Advances in Analysis of Biodistribution of Exosomes by Molecular Imaging
Abstract
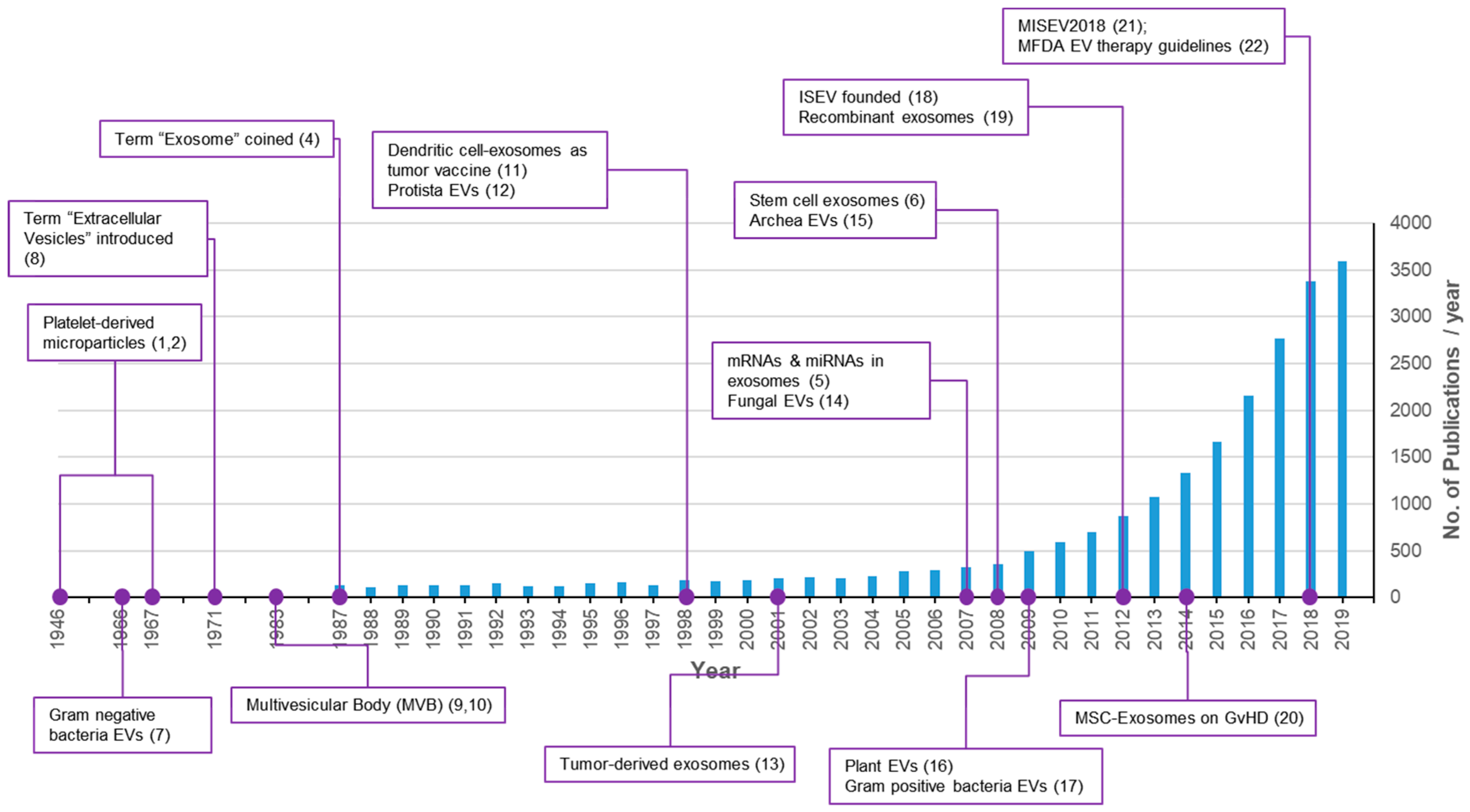
:1. Introduction
2. Exosomes
2.1. Exosomes and Extracellular Vesicles
2.2. Technologies for Isolation of Exosomes
2.3. Quality Control of Exosomes
3. Analysis of Exosomes Biodistribution
3.1. Bioimaging Modalities
3.2. Labeling Methods for Exosomes
3.2.1. Covalent Binding
3.2.2. Surface Modification
3.2.3. Membrane Integration
3.2.4. Encapsulation
3.2.5. Metabolic Labeling
3.3. Analysis of Biodistribution of Exosomes in Literature
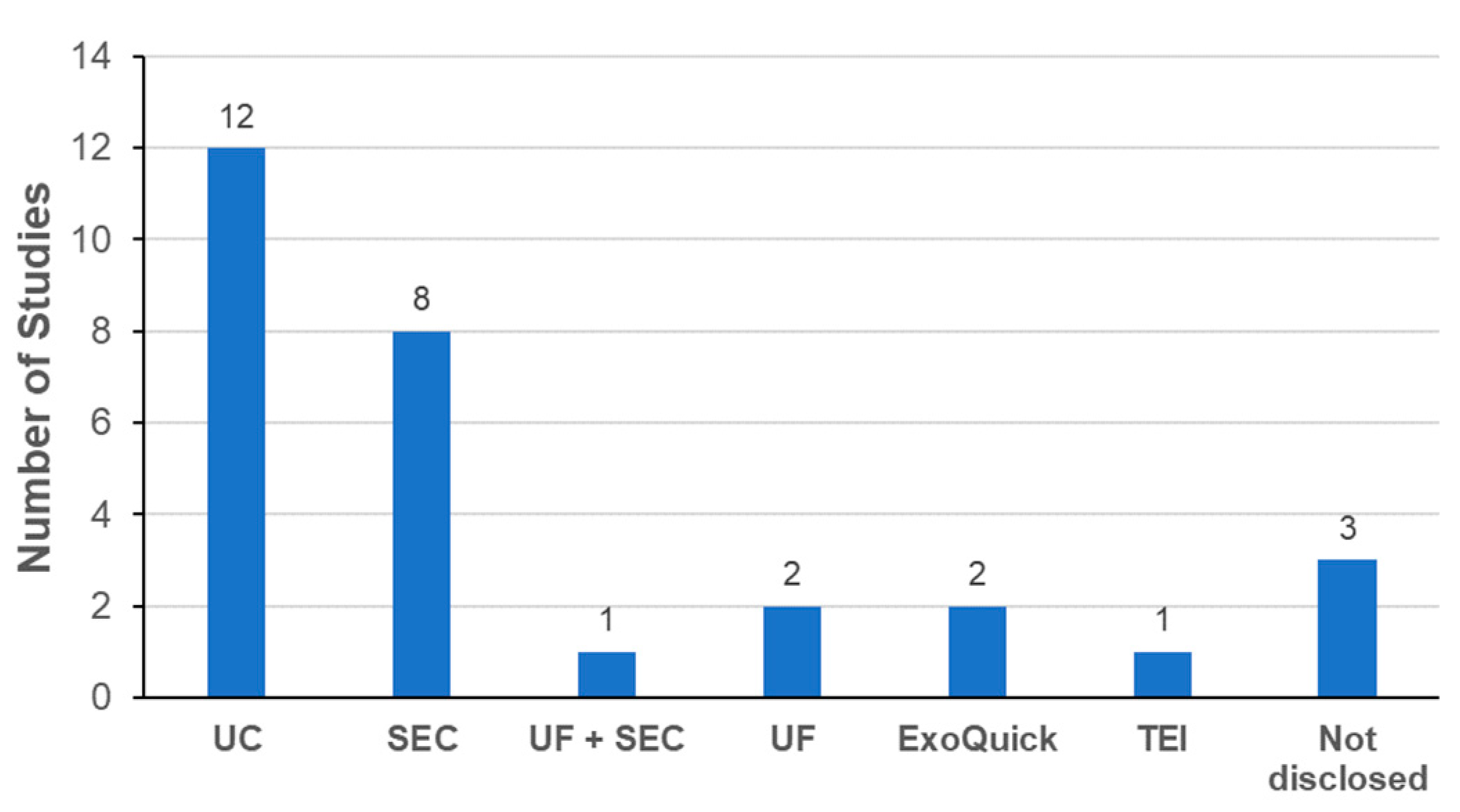
3.3.1. Labeling Methods
3.3.2. Characterization of Exosomes
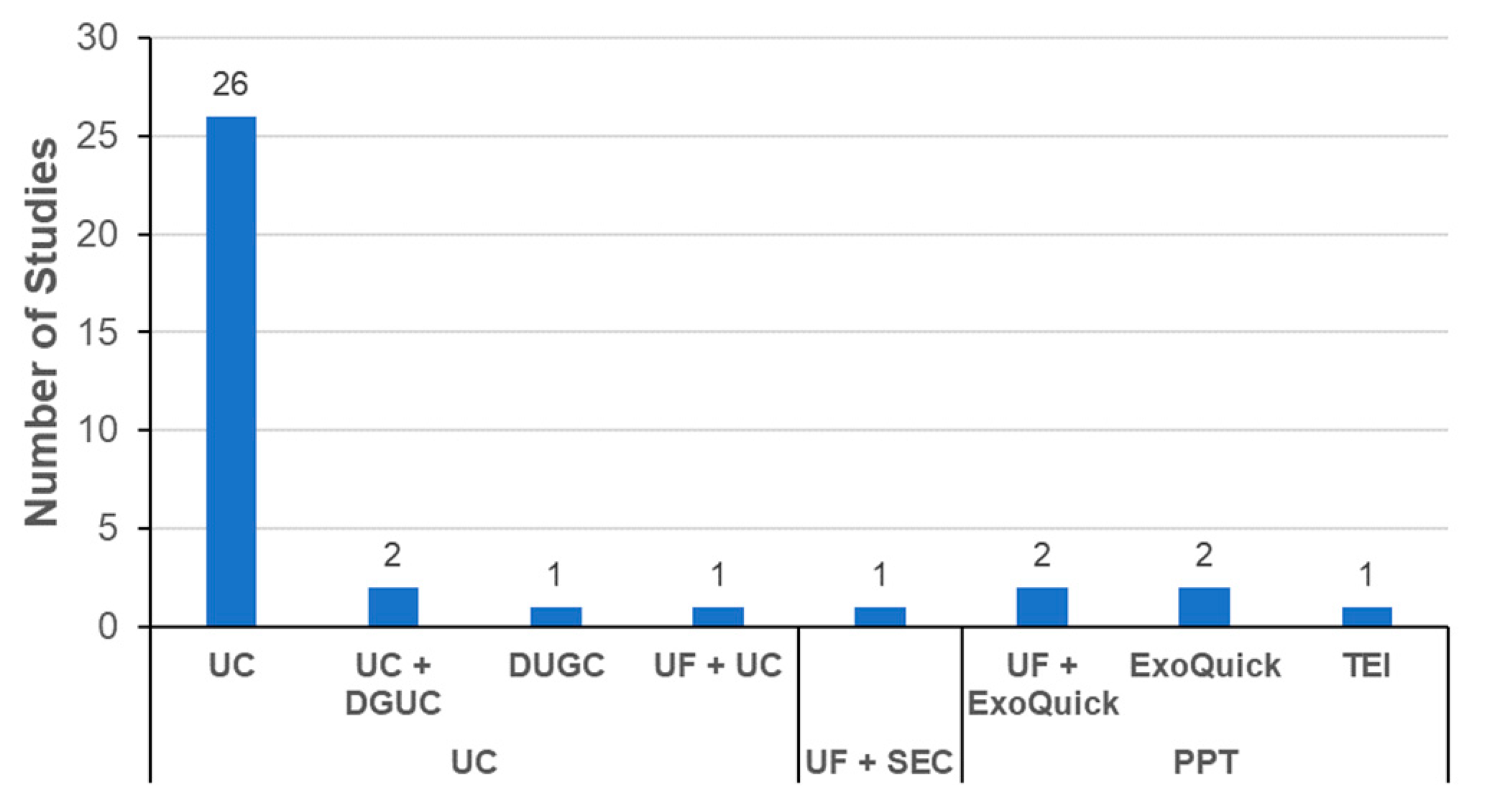
3.3.3. Exosome Isolation Methods
3.3.4. Determination of Exosome Dose
3.3.5. Routes of Administration
3.4. Therapeutic Implication of Exosome Biodistribution
3.4.1. Natural Targeting Properties of Exosomes
3.4.2. Tumor-Homing of Exosomes
3.4.3. Accumulation of MSC-Exosomes in Damaged Tissues
3.4.4. Tissue Targeting by Exosome Engineering
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AKI | acute kidney injury |
| BC | Breast cancer |
| BLI | bioluminescence imaging |
| BMSC | bone marrow stromal cell |
| BW | body weight |
| CAGR | compound annual growth rate |
| CV | cellular vesicle |
| CT | computed tomography |
| DGUC | density-gradient ultracentrifugation |
| DLS | dynamic light scattering |
| EPCs | endothelial progenitor cells |
| EVs | extracellular vesicles |
| FI | fluorescence intensity |
| FP | fluorescence protein |
| GMP | good manufacturing practice |
| GNP | gold nanoparticle |
| GvHD | Graft-versus-host disease |
| ICP-MS | inductively coupled plasma mass spectroscopy |
| IN | intranasal |
| ISEV | International Society for Extracellular Vesicles |
| IV | intravenous |
| IP | intraperitoneal |
| MDSCs | myeloid derived suppressor cells |
| MFDS | Ministry of Food and Drug Safety, Korea |
| MISEV | Minimal Information for Studies of Extracellular Vesicles |
| MHC | Major histocompatibility complex |
| MPI | magnetic particle imaging |
| MRI | magnetic resonance imaging |
| MSCs | mesenchymal stem/stromal cells |
| MVBs | multivesicular bodies |
| NA | not applicable |
| ND | not determined |
| NIR | near infrared |
| NR | nuclear imaging |
| NTA | nanoparticle tracking analysis |
| OMV | outer membrane vesicle |
| PEG | polyethylene glycol |
| PET | position-emission tomography |
| QCs | quality controls |
| RI | radioisotope |
| RLU | relative luminescence unit |
| ROVS | retro-orbital venous sinus |
| RPS | resistive pulse sensing |
| SC | subcutaneous |
| SEC | size exclusion chromatography |
| SPECT | single-photon emission computed tomography |
| SPIO | superparamagnetic iron oxide |
| TEI | total exosome isolation reagent |
| TFF | tangential flow filtration |
| UC | ultracentrifugation |
| UC-MSC | umbilical cord MSC |
| UF | ultrafiltration |
References
- Deatherage, B.L.; Cookson, B.T. Membrane vesicle release in bacterial, eukaryotes, and archeas: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012, 80, 1948–1957. [Google Scholar] [CrossRef] [Green Version]
