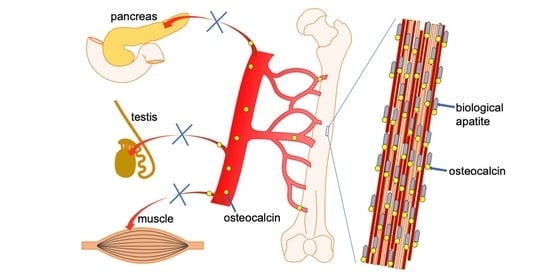
Functions of Osteocalcin in Bone, Pancreas, Testis, and Muscle
Abstract
:1. Introduction
2. Ocn and Runx2
3. Ocn and Bone Formation and Resorption
4. Osteocalcin and Bone Quality
5. Ocn and Bone Strength
6. Ocn and Glucose Metabolism
7. Association of Ocn with Exercise and Glucose Metabolism
8. Function of Ocn in Testosterone Synthesis and Muscle Mass
9. Conclusions
Funding
Conflicts of Interest
References
- Hauschka, P.V.; Lian, J.B.; Gallop, P.M. Direct identification of the calcium-binding amino acid, gamma-carboxyglutamate, in mineralized tissue. Proc. Natl. Acad. Sci. USA 1975, 72, 3925–3929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, P.A.; Otsuka, A.A.; Poser, J.W.; Kristaponis, J.; Raman, N. Characterization of a gamma-carboxyglutamic acid-containing protein from bone. Proc. Natl. Acad. Sci. USA 1976, 73, 1447–1451. [Google Scholar] [CrossRef] [Green Version]
- Hauschka, P.V.; Lian, J.B.; Cole, D.E.; Gundberg, C.M. Osteocalcin and matrix Gla protein: Vitamin K-dependent proteins in bone. Physiol. Rev. 1989, 69, 990–1047. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. What is the function of osteocalcin? J. Oral Biosci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hauschka, P.V.; Reid, M.L. Timed appearance of a calcium-binding protein containing gamma-carboxyglutamic acid in developing chick bone. Dev. Biol. 1978, 65, 426–434. [Google Scholar] [CrossRef]
- Hauschka, P.V.; Reddi, A.H. Correlation of the appearance of gamma-carboxyglutamic acid with the onset of mineralization in developing endochondral bone. Biochem. Biophys. Res. Commun. 1980, 92, 1037–1041. [Google Scholar] [CrossRef]
- Price, P.A.; Lothringer, J.W.; Baukol, S.A.; Reddi, A.H. Developmental appearance of the vitamin K-dependent protein of bone during calcification. Analysis of mineralizing tissues in human, calf, and rat. J. Biol. Chem. 1981, 256, 3781–3784. [Google Scholar] [PubMed]
- Price, P.A.; Williamson, M.K. Effects of warfarin on bone. Studies on the vitamin K-dependent protein of rat bone. J. Biol. Chem. 1981, 256, 12754–12759. [Google Scholar] [PubMed]
- Menanteau, J.; Neuman, W.F.; Neuman, M.W. A study of bone proteins which can prevent hydroxyapatite formation. Metab. Bone Dis. Relat. Res. 1982, 4, 157–162. [Google Scholar] [CrossRef]
- Boskey, A.L.; Wians, F.H., Jr.; Hauschka, P.V. The effect of osteocalcin on in vitro lipid-induced hydroxyapatite formation and seeded hydroxyapatite growth. Calcif. Tissue Int. 1985, 37, 57–62. [Google Scholar] [CrossRef]
- Romberg, R.W.; Werness, P.G.; Riggs, B.L.; Mann, K.G. Inhibition of hydroxyapatite crystal growth by bone-specific and other calcium-binding proteins. Biochemistry 1986, 25, 1176–1180. [Google Scholar] [CrossRef] [PubMed]
- Malone, J.D.; Teitelbaum, S.L.; Griffin, G.L.; Senior, R.M.; Kahn, A.J. Recruitment of osteoclast precursors by purified bone matrix constituents. J. Cell Biol. 1982, 92, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Mundy, G.R.; Poser, J.W. Chemotactic activity of the gamma-carboxyglutamic acid containing protein in bone. Calcif. Tissue Int. 1983, 35, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.B.; Dunn, K.; Key, L.L., Jr. In vitro degradation of bone particles by human monocytes is decreased with the depletion of the vitamin K-dependent bone protein from the matrix. Endocrinology 1986, 118, 1636–1642. [Google Scholar] [CrossRef]
