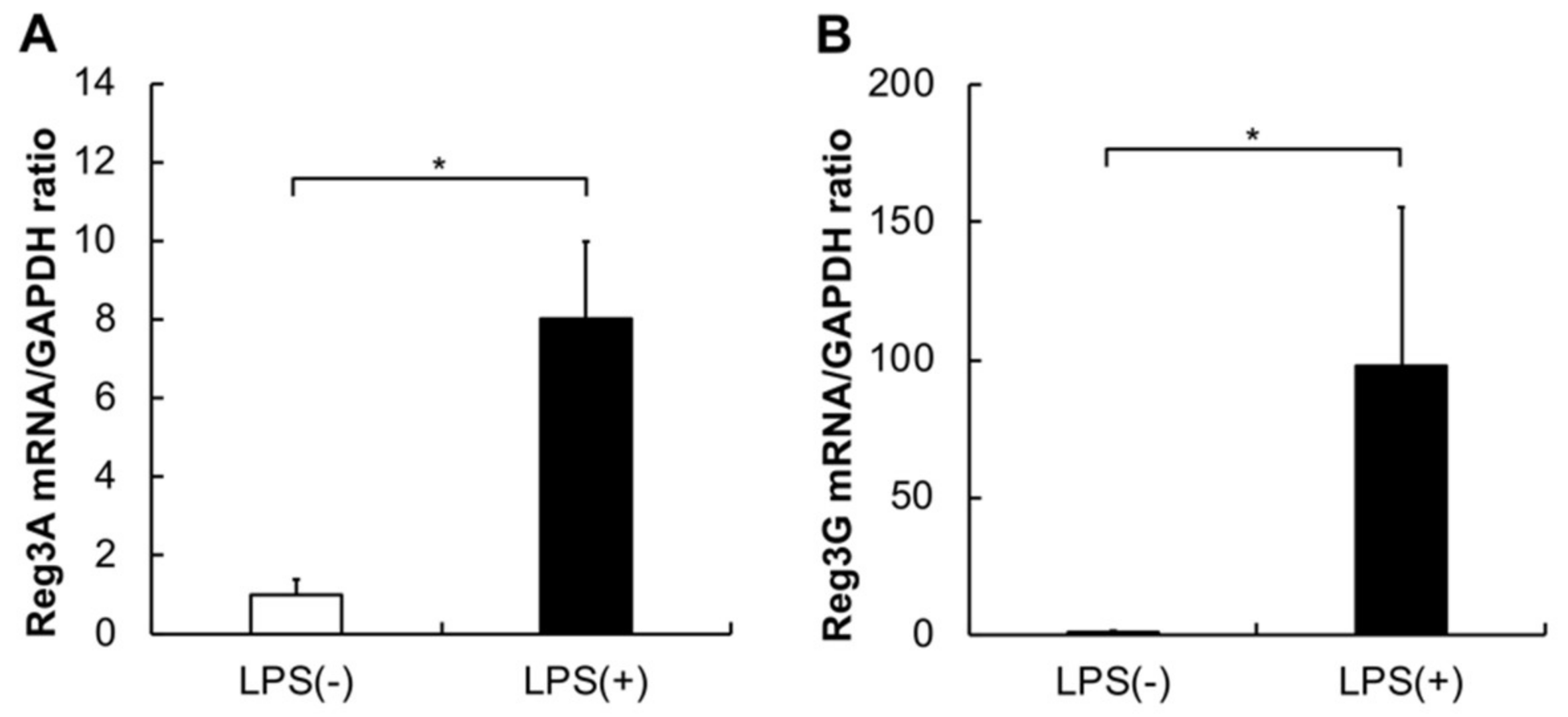
2.2. Gene Expression
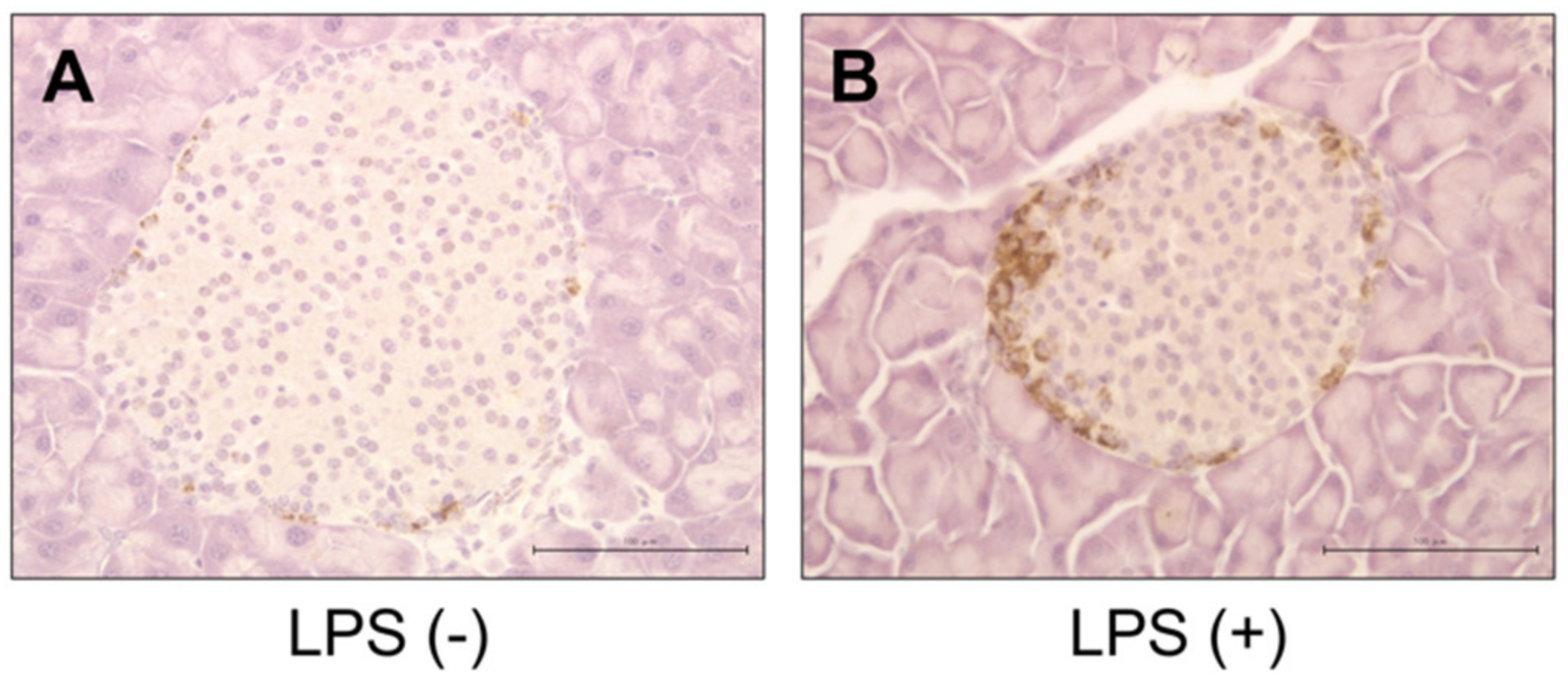
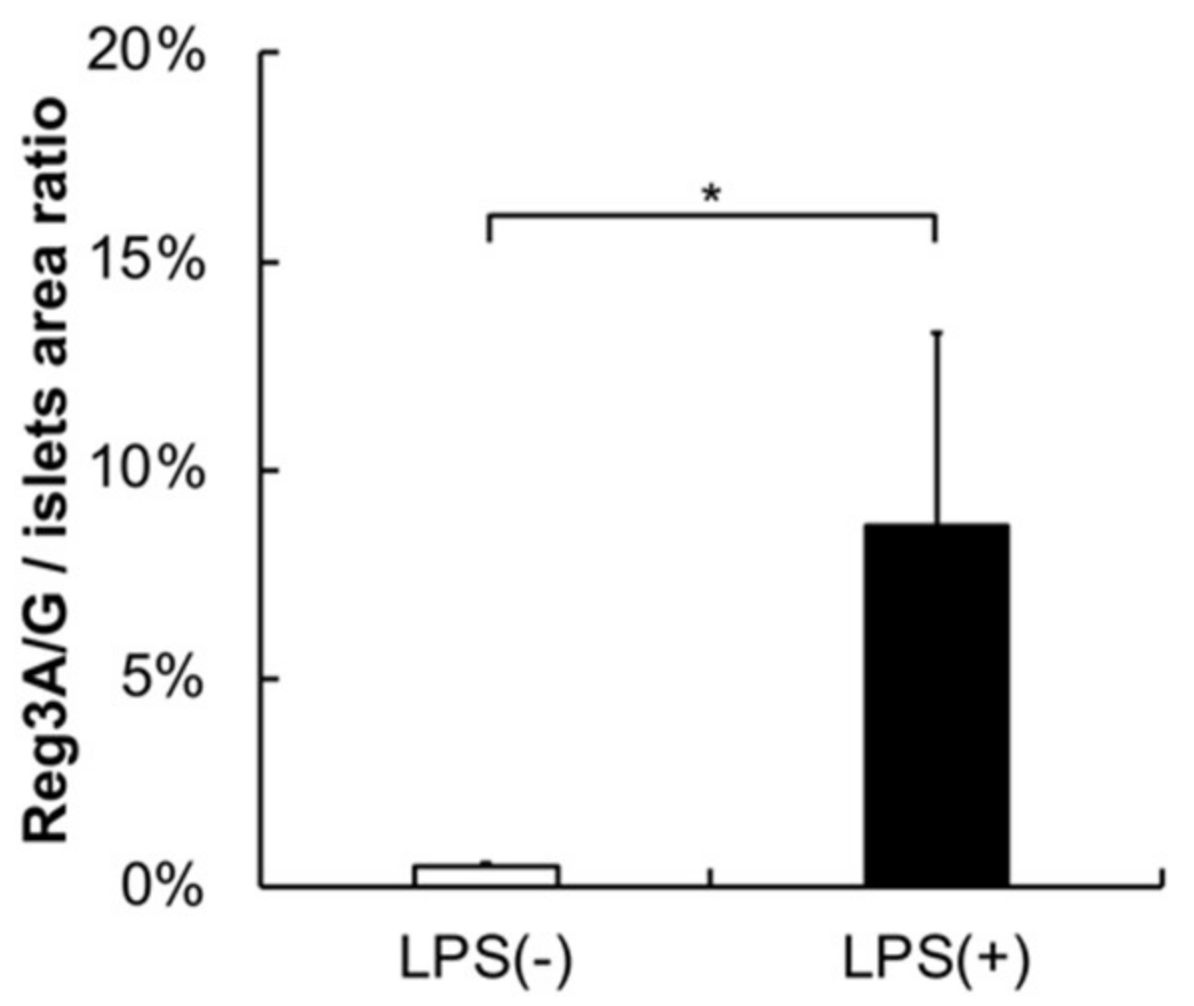
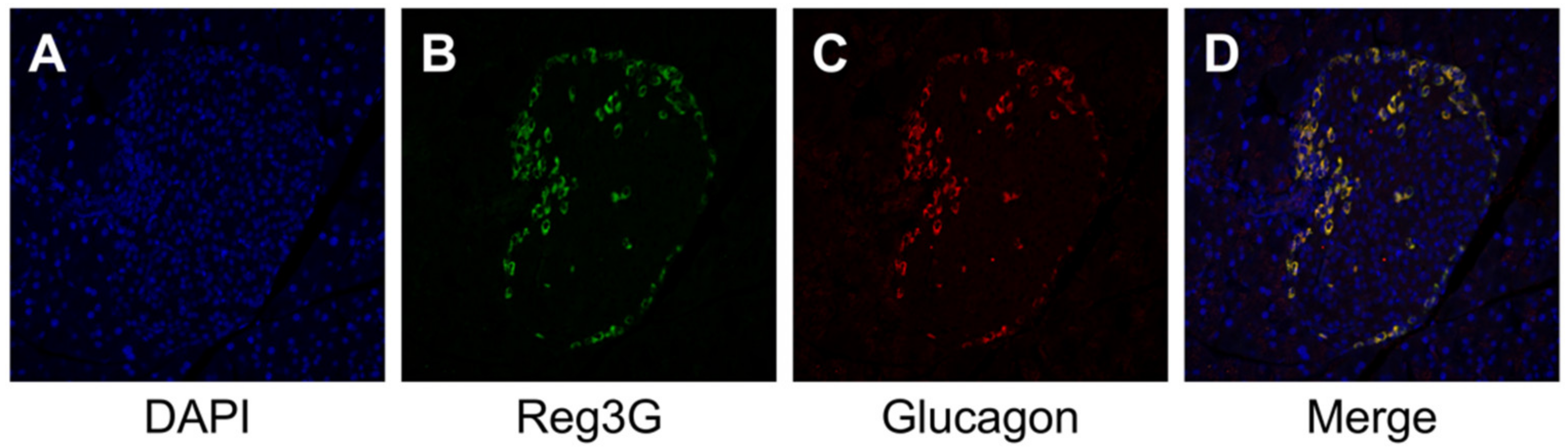
2.3. Histology, Immunohistochemistry, and Morphological Analysis
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. RNA Extraction and Microarray
4.3. Quantitative Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
4.4. Histological, Immunohistochemical, Immunofluorescence, and Morphological Analyses
4.5. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Chil3/Chil4 | Chitinase-like 3 / Chitinase-like 4 |
| E. coli | Escherichia coli |
| H & E | Hematoxylin–eosin |
| HRP | Horseradish peroxidase |
| Ighg3 | Immunoglobulin heavy constant gamma 3 |
| Ighv10-3 | Immunoglobulin heavy variable V10-3 |
| Igk | Immunoglobulin kappa chain |
| Igkv4-53 | Immunoglobulin kappa variable 4-53 |
| Iglv1 | Immunoglobulin lambda variable 1 |
| LPS | Lipopolysaccharides |
| P. gingivalis | Porphyromonas gingivalis |
| PG-LPS | P. gingivalis lipopolysaccharide |
| RT-PCR | Quantitative reverse transcriptase-polymerase chain reaction |
| Reg | Regenerating |
| Reg3G | Regenerating islet-derived 3 gamma |
| S100A8 | S100 calcium binding protein A8 |
| S100A9 | S100 calcium binding protein A9 |
| SD | Standard deviation |
| TLR4 | Toll-like receptor 4 |
References
- Llambés, F.; Arias-Herrera, S.; Caffesse, R. Relationship between diabetes and periodontal infection. World J. Diabetes 2015, 6, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Linden, G.J.; Lyons, A.; Scannapieco, F.A. Periodontal systemic associations: Review of the evidence. J. Periodontol. 2013, 40, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Chambrone, L.; Foz, A.M.; Guglielmetti, M.R.; Pannuti, C.M.; Artese, H.P.; Feres, M.; Romito, G.A. Periodontitis and chronic kidney disease: A systematic review of the association of diseases and the effect of periodontal treatment on estimated glomerular filtration rate. J. Clin. Periodontol. 