- Chargaff, E.; West, R. The biological significance of the thromboplatic protein of blood. J. Biol. Chem. 1946, 166, 189–197. [Google Scholar]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Timmers, L.; Lim, S.K.; Arslan, F.; Armstrong, J.S.; Hoefer, I.E.; Doevendans, P.A.; Piek, J.J.; El Oakley, R.M.; Choo, A.; Lee, C.N.; et al. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem Cell Res. 2008, 1, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Knox, K.W.; Vesk, M.; Work, E. Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol. 1966, 92, 1206–1217. [Google Scholar] [CrossRef] [Green Version]
- Aaronson, S.; Behrens, U.; Orner, R.; Haines, T.H. Ultrastructure of intracellular and extracellular vesicles, membranes, and myelin figures produces by Ochromonas Danica. J. Ultrastruct. Res. 1971, 35, 418–430. [Google Scholar] [CrossRef]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef]
- Pan, B.T.; Johnstone, R.M. Fate of the transferring receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell-derived exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef]
- Tatischeff, I.; Bomsel, M.; de Paillerets, C.; Durand, H.; Geny, B.; Segretain, D.; Turpin, E.; Alfsen, A. Dictyostelium discoideum cells shed vesicles with associated DNA and vital stain Hoeschst 33342. Cell. Mol. Life Sci. 1998, 54, 476–487. [Google Scholar] [CrossRef]
- Wolfers, J.; Lozier, A.; Raposo, G.; Rengnault, A.; Thery, C.; Masurier, C.; Flament, C.; Pousieux, S.; Faure, F.; Tursz, T.; et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Frases, S.; Miranda, K.; Zaragoza, O.; Alvarez, M.; Nakouzi, A.; Feldmesser, M.; Casadevall, A. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 2007, 6, 48–59. [Google Scholar] [CrossRef] [Green Version]
- Soler, N.; Marguet, E.; Verbavatz, J.M.; Forterre, P. Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Res. Microbiol. 2008, 159, 390–399. [Google Scholar] [CrossRef]
- Regente, M.; Corti-Monzon, G.; Maldonado, A.M.; Pinedo, M.; Jorrin, J.; de la Canal, L. Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett. 2009, 583, 3363–3366. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.Y.; Choi, D.Y.; Kim, D.K.; Kim, J.W.; Park, J.O.; Kim, S.; Kim, S.H.; Desiderio, D.M.; Kim, Y.K.; Kim, K.P.; et al. Gram-positive bacteria produce membrane vesicles: Proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 2009, 9, 5425–5436. [Google Scholar] [CrossRef]
- Araldi, E.; Kramer-Albers, E.M.; Hoen, E.N.; Peinado, H.; Psonka-Antonczyk, K.M.; Rao, P.; van Niel, G.; Yanez-Mo, M.; Nazarenko, I. International society for extracellular vesicles: First annual meeting, April 17–21, 2012: ISEV-2012. J. Extracell. Vesicles 2012, 1, 19995. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Kordelas, L.; Rebmann, V.; Ludwig, A.K.; Radtke, S.; Ruesing, J.; Doeppner, T.R.; Epple, M.; Horn, P.A.; Beelen, D.W.; Giebel, B. MSC-derived exosomes: A novel tool to treat therapy-refractory graft-versus-host disease. Leukemia 2014, 28, 970–973. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- National Institute of Food and Drug Safety Evaluation. Guideline on Quality, Non-Clinical and Clinical Assessment of Extracellular Vesicles Therapy Products. 2018. Available online: www.nifds.go.kr/brd/m_15/down.do?brd_id=167&seq=12625&data_tp=A&file_seq=1 (accessed on 13 December 2019).