- Ducy, P.; Desbois, C.; Boyce, B.; Pinero, G.; Story, B.; Dunstan, C.; Smith, E.; Bonadio, J.; Goldstein, S.; Gundberg, C.; et al. Increased bone formation in osteocalcin-deficient mice. Nature 1996, 382, 448–452. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.K.; Sowa, H.; Hinoi, E.; Ferron, M.; Ahn, J.D.; Confavreux, C.; Dacquin, R.; Mee, P.J.; McKee, M.D.; Jung, D.Y.; et al. Endocrine regulation of energy metabolism by the skeleton. Cell 2007, 130, 456–469. [Google Scholar] [CrossRef] [Green Version]
- Oury, F.; Sumara, G.; Sumara, O.; Ferron, M.; Chang, H.; Smith, C.E.; Hermo, L.; Suarez, S.; Roth, B.L.; Ducy, P.; et al. Endocrine regulation of male fertility by the skeleton. Cell 2011, 144, 796–809. [Google Scholar] [CrossRef] [Green Version]
- Mera, P.; Laue, K.; Ferron, M.; Confavreux, C.; Wei, J.; Galán-Díez, M.; Lacampagne, A.; Mitchell, S.J.; Mattison, J.A.; Chen, Y.; et al. Osteocalcin Signaling in Myofibers Is Necessary and Sufficient for Optimum Adaptation to Exercise. Cell Metab. 2016, 23, 1078–1092. [Google Scholar] [CrossRef] [Green Version]
- Oury, F.; Khrimian, L.; Denny, C.A.; Gardin, A.; Chamouni, A.; Goeden, N.; Huang, Y.-Y.; Lee, H.; Srinivas, P.; Gao, X.-B.; et al. Maternal and offspring pools of osteocalcin influence brain development and functions. Cell 2013, 155, 228–241. [Google Scholar] [CrossRef] [Green Version]
- Berger, J.M.; Singh, P.; Khrimian, L.; Morgan, D.A.; Chowdhury, S.; Arteaga-Solis, E.; Horvath, T.L.; Domingos, A.I.; Marsland, A.L.; Yadav, V.K.; et al. Mediation of the Acute Stress Response by the Skeleton. Cell Metab. 2019, 30, 890–902.e8. [Google Scholar] [CrossRef]
- Celeste, A.J.; Rosen, V.; Buecker, J.L.; Kriz, R.; Wang, E.A.; Wozney, J.M. Isolation of the human gene for bone gla protein utilizing mouse and rat cDNA clones. EMBO J. 1986, 5, 1885–1890. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Oberdorf, A.; Montecino, M.; Tanhauser, S.M.; Lian, J.B.; Stein, G.S.; Laipis, P.J.; Stein, J.L. Multiple copies of the bone-specific osteocalcin gene in mouse and rat. Endocrinology 1993, 133, 3050–3053. [Google Scholar] [CrossRef] [PubMed]
- Desbois, C.; Hogue, D.A.; Karsenty, G. The mouse osteocalcin gene cluster contains three genes with two separate spatial and temporal patterns of expression. J. Biol. Chem. 1994, 269, 1183–1190. [Google Scholar] [PubMed]
- Sato, M.; Tada, N. Preferential expression of osteocalcin-related protein mRNA in gonadal tissues of male mice. Biochem. Biophys. Res. Commun. 1995, 215, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of Proliferation, Differentiation and Functions of Osteoblasts by Runx2. Int. J. Mol. Sci. 2019, 20, 1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.; Gao, Y.-H.; Inada, M.; et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, C.; McCabe, L.R.; Choi, J.-Y.; Hiebert, S.W.; Stein, J.L.; Stein, G.S.; Lian, J.B. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. J. Cell. Biochem. 1997, 66, 1–8. [Google Scholar] [CrossRef]
- Ducy, P.; Zhang, R.; Geoffroy, V.; Ridall, A.L.; Karsenty, G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 1997, 89, 747–754. [Google Scholar] [CrossRef] [Green Version]
- Harada, H.; Tagashira, S.; Fujiwara, M.; Ogawa, S.; Katsumata, T.; Yamaguchi, A.; Komori, T.; Nakatsuka, M. Cbfa1 isoforms exert functional differences in osteoblast differentiation. J. Biol. Chem. 1999, 274, 6972–6978. [Google Scholar] [CrossRef] [Green Version]