2013, 40, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Ogrendik, M. Rheumatoid arthritis is an autoimmune disease caused by periodontal pathogens. Int. J. Gen. Med. 2013, 6, 383–386. [Google Scholar] [CrossRef] [Green Version]
- Michaud, D.S.; Izard, J.; Wilhelm-Benartzi, C.S.; You, D.H.; Grote, V.A.; Tjønneland, A.; Dahm, C.C.; Overvad, K.; Jenab, M.; Fedirko, V.; et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 2013, 62, 1764–1770. [Google Scholar] [CrossRef]
- Yadav, D.; Lowenfels, A.B. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology 2013, 144, 1252–1261. [Google Scholar] [CrossRef] [Green Version]
- Terao, K.; Wake, H.; Adachi, N.; Liu, K.; Teshigawara, K.; Takahashi, H.; Mori, S.; Nishibori, M. Histidine-Rich Glycoprotein Suppresses Hyperinflammatory Responses of Lung in a Severe Acute Pancreatitis Mouse Model. Pancreas 2018, 47, 1156–1164. [Google Scholar] [CrossRef]
- Ding, S.P.; Li, J.C.; Jin, C. A mouse model of severe acute pancreatitis induced with caerulein and lipopolysaccharide. World J. Gastroenterol. 2003, 9, 584–589. [Google Scholar] [CrossRef]
- Hoshino, K.; Takeuchi, O.; Kawai, T.; Sanjo, H.; Ogawa, T.; Takeda, Y.; Takeda, K.; Akira, S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: Evidence for TLR4 as the Lps gene product. J. Immunol. 1999, 162, 3749–3752. [Google Scholar]
- Darveau, R.P.; Pham, T.T.T.; Lemley, K.; Reife, R.A.; Bainbridge, B.W.; Coats, S.R.; Howald, W.N.; Way, S.S.; Hajjar, A.M. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect. Immun. 2004, 72, 5041–5051. [Google Scholar] [CrossRef] [Green Version]