- Samuel, M.; Bleackley, M.; Anderson, M.; Mathivanan, S. Extracellular vesicles including exosomes in cross kingdom regulation: A viewpoint from plant-fungal interactions. Front. Plant Sci. 2015, 6, 766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, K.S.; Bandeira, E.; Shelke, G.V.; Lasser, C.; Lotvall, J. Enhancement of therapeutic potential of mesenchymal stem cell-derived extracellular vesicles. Stem Cell Res. Ther. 2019, 10, 288. [Google Scholar] [CrossRef] [PubMed]
- Stemersch, S.; de Smedt, S.C.; Raemdonck, K. Therapeutic and diagnostic applications of extracellular vesicels. J. Contorol. Release 2016, 244, 167–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiklander, O.P.B.; Brennan, M.A.; Lotvall, J.; Breakefield, X.O.; El Andaloussi, S. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 2019, 11, eaav8521. [Google Scholar] [CrossRef]
- Kanada, M.; Bachmann, M.H.; Contag, C.H. Signaling by extracellular vesicles advances cancer hallmarks. Trends Cancer 2016, 2, 84–94. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Cunnane, E.M.; Weinbaum, J.S.; O’Brien, F.J.; Dorp, D.A. Future perspective on the role of stem cells and extracellular vesicles in vascular tissue regeneration. Front. Cardiovasc. Med. 2018, 5, 86. [Google Scholar] [CrossRef] [Green Version]
- Koniusz, S.; Andrzejewska, A.; Muraca, M.; Srivastava, A.K.; Janowski, M.; Lukomska, B. Extracellular vesicles in physiology, pathology, and therapy of the immune and central nervous system, with focus on extracellular vesicles derived from mesenchymal stem cells as therapeutic tools. Front. Cell. Neurosci. 2016, 10, 109. [Google Scholar] [CrossRef]
- Corso, G.; Mager, I.; Lee, Y.; Gorgens, A.; Bultema, J.; Giebel, B.; Wood, M.J.A.; Nordin, J.Z.; El Andaloussi, S. Reproducible and scalable purification of extracellular vesicles using combined bind-elute and size exclusion chromatography. Sci. Rep. 2017, 7, 11561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Yeo, R.W.Y.; Lai, R.C.; Sim, E.W.K.; Chin, K.C.; Lim, S.K. Mesenchymal stromal cell exosome-enhanced regulatory T-cell production through an antigen-presenting cell-mediated pathway. Cytotherapy 2018, 20, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.O.; Ha, D.H.; Kim, J.O.; Crumrine, D.A.; Meyer, J.M.; Kim, H.K.; Lee, J.; Kwon, H.H.; Park, G.H.; Lee, J.H.; et al. Exosomes from human adipose tissue-derived mesenchymal stem cells promote epidermal barrier repair by inducing de novo synthesis of ceramides in atopic dermatitis. Cells. under review.
- Wiklander, O.P.B.; Bostancioglue, R.B.; Welsh, J.A.; Zickler, A.M.; Murke, F.; Corso, G.; Felldin, U.; Hagery, D.W.; Evertsson, B.; Liang, X.M.; et al. Systemic methodological evaluation of a multiplex bead-based flow cytometry assay for detection of extracellular vesicle surface signatures. Front. Immunol. 2018, 9, 1326. [Google Scholar] [CrossRef] [Green Version]
- Ren, K. Exosomes in perspective: A potential surrogate for stem cell therapy. Odontology 2019, 107, 271–284. [Google Scholar] [CrossRef]
- Yin, K.; Wang, S.; Zhao, R.C. Exosomes from mesenchymal stem/stromal cells: A new therapeutic paradigm. Biomark. Res. 2019, 7, 8. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.; Ma, R.; Cai, W.; Huang, W.; Paul, C.; Liang, J.; Wang, Y.; Zhao, Y.; Kim, H.W.; Xu, M.; et al. Exosomes secreted from CXCR4 overexpressing mesenchymal stem cells promote Cardioprotection via Akt signaling pathway following myocardial infarction. Stem Cells Int. 2015, 2015, 659890. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.Y.; Lai, R.C.; Wong, W.; Dan, Y.Y.; Lim, S.K.; Ho, K.K. Mesenchymal stem cell-derived exosomes promote hepatic regeneration in drug-induced liver injury models. Stem Cell Res. Ther. 2014, 5, 76. [Google Scholar] [CrossRef] [Green Version]
- Grange, C.; Iampietro, C.; Bussolati, B. Stem cell extracellular vesicles and kidney injury. Stem Cell Investig. 2017, 4, 90. [Google Scholar] [CrossRef] [Green Version]
- Cho, B.S.; Kim, J.O.; Ha, D.H.; Yi, Y.W. Exosomes derived from human adipose tissue-derived mesenchymal stem cells alleviate atopic dermatitis. Stem Cell Res. Ther. 2018, 9, 187. [Google Scholar] [CrossRef] [Green Version]
- Boltze, J.; Arnold, A.; Walczak, P.; Jolkkonen, J.; Cui, L.; Wagner, D.C. The dark side of the force—Constraints and complications of cell therapies for stroke. Front. Neurol. 2015, 6, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, G.; Chen, Z.; Zheng, M.; Liu, Y. Mesenchymal stem cell-derived exosomes as a new therapeutic strategy for liver diseases. Exp. Mol. Med. 2017, 49, e346. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.H.; Kim, S.D.; Cho, B.S.; Lee, J.; Lee, J.H.; Park, S.R.; Youn, J.; Lee, S.H.; Kim, J.E.; Lim, J.; et al. Toxicological evaluation of exosomes derived from human adipose tissue-derived mesenchymal stem/stromal cells. Regul. Toxicol. Pharm. under review.