- Ducy, P.; Starbuck, M.; Priemel, M.; Shen, J.; Pinero, G.; Geoffroy, V.; Amling, M.; Karsenty, G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999, 13, 1025–1036. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, Z.; Yoshida, C.A.; Furuichi, T.; Amizuka, N.; Ito, M.; Fukuyama, R.; Miyazaki, T.; Kitaura, H.; Nakamura, K.; Fujita, T.; et al. Runx2 determines bone maturity and turnover rate in postnatal bone development and is involved in bone loss in estrogen deficiency. Dev. Dyn. 2007, 236, 1876–1890. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Animal models for osteoporosis. Eur. J. Pharmacol. 2015, 759, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Drissi, H.; Luc, Q.; Shakoori, R.; Chuva De Sousa Lopes, S.; Choi, J.Y.; Terry, A.; Hu, M.; Jones, S.; Neil, J.C.; Lian, J.B.; et al. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell. Physiol. 2000, 184, 341–350. [Google Scholar] [CrossRef]
- Yoshida, C.A.; Furuichi, T.; Fujita, T.; Fukuyama, R.; Kanatani, N.; Kobayashi, S.; Satake, M.; Takada, K.; Komori, T. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat. Genet. 2002, 32, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Takarada, T.; Hinoi, E.; Nakazato, R.; Ochi, H.; Xu, C.; Tsuchikane, A.; Takeda, S.; Karsenty, G.; Abe, T.; Kiyonari, H.; et al. An analysis of skeletal development in osteoblast-specific and chondrocyte-specific runt-related transcription factor-2 (Runx2) knockout mice. J. Bone Miner. Res. 2013, 28, 2064–2069. [Google Scholar] [CrossRef] [PubMed]
- Adhami, M.D.; Rashid, H.; Chen, H.; Javed, A. Runx2 activity in committed osteoblasts is not essential for embryonic skeletogenesis. Connect. Tissue Res. 2014, 55 (Suppl. S1), 102–106. [Google Scholar] [CrossRef] [Green Version]
- Bailey, S.; Karsenty, G.; Gundberg, C.; Vashishth, D. Osteocalcin and osteopontin influence bone morphology and mechanical properties. Ann. N. Y. Acad. Sci. 2017, 1409, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Berezovska, O.; Yildirim, G.; Budell, W.; Yagerman, S.; Pidhaynyy, B.; Bastien, C.; Van Der Meulen, M.; Dowd, T.L. Osteocalcin affects bone mineral and mechanical properties in female mice. Bone 2019, 128, 115031. [Google Scholar] [CrossRef]
- Diegel, C.R.; Hann, S.; Ayturk, U.M.; Hu, J.C.W.; Lim, K.-E.; Droscha, C.J.; Madaj, Z.B.; Foxa, G.E.; Izaguirre, I.; Core, V.V.A.T.; et al. An osteocalcin-deficient mouse strain without endocrine abnormalities. PLoS Genet. 2020, 16, e1008361. [Google Scholar] [CrossRef]
- Moriishi, T.; Ozasa, R.; Ishimoto, T.; Nakano, T.; Hasegawa, T.; Miyazaki, T.; Liu, W.; Fukuyama, R.; Wang, Y.; Komori, H.; et al. Osteocalcin is necessary for the alignment of apatite crystallites, but not glucose metabolism, testosterone synthesis, or muscle mass. PLoS Genet. 2020, 16, e1008586. [Google Scholar] [CrossRef]
- Lambert, L.J.; Challa, A.K.; Niu, A.; Zhou, L.; Tucholski, J.; Johnson, M.S.; Nagy, T.R.; Eberhardt, A.W.; Estep, P.N.; Kesterson, R.A.; et al. Increased trabecular bone and improved biomechanics in an osteocalcin-null rat model created by CRISPR/Cas9 technology. Dis. Models Mech. 2016, 9, 1169–1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boskey, A.L.; Gadaleta, S.; Gundberg, C.; Doty, S.B.; Ducy, P.; Karsenty, G. Fourier transform infrared microspectroscopic analysis of bones of osteocalcin-deficient mice provides insight into the function of osteocalcin. Bone 1998, 23, 187–196. [Google Scholar] [CrossRef]