- Lanki, M.A.; Seppänen, H.E.; Mustonen, H.K.; Böckelman, C.; Juuti, A.T.; Hagström, J.K.; Haglund, C.H. Toll-like receptor 2 and Toll-like receptor 4 predict favorable prognosis in local pancreatic cancer. Tumor Biol. 2018, 40, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, F.; Uehara, O.; Morikawa, T.; Hiraki, D.; Onishi, A.; Toraya, S.; Adhikari, B.R.; Takai, R.; Yoshida, K.; Sato, J.; et al. Effect of systemic administration of lipopolysaccharides derived from Porphyromonas gingivalis on gene expression in mice kidney. Med. Mol. Morphol. 2018, 51, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhou, Z.; Cheng, Q.; Wang, H.; Cao, H.; Xu, Q.; Tuo, Y.; Jiang, L.; Zou, Y.; Ren, H.; et al. Acceleration of pancreatic tumorigenesis under immunosuppressive microenvironment induced by Reg3g overexpression. Cell Death Dis. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, G.; Du, J.; Cao, H.; Liu, X.; Xu, Q. Reg3g Promotes Pancreatic Carcinogenesis in a Murine Model of Chronic Pancreatitis. Dig. Dis. Sci. 2015, 60, 3656–3668. [Google Scholar] [CrossRef] [PubMed]
- Hartupee, J.C.; Zhang, H.; Bonaldo, M.F.; Soares, M.B.; Dieckgraefe, B.K. Isolation and characterization of a cDNA encoding a novel member of the human regenerating protein family: Reg IV. Biochim. Biophys. Acta Gene Struct. Expr. 2001, 1518, 287–293. [Google Scholar] [CrossRef]
- Parikh, A.; Stephan, A.-F.; Tzanakakis, E.S. Regeneratin proteins and their expression, regulation and signalling. Biomol. Concepts 2012, 3, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Wang, H.; Zogopoulos, G.; Shao, Q.; Dong, K.; Lv, F.; Nwilati, K.; Gui, X.Y.; Cuggia, A.; Liu, J.L.; et al. Reg proteins promote acinar-to-ductal metaplasia and act as novel diagnostic and prognostic markers in pancreatic ductal adenocarcinoma. Oncotarget 2016, 7, 77838–77853. [Google Scholar] [CrossRef]