- Mendt, M.; Kamerakar, S.; Sugimoto, H.; McAndrews, K.M.; Wu, C.C.; Gagea, M.; Yang, S.; Blanko, E.V.R.; Peng, Q.; Ma, X.; et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight 2018, 3, e99263. [Google Scholar] [CrossRef]
- Zhu, X.; Badawi, M.; Pomeroy, S.; Sutaria, D.S.; Xie, Z.; Baek, A.; Jiang, J.; Elgamal, O.A.; Mo, X.; La Perle, K.; et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicels derived from HEK293T cells. J. Extracell. Vesicles 2017, 6, 1324730. [Google Scholar] [CrossRef]
- Saleh, A.F.; Lazaro-Ibanez, E.; Forsgard, M.A.; Shatnyeva, O.; Osteikoetxea, X.; Karlsson, F.; Heath, N.; Ingelsten, M.; Rose, J.; Harris, J.; et al. Extracellular vesicles induce minimal hepatotoxicity and immunogenicity. Nanoscale 2019, 11, 6990–7001. [Google Scholar] [CrossRef]
- Maji, S.; Yan, I.K.; Parasramka, M.; Mohankumar, S.; Matsuda, A.; Patel, T. In vitro toxicology studies of extracellular vesicles. J. Appl. Toxicol. 2017, 37, 310–318. [Google Scholar] [CrossRef]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Moller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Gimona, M.; Pachler, K.; Laner-Plamberger, S.; Schallmoser, K.; Rohde, E. Manufacturing of human extracellular vesicle-based therapeutics for clinical use. Int. J. Mol. Sci. 2017, 18, 1190. [Google Scholar] [CrossRef]
- Reiner, A.; Witwer, K.W.; van Balkom, B.W.M.; de Beer, J.; Brondie, C.; Corteling, R.L.; Gabrielsson, S.; Gimona, M.; Ibrahim, A.G.; de Kleijn, D.; et al. Concise review: Developing best-practice models for the therapeutic use of extracellular vesicles. Stem Cells Transl. Med. 2017, 6, 1730–1739. [Google Scholar] [CrossRef] [Green Version]
- Cheyuo, C.; Aziz, M.; Wang, P. Neurogenesis in neurodegenerative diseases: Role of MFG-E8. Front. Neurosci. 2019, 13, 569. [Google Scholar] [CrossRef] [PubMed]
- Shelke, G.V.; Yin, Y.; Jang, S.C.; Lasser, C.; Wennmalm, S.; Hoffmann, H.J.; Li, L.; Gho, Y.S.; Nilsson, J.A.; Lotvall, J. Endosomal signalling via exosome surface TGFβ-1. J. Extracell. Vesicles 2019, 8, 1650458. [Google Scholar] [CrossRef] [Green Version]
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Soekmadji, C.; Hill, A.F.; Wauben, M.H.; Buzas, E.I.; Di Vizio, D.; Falcon-Perez, J.M.; Gardiner, C.; Hochberg, F.; Kurochkin, I.V.; et al. Updating the minimal requirements for extracellular vesicle studies: Building bridges to reproducibility. J. Extracell. Vesicles 2017, 6, 1396823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andriolo, G.; Provasi, E.; Lo Cicero, V.; Brambilla, A.; Soncin, S.; Torre, T.; Milano, G.; Biemmi, V.; Vassalli, G.; Turchetto, L.; et al. Exosomes from human cardiac progenitor cells for therapeutic applications: Development of a GMP-grade manufacturing method. Front. Physiol. 2018, 9, 1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bari, E.; Perteghella, S.; Di Silvestre, D.; Sorlini, M.; Catenacci, L.; Sorrenti, M.; Marrubini, G.; Rossi, R.; Tripodo, G.; Mauri, P.; et al. Pilot production of mesenchymal stem/stromal freeze-dried secretome for cell-free regenerative nanomedicine: A validated GMP-compliant process. Cells 2018, 7, 190. [Google Scholar] [CrossRef] [Green Version]
- Bari, E.; Perteghella, S.; Catenacci, L.; Sorlini, M.; Croce, S.; Mantelli, M.; Avanzini, M.A.; Sorrenti, M.; Torre, M.L. Freeze-dried and GMP-compliant pharmaceuticals containing exosomes for acellular mesenchymal stromal cell immunomodulant therapy. Nanomedicine 2019, 14, 753–765. [Google Scholar] [CrossRef]
- Navabi, H.; Croston, D.; Hobot, J.; Clayton, A.; Zitvogel, L.; Jasani, B.; Bailey-Wood, R.; Wilson, K.; Tabi, Z.; Mason, M.D.; et al. Preparation of human ovarian cancer ascites-derived exosomes for a clinical trial. Blood Cells Mol. Dis. 2005, 35, 149–152. [Google Scholar] [CrossRef]
- Pachler, K.; Lener, T.; Streif, D.; Dunai, Z.A.; Desgeorges, A.; Feichtner, M.; Oller, M.; Schallmoser, K.; Rohde, E.; Gimona, M. A good manufacturing practice-grade standard protocol for exclusively human mesenchymal stromal cell-derived extracellular vesicles. Cytotherapy 2017, 19, 458–472. [Google Scholar] [CrossRef] [Green Version]
- Rohde, E.; Pachler, K.; Gimona, M. Manufacturing and characterization of extracellular vesicles from umbilical cord-derived mesenchymal stromal cells for clinical testing. Cytotherapy 2019, 21, 581–592. [Google Scholar] [CrossRef]
- Roy, S.; Hochberg, F.H.; Jones, P.S. Extracellular vesicles: The growth as diagnostics and therapeutics; a survey. J. Extracell. Vesicles 2018, 7, 1438720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassidy, P.J.; Radda, G.K. Molecular imaging perspectives. J. R. Soc. Interface 2005, 2, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youn, H.; Hong, K.J. In vivo non invasive molecular imaging for immune cell tracking in small animals. Immune Netw. 2012, 12, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Gangadaran, P.; Hong, C.M.; Oh, J.M.; Rajendran, R.L.; Kalimuthu, S.; Son, S.H.; Gopal, A.; Zhu, L.; Baek, S.H.; Jeong, S.Y.; et al. In vivo non-invasive imaging of radio-labeled exosome-mimetics derived from red blood cells in mice. Front. Pharmacol. 2018, 9, 817. [Google Scholar] [CrossRef] [Green Version]
- Royo, F.; Cossio, U.; de Angulo, A.R.; Llop, J.; Falcon-Perez, J.M. Modification of the glycosylation of extracellular vesicles alters their biodistribution in mice. Nanoscale 2019, 11, 1531. [Google Scholar] [CrossRef] [Green Version]
- Gangadaran, P.; Li, X.J.; Lee, H.W.; Oh, J.M.; Kalimuthu, S.; Rajendran, R.L.; Son, S.H.; Baek, S.H.; Signh, T.D.; Zhu, L.; et al. A new bioluminescent reporter system to study the biodistribution of systematically injected tumor-derived bioluminescent extracellular vesicles in mice. Oncotarget 2017, 8, 109894–109914. [Google Scholar] [CrossRef] [Green Version]
- Wiklander, O.P.B.; Nordin, J.Z.; O’Loughlin, A.; Gustafsson, Y.; Corso, G.; Mäger, I.; Vader, P.; Lee, Y.; Sork, H.; Seow, Y.; et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extcell. Vesicles 2015, 4, 26316. [Google Scholar] [CrossRef] [Green Version]
- Corso, G.; Heusermann, W.; Trojer, D.; Gorgens, A.; Steib, E.; Voshol, J.; Graff, A.; Genoud, C.; Lee, Y.; Hean, J.; et al. Systemic characterization of extracellular vesicle sorting domains and quantification at the single molecule—Single vesicle level by fluorescent correlation spectroscopy and single particle imaging. J. Extracell. Vesicles 2019, 8, 1663043. [Google Scholar] [CrossRef]