- Kavukcuoglu, N.B.; Patterson-Buckendahl, P.; Mann, A.B. Effect of osteocalcin deficiency on the nanomechanics and chemistry of mouse bones. J. Mech. Behav. Biomed. Mater. 2009, 2, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Poundarik, A.A.; Boskey, A.; Gundberg, C.; Vashishth, D. Biomolecular regulation, composition and nanoarchitecture of bone mineral. Sci. Rep. 2018, 8, 1191. [Google Scholar] [CrossRef] [Green Version]
- Ishimoto, T.; Sato, B.; Lee, J.W.; Nakano, T. Co-deteriorations of anisotropic extracellular matrix arrangement and intrinsic mechanical property in c-src deficient osteopetrotic mouse femur. Bone 2017, 103, 216–223. [Google Scholar] [CrossRef]
- Sekita, A.; Matsugaki, A.; Ishimoto, T.; Nakano, T. Synchronous disruption of anisotropic arrangement of the osteocyte network and collagen/apatite in melanoma bone metastasis. J. Struct. Biol. 2017, 197, 260–270. [Google Scholar] [CrossRef]
- Wang, J.; Ishimoto, T.; Nakano, T. Unloading-Induced Degradation of the Anisotropic Arrangement of Collagen/Apatite in Rat Femurs. Calcif. Tissue Int. 2017, 100, 87–94. [Google Scholar] [CrossRef]
- Raghavan, M.; Sahar, N.D.; Wilson, R.H.; Mycek, M.-A.; Pleshko, N.; Kohn, D.H.; Morris, M.D. Quantitative polarized Raman spectroscopy in highly turbid bone tissue. J. Biomed. Opt. 2010, 15, 037001. [Google Scholar] [CrossRef] [Green Version]
- Ishimoto, T.; Nakano, T.; Umakoshi, Y.; Yamamoto, M.; Tabata, Y. Degree of biological apatite c-axis orientation rather than bone mineral density controls mechanical function in bone regenerated using recombinant bone morphogenetic protein-2. J. Bone Miner. Res. 2013, 28, 1170–1179. [Google Scholar] [CrossRef]
- Nikel, O.; Poundarik, A.A.; Bailey, S.; Vashishth, D. Structural role of osteocalcin and osteopontin in energy dissipation in bone. J. Biomech. 2018, 80, 45–52. [Google Scholar] [CrossRef]
- Lee, K.; Nichols, J.; Smith, A. Identification of a developmentally regulated protein tyrosine phosphatase in embryonic stem cells that is a marker of pluripotential epiblast and early mesoderm. Mech. Dev. 1996, 59, 153–164. [Google Scholar] [CrossRef]
- Mauro, L.J.; Olmsted, E.A.; Davis, A.R.; Dixon, J.E. Parathyroid hormone regulates the expression of the receptor protein tyrosine phosphatase, OST-PTP, in rat osteoblast-like cells. Endocrinology 1996, 137, 925–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferron, M.; Wei, J.; Yoshizawa, T.; Del Fattore, A.; DePinho, R.A.; Teti, A.; Ducy, P.; Karsenty, G. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell 2010, 142, 296–308. [Google Scholar] [CrossRef] [Green Version]
- Rached, M.T.; Kode, A.; Silva, B.C.; Jung, D.Y.; Gray, S.; Ong, H.; Paik, J.H.; DePinho, R.A.; Kim, J.K.; Karsenty, G.; et al. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. J. Clin. Investig. 2010, 120, 357–368. [Google Scholar] [CrossRef] [Green Version]
- Yoshizawa, T.; Hinoi, E.; Jung, D.Y.; Kajimura, D.; Ferron, M.; Seo, J.; Graff, J.M.; Kim, J.K.; Karsenty, G. The transcription factor ATF4 regulates glucose metabolism in mice through its expression in osteoblasts. J. Clin. Investig. 2009, 119, 2807–2817. [Google Scholar] [CrossRef]
- Zee, T.; Settembre, C.; Levine, R.L.; Karsenty, G. T-cell protein tyrosine phosphatase regulates bone resorption and whole-body insulin sensitivity through its expression in osteoblasts. Mol. Cell. Biol. 2012, 32, 1080–1088. [Google Scholar] [CrossRef] [Green Version]
- Manolagas, S.C. Osteocalcin promotes bone mineralization but is not a hormone. PLoS Genet. 2020, 16, e1008714. [Google Scholar] [CrossRef] [PubMed]
- Moriishi, T.; Komori, T. Lack of reproducibility in osteocalcin-deficient mice. PLoS Genet. 2020, 16, e1008939. [Google Scholar] [CrossRef]