- Gironella, M.; Calvo, C.; Fernández, A.; Closa, D.; Iovanna, J.L.; Rosello-Catafau, J.; Folch-Puy, E. Reg3b deficiency impairs pancreatic tumor growth by skewing macrophage polarization. Cancer Res. 2013, 73, 5682–5694. [Google Scholar] [CrossRef] [Green Version]
- Porterfield, M.; Zhao, P.; Han, H.; Cunningham, J.; Aoki, K.; von Hoff, D.D.; Demeure, M.J.; Pierce, J.M.; Tiemeyer, M.; Wells, L. Discrimination between adenocarcinoma and normal pancreatic ductal fluid by proteomic and glycomic analysis. J. Proteome Res. 2014, 13, 395–407. [Google Scholar] [CrossRef] [Green Version]
- Cavard, C.; Terris, B.; Grimber, G.; Christa, L.; Audard, V.; Radenen-Bussiere, B.; Simon, M.T.; Renard, C.A.; Buendia, M.A.; Perret, C. Overexpression of regenerating islet-derived 1 alpha and 3 alpha genes in human primary liver tumors with β-catenin mutations. Oncogene 2006, 25, 599–608. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Xiao, L.; Wang, S.-J.; Yue, W.; Yin, Q.-S.; Sun, M.-Y.; Xia, W.; Shao, Z.-Y.; Zhang, H. Up-regulation of REG3A in colorectal cancer cells confers proliferation and correlates with colorectal cancer risk. Oncotarget 2015, 7, 3921–3933. [Google Scholar] [CrossRef] [PubMed]
- Nata, K.; Liu, Y.; Xu, L.; Ikeda, T.; Akiyama, T.; Noguchi, N.; Kawaguchi, S.; Yamauchi, A.; Takahashi, I.; Shervani, N.J.; et al. Molecular cloning, expression and chromosomal localization of a novel human REG family gene, REG III. Gene 2004, 340, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Fishwick, D.A.; Bowman, A.; Korngiebel-Rosique, M.C.; Vinik, A.I. Pancreatic islet immunoreactivity to the Reg protein INGAP. J. Histochem. Cytochem. 2008, 56, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Gurr, W.; Yavari, R.; Wen, L.; Shaw, M.; Mora, C.; Christa, L.; Sherwin, R.S. A Reg family protein is overexpressed in islets from a patient with new-onset type 1 diabetes and acts as T-cell autoantigen in NOD mice. Diabetes 2002, 51, 339–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norkina, O.; Graf, R.; Appenzeller, P.; de Lisle, R.C. Caerulein-induced acute pancreatitis in mice that constitutively overexpress Reg/PAP genes. BMC Gastroenterol. 