- Nagyova, M.; Slovinska, L.; Blasko, J.; Grulova, I.; Kuricova, M.; Cigankova, V.; Harvanova, D.; Cizkova, D. A comparative study of PKH67, Dil, and BrdU labeling techniques for tracing rat mesenchymal stem cells. In Vitro Cell. Dev. Biol. Anim. 2014, 50, 656–663. [Google Scholar] [CrossRef]
- Horan, P.K.; Slezak, S.E. Stable cell membrane labeling. Nature 1989, 340, 167–168. [Google Scholar] [CrossRef]
- Kuffler, D.P. Long-term survival and sprouting in culture by motoneurons isolated from the spinal cord of adult frogs. J. Comp. Neurol. 1990, 302, 72–738. [Google Scholar] [CrossRef]
- Morales-Kastresana, A.; Telford, B.; Musich, T.A.; McKinnon, K.; Clayborne, C.; Braig, Z.; Rosner, A.; Demberg, T.; Watson, D.C.; Karpova, T.S.; et al. Labeling extracellular vesicles for nanoscale flow cytometry. Sci. Rep. 2017, 7, 1878. [Google Scholar] [CrossRef]
- Lee, T.S.; Kim, Y.; Zhang, W.; Song, I.H.; Tung, C.H. Facile metabolic glycan labeling strategy for exosome tracking. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.O.; Jo, H.; Yu, J.H.; Gambhir, S.S.; Pratx, G. Development and MPI tracking of novel hypoxia-targeted theranostic exosomes. Biomaterials 2018, 177, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Park, H.; Noh, G.J.; Lee, E.S. pH-responsive hyaluronate-anchored extracellular vesicles to promote tumor-targeted drug delivery. Carbohydr. Polym. 2018, 202, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Goh, W.J.; Zou, S.; Ong, W.; Torta, F.; Alexandra, A.F.; Schiffelers, R.M.; Storm, G.; Wang, J.W.; Czarny, B.; Pastorin, G. Bioinspired cell-derived nanovesicles versus exosomes as drug delivery systems: A cost-effective alternative. Sci. Rep. 2017, 7, 14322. [Google Scholar] [CrossRef]
- Choi, H.I.; Choi, J.P.; Seo, J.; Kim, B.J.; Rho, M.; Han, J.K.; Kim, J.G. Helicobacter pylori-derived extracellular vesicles increased in the gastric juices of gastric adenocarcinoma patients and induced inflammation mainly via specific targeting of gastric epithelial cells. Exp. Mol. Med. 2017, 49, e330. [Google Scholar] [CrossRef]
- Jang, S.C.; Kim, S.R.; Yoon, Y.J.; Park, K.S.; Kim, J.H.; Lee, J.; Kim, O.Y.; Choi, E.J.; Kim, D.K.; Choi, D.S.; et al. In vivo kinetic biodistribution of nano-sized outer membrane vesicles derived from bacteria. Small 2015, 11, 456–461. [Google Scholar] [CrossRef]
- Faruqu, F.N.; Wang, J.T.W.; Xu, L.; McNickle, L.; Chong, E.M.Y.; Walters, A.; Gurney, M.; Clayton, A.; Smyth, L.A.; Hider, R.; et al. Membrane radiolabeling of exosomes for comparative biodistribution analysis in immunocompetent and immunodeficient mice—A novel and universal approach. Theranostics 2019, 9, 1666–1682. [Google Scholar] [CrossRef]
- Rashid, M.H.; Borin, T.F.; Ara, R.; Angara, K.; Cai, J.; Achyut, B.R.; Liu, Y.; Arbab, A.S. Differential in vivo biodistribution of 131I-labeled exosomes from diverse cellular origins and its implication for theranostic application. Nanomedicine 2019, 21, 102072. [Google Scholar] [CrossRef]
- Varga, Z.; Gyurkó, I.; Pálóczi, K.; Buzás, E.I.; Horváth, I.; Hegedűs, N.; Máthé, D.; Szigeti, K. Radiolableling of extracellular vesicles with 99mTc for quantitative in vivo imaging studies. Cancer Biother. Radiopharm. 2016, 31, 168–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hikita, T.; Miyata, M.; Watanabe, R.; Oneyama, C. Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Sci. Rep. 2018, 8, 14035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, C.P.; Mardini, O.; Ericsson, M.; Prabhakar, S.; Maguire, C.A.; Chen, J.W.; Tannous, B.A.; Breakefield, X.O. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano 2014, 8, 483–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, Y.; Nishikawa, M.; Shinotsuka, H.; Matsui, Y.; Ohara, S.; Imai, T.; Takakura, Y. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 in mice after intravenous injection. J. Biotechnol. 2013, 165, 77–84. [Google Scholar] [CrossRef]
- Abello, J.; Nguyen, T.D.T.; Marasini, R.; Aryal, S.; Weiss, M.L. Biodistribution of gadolinium- and near infrared-labeled human umbilical cord mesenchymal stromal cell-derived exosomes in tumor bearing mice. Theranostics 2019, 9, 2325–2345. [Google Scholar] [CrossRef]
- Banfai, K.; Garai, K.; Ernszt, D.; Pongracz, J.E.; Kvell, K. Transgenic exosomes for thymus regeneration. Front. Immunol. 2019, 10, 862. [Google Scholar] [CrossRef] [PubMed]
- Marazioti, A.; Papadia, K.; Kannavou, M.; Spella, M.; Basta, A.; de Lastic, A.L.; Mouzaki, R.A.; Samiotaki, M.; Panayotou, G.; Stathopoulos, G.T.; et al. Cellular vesicles: New insights in engineering methods, interaction with cells and potential for brain targeting. J. Pharmacol. Exp. Ther. 2019, 370, 772–785. [Google Scholar] [CrossRef]
- Gao, X.; Ran, N.; Dong, X.; Zuo, B.; Yang, R.; Zhuo, Q.; Moulton, H.M.; Seow, Y.; Yin, H. Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci. Transl. Med. 2018, 10, eaat0195. [Google Scholar] [CrossRef] [Green Version]
- Naseri, Z.; Oskuee, R.K.; Jaafari, M.R.; Moghadam, M.F. Exosome-mediated delivery of functionally active miRNA-142-3p inhibitor reduces tumorigenicity of breast cancer in vitro and in vivo. Int. J. Nanomed. 2018, 13, 7727–7747. [Google Scholar] [CrossRef] [Green Version]
- Viñas, J.L.; Spence, M.; Gutsol, A.; Knoll, W.; Burger, D.; Zimplelmann, J.; Allan, D.S.; Burns, K.D. Receptor-ligand interaction mediates targeting of endothelial colony forming cell-derived exosomes to the kidney after ischemic injury. Sci. Rep. 2018, 8, 16320. [Google Scholar] [CrossRef]
- Wen, S.W.; Sceneay, J.; Lima, L.G.; Wong, C.S.F.; Becker, M.; Krumeich, S.; Lobb, R.J.; Castillo, V.; Wong, K.N.; Ellis, S.; et al. The biodistribution and immune suppressive effects of breast cancer-derived exosomes. Cancer Res. 2016, 76, 6816–6827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordin, J.Z.; Lee, Y.; Vader, P.; Mäger, I.; Johansson, H.J.; Heusermann, W.; Wiklander, O.P.B.; Hällbrink, M.; Seow, Y.; Bultema, J.J.; et al. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine 2015, 11, 879–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Hendrix, A.; Hernot, S.; Lemarire, M.; Bruyne, E.D.; Valckenborgh, E.V.; Lahoutte, T.; Wever, O.D.; Vanderkerken, K.; Menu, E. Bone marrow stromal cell-derived exosomes as communicators in drug resistance in multiple myeloma cells. Blood 2014, 124, 555–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grange, C.; Tapparo, M.; Bruno, S.; Chatterjee, D.; Quesenberry, P.J.; Tetta, C.; Camussi, G. Biodistribution of mesenchymal stem cell-derived extracellular vesicles in a model of acute kidney injury monitored by optical imaging. Int. J. Mol. Med. 2014, 33, 1055–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betzer, O.; Perets, N.; Angel, A.; Motiei, M.; Sadan, T.; Yadid, G.; Offen, D.; Popovtzer, R. In vivo neuroimaging of exosomes using gold nanoparticles. ASC Nano 2017, 11, 10883–10893. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.W.; Choi, H.; Jang, S.C.; Yoo, M.Y.; Park, J.Y.; Choi, N.A.; Oh, H.J.; Ha, S.; Lee, Y.S.; Jeong, J.M.; et al. Noninvasive imaging of radiolabeled exosome-mimetic nanovesicle using 99mTc-HMPAO. Sci. Rep. 2015, 5, 15636. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bloch, S.; Akers, W.; Achilefu, S. Near-infrared molecular probes for in vivo imaging. Curr. Protoc. Cytom. 2012, 60, 12–27. [Google Scholar]