- Diegel, C.R.; Hann, S.; Ayturk, U.M.; Hu, J.C.W.; Lim, K.-E.; Droscha, C.J.; Madaj, Z.B.; Foxa, G.E.; Izaguirre, I.; Robling, A.G.; et al. Independent validation of experimental results requires timely and unrestricted access to animal models and reagents. PLoS Genet. 2020, 16, e1008940. [Google Scholar] [CrossRef]
- Funkat, A.; Massa, C.M.; Jovanovska, V.; Proietto, J.; Andrikopoulos, S. Metabolic adaptations of three inbred strains of mice (C57BL/6, DBA/2, and 129T2) in response to a high-fat diet. J. Nutr. 2004, 134, 3264–3269. [Google Scholar] [CrossRef] [Green Version]
- Goren, H.J.; Kulkarni, R.N.; Kahn, C.R. Glucose homeostasis and tissue transcript content of insulin signaling intermediates in four inbred strains of mice: C57BL/6, C57BLKS/6, DBA/2, and 129X1. Endocrinology 2004, 145, 3307–3323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrikopoulos, S.; Massa, C.M.; Aston-Mourney, K.; Funkat, A.; Fam, B.C.; Hull, R.L.; Kahn, S.E.; Proietto, J. Differential effect of inbred mouse strain (C57BL/6, DBA/2, 129T2) on insulin secretory function in response to a high fat diet. J. Endocrinol. 2005, 187, 45–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bock, T.; Pakkenberg, B.; Buschard, K. Genetic background determines the size and structure of the endocrine pancreas. Diabetes 2005, 54, 133–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgess, S.; Jeffrey, F.M.H.; Storey, C.; Milde, A.; Hausler, N.; Merritt, M.E.; Mulder, H.; Holm, C.; Sherry, A.D.; Malloy, C.R. Effect of murine strain on metabolic pathways of glucose production after brief or prolonged fasting. Am. J. Physiol. Endocrinol. Metab. 2005, 289, E53–E61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berglund, E.D.; Li, C.Y.; Poffenberger, G.; Ayala, J.E.; Fueger, P.T.; Willis, S.E.; Jewell, M.M.; Powers, A.C.; Wasserman, D.H. Glucose metabolism in vivo in four commonly used inbred mouse strains. Diabetes 2008, 57, 1790–1799. [Google Scholar] [CrossRef] [Green Version]
- Toye, A.A.; Lippiat, J.D.; Proks, P.; Shimomura, K.; Bentley, L.; Hugill, A.; Mijat, V.; Goldsworthy, M.; Moir, L.; Haynes, A.; et al. A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia 2005, 48, 675–686. [Google Scholar] [CrossRef] [Green Version]
- Freeman, H.C.; Hugill, A.; Dear, N.T.; Ashcroft, F.M.; Cox, R.D. Deletion of nicotinamide nucleotide transhydrogenase: A new quantitive trait locus accounting for glucose intolerance in C57BL/6J mice. Diabetes 2006, 55, 2153–2156. [Google Scholar] [CrossRef] [Green Version]
- Fontaine, D.A.; Davis, D.B. Attention to Background Strain Is Essential for Metabolic Research: C57BL/6 and the International Knockout Mouse Consortium. Diabetes 2016, 65, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, A.; Reifsnyder, P.C.; Malcolm, R.D.; Lucas, C.A.; MacGregor, G.R.; Zhang, W.; Leiter, E.H. Diet-induced obesity in two C57BL/6 substrains with intact or mutant nicotinamide nucleotide transhydrogenase (Nnt) gene. Obesity 2010, 18, 1902–1905. [Google Scholar] [CrossRef] [Green Version]
- Aston-Mourney, K.; Wong, N.; Kebede, M.; Zraika, S.; Balmer, L.; McMahon, J.M.; Fam, B.C.; Favaloro, J.; Proietto, J.; Morahan, G.; et al. Increased nicotinamide nucleotide transhydrogenase levels predispose to insulin hypersecretion in a mouse strain susceptible to diabetes. Diabetologia 2007, 50, 2476–2485. [Google Scholar] [CrossRef] [Green Version]