2006, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaz, J.; Akbarshahi, H.; Andersson, R. Controversial role of toll-like receptors in acute pancreatitis. World J. Gastroenterol. 2013, 19, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Ghosh, S. Toll-like receptor-mediated NF-κB activation: A phylogenetically conserved paradigm in innate immunity. J. Clin. Invest. 2001, 107, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Groeger, S.; Jarzina, F.; Domann, E.; Meyle, J. Porphyromonas gingivalis activates NFΚB and MAPK pathways in human oral epithelial cells. BMC Immunol. 2017, 18, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Säkkinen, H.; Aro, J.; Kaikkonen, L.; Ohukainen, P.; Näpänkangas, J.; Tokola, H.; Ruskoaho, H.; Rysä, J. Mitogen-activated protein kinase p38 target regenerating islet-derived 3γ expression is upregulated in cardiac inflammatory response in the rat heart. Physiol. Rep. 2016, 4, e12996. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Li, Y.; Gao, Y. Early changes in the urine proteome in a diethyldithiocarbamate-induced chronic pancreatitis rat model. J. Proteom. 2018, 186, 8–14. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.J.; Walker, F.R. Strategies to improve quantitative assessment of immunohistochemical and immunofluorescent labelling. Sci. Rep. 2015, 5, 1–4. [Google Scholar] [CrossRef] [PubMed]

| Gene Symbol | Protein Name | Fold Change (Control vs LPS) |
|---|---|---|
| Ighg3 | Immunoglobulin heavy constant gamma 3 | 4123.958 |
| S100A8 | S100 calcium-binding protein A8 | 523.053 |
| S100A9 | S100 calcium-binding protein A9 | 336.474 |
| LOC102642252 | Immunoglobulin heavy chain variable region | 263.940 |