- Boddington, S.; Henning, T.D.; Sutton, E.J.; Daldrup-Link, H.E. Labeling stem cells with fluorescent dyes for non-invasive detection with optical imaging. J. Vis. Exp. 2008, 2, 686. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Jung, H.; Mok, H. Idocayanine green-incorporated exosomes for improved in vivo imaging of sentinel lymph node. Appl. Biol. Chem. 2016, 59, 71–76. [Google Scholar] [CrossRef]
- Verweij, F.J.; Hyenne, V.; Van Niel, G.; Goetz, J.G. Extracellular vesicles: Catching the light in zebrafish. Trands Cell. Biol. 2019, 29, 770–776. [Google Scholar] [CrossRef]
- Murphy, D.E.; de Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Exp. Mol. Ther. 2019, 51, 32. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumor exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Wrana, J.L. The emerging role of exosomes in Wnt secretion and transport. Curr. Opin. Genet. Dev. 2014, 27, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, S.; Teixeira, V.H.; Kalber, T.; Jose, R.J.; Floto, R.A.; Janes, S.M. Macrophage migration inhibitory factor-CXCR4 is the dominant chemotaxic axis in human mesenchymal stem cell recruitment to tumors. J. Immunol. 2015, 194, 3463–3474. [Google Scholar] [CrossRef] [Green Version]
- Garofalo, M.; Villa, A.; Crescenti, D.; Marzagalli, M.; Kuryk, L.; Limonta, P.; Mazzaferro, V.; Ciana, P. Heterologous and cross-species tropism of cancer-derived extracellular vesicles. Theranostics 2019, 9, 5681–5693. [Google Scholar] [CrossRef]
- Yun, C.W.; Lee, S.H. Potential and therapeutic efficacy of cell-based therapy using mesenchymal stem cells for acute/chronic kidney disease. Int. J. Mol. Sci. 2019, 20, 1619. [Google Scholar] [CrossRef] [Green Version]
- Lv, L.L.; Wu, W.J.; Feng, Y.; Li, Z.L.; Tang, T.T.; Liu, B.C. Therapeutic application of extracellular vesicles in kidney disease: Promises and challenges. J. Cell. Mol. Med. 2018, 22, 728–737. [Google Scholar] [CrossRef]
- Herrera, M.B.; Bussolati, B.; Bruno, S.; Morando, L.; Mauriello-Romanazzi, G.; Sanavio, F.; Stamenkovic, I.; Biancore, L.; Camussi, G. Exogenous mesenchymal stem cells localize to the kidney by means of CD44 following acute tubular injury. Kidney Int. 2007, 72, 430–441. [Google Scholar] [CrossRef] [Green Version]
- Ullar, M.; Liu, D.D.; Thakor, A.S. Mesenchymal stromal cell homing: Mechanisms and strategies for improvement. iScience 2019, 15, 421–438. [Google Scholar]
- Antes, T.J.; Middleton, R.C.; Luther, K.M.; Ijichi, T.; Peck, K.A.; Liu, W.J.; Valle, J.; Echavez, A.K.; Marban, E. Targeting extracellular vesicles to injured tissue using membrane cloaking and surface display. J. Nanotechnol. 2018, 16, 61. [Google Scholar] [CrossRef]
- Tian, T.; Zhang, H.X.; He, C.P.; Fan, S.; Zhu, Y.L.; Qi, C.; Huang, N.P.; Xiao, Z.D.; Lu, Z.H.; Tannous, B.A.; et al. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials 2018, 150, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Kooijmans, S.A.; Aleza, C.G.; Roffler, S.R.; van Solinge, W.W.; Vader, P.; Schiffelers, R.M. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumor cell targeting. J. Extracell. Vesicles 2016, 5, 31053. [Google Scholar] [CrossRef] [PubMed]
- Fais, S.; O’Driscoll, L.; Borras, F.E.; Buzas, E.; Camussi, G.; Cappello, F.; Carvalho, J.; Cordeiro da Silva, A.; Del Portillo, H.; El Andaloussi, S.; et al. Evidence-based clinical use of nanoscale extracellular vesicles in nanomedicine. ASC Nano 2016, 10, 3886–3899. [Google Scholar] [CrossRef] [PubMed]
- Fais, S.; Logozzi, M.; Lugini, L.; Federici, C.; Azzarito, T.; Zarovni, N.; Chiesi, A. Exosomes: The ideal nanovectors for biodelivery. Biol. Chem. 2013, 394, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Villa, A.; Rizzi, N.; Kuryk, L.; Rinner, B.; Cerullo, V.; Yliperttula, M.; Mazzaferro, V.; Ciana, P. Extracellular vesicles enhance the targeted delivery of immunogenic oncolytic adenovirus and paclitaxel in immunocompetent mice. J. Control. Release 2019, 294, 1650175. [Google Scholar] [CrossRef]
- Schindler, C.; Collinson, A.; Mattews, C.; Pointon, A.; Jenkinson, L.; Minter, R.R.; Vaughan, T.; Tigue, N.J. Exosomal delivery of doxorubicin enables rapid cell entry and enhanced in vitro potency. PLoS ONE 2019, 14, e021454. [Google Scholar] [CrossRef] [Green Version]
- Ran, L.; Tan, X.; Li, Y.; Zhang, H.; Ma, R.; Ji, T.; Dong, W.; Tong, T.; Liu, Y.; Chen, D.; et al. Delivery of oncolytic adenovirus into the nucleus of tumorigenic cells by tumor microparticles for virotherapy. Biomaterials 2016, 89, 55–66. [Google Scholar] [CrossRef]
- Garofalo, M.; Villa, A.; Rizzi, N.; Kyryk, L.; Mazzaferro, V.; Ciana, P. Systemic administration and targeted delivery of immunogenic oncolytic adenovirus encapsulated in extracellular vesicles for cancer therapies. Viruses 2018, 10, 558. [Google Scholar] [CrossRef] [Green Version]





| QC Criteria | MISEV2018 Recommendation | MFDS Guideline (2018) | Examples |
|---|---|---|---|
| Exosome Number (or quantification) | Global quantification by at least two methods: protein amount, particle number, lipid amount, etc. | Number of vesicles (or particles) and total protein amount or others | Nanoparticle tracking analysis (NTA) Protein quantification |
| Exosome Size | RPS, NTA, DLS, etc. | NTA, DLS, RPS, fluorescence correlation spectroscopy, etc. | NTA |
| Identity | Protein markers; Phospholipids | At least semi-quantitative method to detect proteins, RNAs, or lipids enriched in exosome | Western blot: CD9, CD63, CD81, ALIX, TSG101 FCM: CD9, CD63, CD81, and more ELISA |
| Purity | Ratios of two quantification figures (e.g., protein:particle) Assessment of absence of expected contamination | For proteins which are not expected to enrich in exosomes; For process impurities: serum albumin, antibiotics, etc. | ELISA for Calnexin or GM130 ELISA for impurities |