| Igk | Immunoglobulin kappa chain | 241.758 |
| Iglv1 | Immunoglobulin lambda variable 1 | 112.254 |
| Reg3G | Regenerating islet-derived 3 gamma | 73.305 |
| Igkv4-53 | Immunoglobulin kappa variable 4-53 | 66.337 |
| Ighv10-3 | Immunoglobulin heavy variable V10-3 | 51.897 |
| Chil3/Chil4 | Chitinase-like 3/Chitinase-like 4 | 48.847 |
| Gene | Forward | Reverse |
|---|---|---|
| GAPDH | AGAACATCATCCCTGCATCC | CACATTGGGGGTAGGAACAC |
| Reg3A | TTCCTTTGTGTCCTCCTTGG | ACCTCCATTGGGTTGTTGAC |
| Reg3G | AACAGAGGTGGATGGGAGTG | GTGATTGCCTGAGGAAGAGG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hiraki, D.; Uehara, O.; Kuramitsu, Y.; Morikawa, T.; Harada, F.; Yoshida, K.; Akino, K.; Chiba, I.; Asaka, M.; Abiko, Y. P. gingivalis Lipopolysaccharide Stimulates the Upregulated Expression of the Pancreatic Cancer-Related Genes Regenerating Islet-Derived 3 A/G in Mouse Pancreas. Int. J. Mol. Sci. 2020, 21, 7351. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21197351
Hiraki D, Uehara O, Kuramitsu Y, Morikawa T, Harada F, Yoshida K, Akino K, Chiba I, Asaka M, Abiko Y. P. gingivalis Lipopolysaccharide Stimulates the Upregulated Expression of the Pancreatic Cancer-Related Genes Regenerating Islet-Derived 3 A/G in Mouse Pancreas. International Journal of Molecular Sciences. 2020; 21(19):7351. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21197351
Chicago/Turabian StyleHiraki, Daichi, Osamu Uehara, Yasuhiro Kuramitsu, Tetsuro Morikawa, Fumiya Harada, Koki Yoshida, Kozo Akino, Itsuo Chiba, Masahiro Asaka, and Yoshihiro Abiko. 2020. "P. gingivalis Lipopolysaccharide Stimulates the Upregulated Expression of the Pancreatic Cancer-Related Genes Regenerating Islet-Derived 3 A/G in Mouse Pancreas" International Journal of Molecular Sciences 21, no. 19: 7351. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21197351