| Potency Assays | Dose-response assessment | Biological assay which can represent MoA | Various methods: immune-modulation, proliferation, collagen, etc. |
| Others | not mentioned | Mycoplasma, Sterility, Endotoxin, and Virus tests |
| Modality | Examples | Pros | Cons |
|---|---|---|---|
| Bioluminescence Imaging [63,64] | Luciferase | Highest sensitivity (10−15−10−17 mole/L) Medium cost High signal-to-noise (compared to fluorescence) | Substrate needed Medium penetration (mm−cm) Low spatial resolution (mm) Low temporal resolution (sec−min) |
| Nuclear Imaging (PET/SPECT) [63,64,65] | 99 mTc | Highest penetration (m) High sensitivity (10−10−10−12 mole/L) Medium temporal resolution (10 s−min) | Hazardous Low spatial resolution (mm) High cost |
| NIR Fluorescence Imaging [63,64] | DiR | Medium penetration (mm−cm) Medium sensitivity (10−9−10−12 mole/L) Low cost | Low spatial resolution (mm) Low temporal resolution (s−min) |
| Fluorescent Protein Imaging [63,64] | GFP | Highest spatial resolution (nm) Medium sensitivity | Lowest penetration (mm): does not allow noninvasive in vivo imaging |
| Magnetic Resonance Imaging (MRI) [63,64] | SPIO | Highest penetration (m) High spatial resolution (μm) Highest temporal resolution (min−h) | Lowest sensitivity (10−3−10−5 mole/L) High cost |
| Labeling Methods | Pros | Cons | Reference |
|---|---|---|---|
| Covalent biding | Tight binding of probes to proteins | Cannot distinguish between exosomes vs. non-exosome proteins May change membrane protein functions which affect the interaction of exosomes with target cells | [66] |
| Genetic modification | Can avoid surface modification | Genetic change of cells may change the property of cells and/or exosomes Uneven loading into exosomes | [68] |
| Membrane integration (lipophilic fluorescent dyes) | Simple and easy | May cause clumping of exosomes Cannot distinguish between lipid proteins and micelle May cause background signals from dissociated probes May cause pseudo signals even after clearance of exosomes May affect the interaction of exosomes with target cells | [65] |
| Encapsulation by electroporation | May avoid surface modification | May cause aggregation or fusion of exosomes | [65] |
| Encapsulation by lipophilic agents | Simple and easy | May cause background signals from released probes | [65] |
| Transporter-dependent encapsulation | Simple and easy | Depends on transporter (e.g., GLUT1) Un-even encapsulation May cause background signals from released probes | [75] |
| Metabolic labeling | Covalent biding of probes by click chemistry | May change the property of cells and/or exosomes May change membrane protein functions which affect the interaction of exosomes with target cells | [74,76] |
| Labeling Method | Modality | Nomenclature (Markers) | Cell Source | Isolation Method | Purification after Labeling | Dose (/Head) | Animal Model | Admin. Route | Imaging Method | Tissue Distribution | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Covalent binding | RI (124I) | EVs | MLP29 (murine liver-derived progenitor cell line) | UC (100,000 g, 70 min) | SEC (Sephadex G-25) | 0.6–1.8 MBq (40–120 ng) | Mouse (BALB/cJRj) | IV hock | PET | Bladder > liver > thyroid > lung > kidney > brain | [66] |
| Covalent binding | Fluorescence (Cy7-NHS) | Exosomes (CD9, ALIX, TSG101) | Human U937 leukemia cells | UC (100,000 g, 2 h) | SEC (Sephadex G50) | 40 μg | Mouse (BALB/c) with syngeneic CT26 colon adenocarcinoma | IV | IVIS | Liver > kidney, tumor, spleen, heart, lung, colon, brain, bladder, blood | [77] |
| Covalent binding | Fluorescence (Cy7-NHS) | EVs | Helicobacter pylori | UC (150,000 g, 3 h) and DGUC (100,000 g, 2 h) | Not disclosed | Not disclosed | Mouse (C57BL/6) | Oral | IVIS | Mouse, stomach | [78] |
| Covalent binding | Fluorescence (Cy7-NHS) | Bacterial EVs (OMVs) | E. coli | UC (150,000 g, 3 h) | UC (150,000 g, 3 h) | 15 μg | Mouse (C57BL/6 and SKH1-E) | IP | IVIS | (3 h) liver > kidney > lung > spleen > small intestine (24 h) liver | [79] |
| Covalent binding | RI (111Indium) | Exosomes (CD81, CD9) | Murine B16F10 melanoma | UC (100,000 g, 90 min) | SEC (Sepharose CL-2B) | 1 × 1011 | Mouse (C57BL/6 and NSG), melanoma-bearing | IV | SPECT/CT | Liver > spleen > bone, kidney, lung | [80] |
| Covalent binding | RI (131I) | Exosomes (CD9, CD63) | Mouse MDSCs and EPCs, HEK293 | UF (100 kDa) and UC (100,000 g, 70 min) | UF (100 kDa) | 350 ± 50 μCi | Mouse (BALB/c or C57BL/J6) Xenograft bearing 4T1 or AT3 | IV | SPECT/CT | (Tumor exosomes) tumor > liver > lung, spleen, kidney, brain, heart (MDSC-exosome) liver, lung, tumor > kidney, spleen, brain, heart (EPC-exosomes) tumor > liver > lung, kidney, brain, spleen, heart | [81] |
| Covalent binding | RI [99mTc(CO)3(−H2O)3]+ | EVs | Erythrocyte | UC (130,000 g, 30 min) and SEC | SEC (Desalting Column) | 15 ± 2 Mbq | Mouse (BALB/c) | IV | SPECT/CT | Liver, bladder, spleen > kidney > lung, heart, bone | [82] |
| Metabolic labeling | Fluorescence (Cy3 or Cy5.5) | Exosomes (CD63) | Human MDA-MD-231 and MCF7 breast cancer cells | ExoQuick | Gel filtration (G-25) | 10 μg | Mouse, athymic MDA-MB-231 or MCF7 tumor bearing | IV | IVIS | (MCF7 exosomes) liver > large and small intestines > kidney, tumor, spleen, lung, muscle, blood (MDA-MD-231 exosomes) liver > large and small intestines > lung > tumor, spleen, kidney > muscle, blood | [74] |
| Genetic Engineering | Luminescence (CD63-NanoLuc) | Exosomes (CD63) | HT29/CD63Nluc and HCD116/CD63Nluc | UC (110,000 g, 70 min) | NA | NA | Female mouse (Balb/c-nu/nu) | NA (SC implant of cells) | BLI (IVIS) | Stomach, intestine | [83] |
| Genetic Engineering | Luminescence (Renilla Luciferase; Rluc) | EVs (CD63, Alix) | CAL-62 thyroid cancer cell and MDA-MB-231 breast cancer cells | UC (100,000 g, 60 min) | NA | 25 μg | Mouse (BALB/c, female) N = 3 | IV | BLI (IVIS) | 62/Rluc: lung> liver > spleen > kidney 62/Rluc/DiR: liver > lung, spleen 231Rluc: lung, liver > spleen > kidney | [67] |
| Genetic Engineering | Luminescence (Gaussia Luciferase) | EVs (CD63, ALIX) | HEK293T cells | UC (100,000 g, 90 min) | NA | 100 μg | Mouse (athymic nude) | IV | BLI | Spleen, liver > lung, kidney, brain, heart, muscle | [84] |
| Genetic Engineering | Luminescence (Gaussia Luciferase) | Exosomes | B16-Bl6 murine melanoma cells | UC (100,000 g, 1 h) | NA | 1 × 1010 RLU (5 μg) | Mouse (BALB/c) | IV | BLI (LAS3000) | Lung > spleen > kidney, liver, heart, brain, intestine | [85] |
| Membrane integration | MR (gadolinium) | Exosomes (CD9, CD63, CD81) | Human UC-MSCs | UC (120,000 g, 90 min) | UF (10 kDa) | 0.015 mmol/kg | Mouse, K7M2 (human osteosarcoma) xenograft (NU/NU) | IV | MRI | Liver, spleen > tumor > lung, kidney, heart, brain | [86] |
| Membrane integration | Fluorescence (DiR) | Exosomes (CD9, CD63, CD81) | Human UC-MSCs | UC 120,000 g, 90 min) | Not disclosed | 5 mg/kg | Mouse, K7M2 (human osteosarcoma) xenograft (NU/NU) | IV | LI-COR | Spleen > liver > tumor, lung > kidney, brain, heart | [86] |
| Membrane integration | Fluorescence (Dil) | Wnt4-exosomes | Mouse TEP1 (primary thymic epithelial cell) | TEI (Invitrogen) | TEI (pre-labeling) | Not disclosed | Mouse (BALB/c) | IV | IVIS | Thymus > lung, liver, spleen | [87] |
| Membrane integration | Fluorescence (DiR) | CVs (by sonication) | hCMEC/D3 B16 | UC (60,000 rpm, 24 h) | SEC | 200 μg of lipid | Mouse (FVB albino) | ROVS | IVIS | Liver > spleen, lung > brain | [88] |
| Membrane integration | Fluorescence (DiR) | Exosomes (ALIX, CD63, CD81, CD9, TSG101) | C2C12 murine myoblast cell | UC (100,000 g, 1 h) | Not disclosed | 30 μg | Mouse (C57BL/6) | IV | IVIS | Liver > spleen > lung | [89] |
| Membrane integration | Fluorescence (DiR) | Exosomes | BM-MSC | UC (100,000 g, 3 h) | UC (100,000 g, ND) | 8 × 109 | Mouse (C57BL/6) Tumor vs. non tumor | IP | IVIS | Liver, spleen, pancreas | [45] |
| Membrane integration | Fluorescence (PKH67) | Exosomes (CD63) | Mouse BM-MSC | UF + ExoQuick | ExoQuick | 30 μg | Mouse (BALB/c) TUBO tumor | IV | IVIS | (24 h) Tumor > spleen > kidney, liver, lung | [90] |
| Membrane integration | Fluorescence (DiR) | Exosomes (TSG101, CD81) | Endothelial colony forming cell (ECFC) | UC (100,000 g, 90 min) | UC (100,000 g) | 20 μg | Male FVB mice | IV | IVIS | (4 h) kidney > liver, heart, spleen, lung | [91] |
| Membrane integration | Fluorescence (DiD) | Exosomes (TSG101, CD9, HSP70; GM130-) | Murine EO771 BC cells | Combination of UF (100 kDa) and SEC | UC (100,000 g, 90 min) | 20 μg (1.6 × 1011) | Mouse (C57BL/6 or BALB/C) | IV | IVIS organ imaging | Lung, > liver > spleen, kidney > heart > bone marrow | [92] |
| Membrane integration | Fluorescence (DiD) | Exosomes (TSG101, CD9, HSP70; GM130-) | Murine 4T1 BC cells | UC (100,000× g, 90 min) | UC (100,000 g, 90 min) | 20 μg (1.2 × 1011) | Mouse (C57BL/6 or BALB/C) | IV | IVIS organ imaging | Lung > liver > kidney > spleen, heart, bone marrow | [92] |
| Membrane integration | Fluorescence (DiD) | Exosomes | Murine 67NR BC cells | UC (100,000× g, 90 min) | UC (100,000 g, 90 min) | 20 μg (1.2 × 1011) | Mouse (C57BL/6 or BALB/C) | IV | IVIS organ imaging | Lung > liver > kidney > spleen, heart, bone marrow | [92] |
| Membrane integration | Fluorescence (DiR) | EVs | Undisclosed | NA | UC (120,000 g, 70 min) vs. UF (100 kDa)–SEC (S-400) | Undisclosed | Mouse (BALB/c) | IV | IVIS organ imaging | (UC) liver > lung, spleen > kidney (UF-SEC) liver > spleen > lung > kidney | [93] |
| Membrane integration | Fluorescence (DiR) | EVs (ALIX, TSG101) | HEK293T cells | UC (110,000 g, 70 min) | NA (pre-labeling before UC) | 1.5 × 1010, 1,0 × 1010, 0.25 × 1010 p/g BW | Mouse (NMRI or C57BL/6) | IV IP SC | IVIS | (IV) liver > GI-tract, spleen > lung > pancreas (IP) liver, GI-tract, pancreas > spleen, lung (SC) GI-tract > liver > pancreas, lung > spleen | [68] |
| Membrane integration | Fluorescence (DiR) | EVs (ALIX, TSG101) | DC cells | UC (110,000 g, 70 min) | NA (pre-labeling before UC) | 1.0 × 1010 p/g BW | Mouse (NMRI or C57BL/6) | IV | IVIS | Liver > spleen > GI-tract, lung > pancreas | [68] |
| Membrane integration | Fluorescence (DiR) | EVs (ALIX, TSG101) | C2C12 cells | UC (110,000 g, 70 min) | NA (pre-labeling before UC) | 1.0 × 1010 p/g BW | Mouse (NMRI or C57BL/6) | IV | IVIS | Liver > spleen > GI-tract > lung > pancreas | [68] |
| Membrane integration | Fluorescence (DiR) | EVs (ALIX, TSG101) | B16F10 cells | UC (110,000 g, 70 min) | NA (pre-labeling before UC) | 1.0 × 1010 p/g BW | Mouse (NMRI or C57BL/6) | IV | IVIS | Liver > GI-tract, spleen, lungs > pancreas | [68] |
| Membrane integration | Fluorescence (DiR) | Exosome (CD63, flotillin-1) | BMSCs | UF (3 kDa)-ExoQuick-TC | ExoQuick-TC | 500 μg | C57BL/KaLwRij | IV | Fluobean 800 | BM, spleen, liver | [94] |
| Membrane integration | Fluorescence (DiD) | EVs (CD44, CD105, CD90, α5-integrin) | MSCs | UC (100,000 g, 1 h) | UC | 200 μg | Mouse CD1 with or without glycerol-induced AKI | IV | IVIS organ imaging | (24 h) liver > spleen > lung | [95] |
| Encapsulation | RI 99mTc | Exosome mimetics | Rat RBCs | UC (100,000 g, 1 h) + DGUC | Centrifugation (not disclosed in detail) | 37 Mbq | Mouse (C57BL/6, male) | IV | Gamma camera imaging | Liver, spleen, kidney > thyroid, stomach, lung, blood, intestine > heart, muscle, bone | [65] |
| Encapsulation | MR gold nanoparticles | Exosomes (CD9) | Human MSCs | UC (100,000 g, 70 min) | UC (100,000 g, 2 h) | 2.8 × 109 | Mouse (C57bl/6, male) | IV IN | CT | (IV) lung, liver > spleen > kidney, brain, blood (IN) lung > spleen > kidney, brain, blood, liver | [96] |
| Encapsulation | RI 99mTc-HMPAO | Exosome mimetic | RAW264.7 | DGUC (100,000 g, 2 h) | SEC (MW3000) | 7.4–14.8 Mbq (29–64 μg) | Mouse (BALB/c) | IV | SPECT/CT | (5 h) liver > kidney > spleen > intestine > lung, heart, stomach, heart > bone, muscle, blood | [97] |
| Encapsulation by transfection | MR SPIO | Exosomes (CD9, CD63) | MDA-MB-231 | ExoQuick | NA | 100 μg | Mouse | IV | MPI CT | Liver | [75] |
| Encapsulation by Sonication | Fluorescence Chlorin e6 (Ce6) | Tumor targeting EVs | RAW264.7 | UC (100,000 g, 70 min) | UC (100,000 g, 70 min) | 10 mg/kg | Mouse (BALB/c nu/nu) with HCT116 tumor | IV | Image Station 4000 MM | Tumor > liver > lung, kidney, spleen, brain, heart | [76] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, Y.W.; Lee, J.H.; Kim, S.-Y.; Pack, C.-G.; Ha, D.H.; Park, S.R.; Youn, J.; Cho, B.S. Advances in Analysis of Biodistribution of Exosomes by Molecular Imaging. Int. J. Mol. Sci. 2020, 21, 665. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21020665
Yi YW, Lee JH, Kim S-Y, Pack C-G, Ha DH, Park SR, Youn J, Cho BS. Advances in Analysis of Biodistribution of Exosomes by Molecular Imaging. International Journal of Molecular Sciences. 2020; 21(2):665. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21020665
Chicago/Turabian StyleYi, Yong Weon, Jun Ho Lee, Sang-Yeob Kim, Chan-Gi Pack, Dae Hyun Ha, Sang Rae Park, Jinkwon Youn, and Byong Seung Cho. 2020. "Advances in Analysis of Biodistribution of Exosomes by Molecular Imaging" International Journal of Molecular Sciences 21, no. 2: 665. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21020665