- Simon, M.M.; Greenaway, S.; White, J.K.; Fuchs, H.; Gailus-Durner, V.; Wells, S.; Sorg, T.; Wong, K.; Bedu, E.; Cartwright, E.J.; et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 2013, 14, R82. [Google Scholar] [CrossRef] [PubMed]
- Fergusson, G.; Ethier, M.; Guévremont, M.; Chrétien, C.; Attané, C.; Joly, E.; Fioramonti, X.; Prentki, M.; Poitout, V.; Alquier, T. Defective insulin secretory response to intravenous glucose in C57Bl/6J compared to C57Bl/6N mice. Mol. Metab. 2014, 3, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Dacquin, R.; Starbuck, M.; Schinke, T.; Karsenty, G. Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev. Dyn. 2002, 224, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Im, J.A.; Yu, B.P.; Jeon, J.Y.; Kim, S.H. Relationship between osteocalcin and glucose metabolism in postmenopausal women. Clin. Chim. Acta 2008, 396, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Real, J.M.; Izquierdo, M.; Ortega, F.J.; Gorostiaga, E.; Gómez-Ambrosi, J.; Moreno-Navarrete, J.M.; Frühbeck, G.; Martínez, C.; Idoate, F.; Salvador, J.; et al. The relationship of serum osteocalcin concentration to insulin secretion, sensitivity, and disposal with hypocaloric diet and resistance training. J. Clin. Endocrinol. Metab. 2009, 94, 237–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanazawa, I.; Yamaguchi, T.; Yamamoto, M.; Yamauchi, M.; Kurioka, S.; Yano, S.; Sugimoto, T. Serum osteocalcin level is associated with glucose metabolism and atherosclerosis parameters in type 2 diabetes mellitus. J. Clin. Endocrinol. Metab. 2009, 94, 45–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kindblom, J.M.; Ohlsson, C.; Ljunggren, Ö.; Karlsson, M.K.; Tivesten, Å.; Smith, U.; Mellström, D. Plasma osteocalcin is inversely related to fat mass and plasma glucose in elderly Swedish men. J. Bone Miner. Res. 2009, 24, 785–791. [Google Scholar] [CrossRef]
- Pittas, A.G.; Harris, S.S.; Eliades, M.; Stark, P.; Dawson-Hughes, B. Association between serum osteocalcin and markers of metabolic phenotype. J. Clin. Endocrinol. Metab. 2009, 94, 827–832. [Google Scholar] [CrossRef]
- Zhou, M.; Ma, X.; Li, H.; Pan, X.; Tang, J.; Gao, Y.; Hou, X.; Lu, H.; Bao, Y.; Jia, W. Serum osteocalcin concentrations in relation to glucose and lipid metabolism in Chinese individuals. Eur. J. Endocrinol. 2009, 161, 723–729. [Google Scholar] [CrossRef] [Green Version]
- Hwang, Y.C.; Jeong, I.K.; Ahn, K.J.; Chung, H.Y. The uncarboxylated form of osteocalcin is associated with improved glucose tolerance and enhanced β-cell function in middle-aged male subjects. Diabetes/Metab. Res. Rev. 2009, 25, 768–772. [Google Scholar] [CrossRef]
- Kanazawa, I.; Yamaguchi, T.; Yamauchi, M.; Yamamoto, M.; Kurioka, S.; Yano, S.; Sugimoto, T. Serum undercarboxylated osteocalcin was inversely associated with plasma glucose level and fat mass in type 2 diabetes mellitus. Osteoporos. Int. 2011, 22, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Iki, M.; Tamaki, J.; Fujita, Y.; Kouda, K.; Yura, A.; Kadowaki, E.; Sato, Y.; Moon, J.S.; Tomioka, K.; Okamoto, N.; et al. Serum undercarboxylated osteocalcin levels are inversely associated with glycemic status and insulin resistance in an elderly Japanese male population: Fujiwara-kyo Osteoporosis Risk in Men (FORMEN) Study. Osteoporos. Int. 2012, 23, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Díaz-López, A.; Bulló, M.; Juanola-Falgarona, M.; Martínez-González, M.A.; Estruch, R.; Covas, M.-I.; Arós, F.; Salas-Salvadó, J. Reduced serum concentrations of carboxylated and undercarboxylated osteocalcin are associated with risk of developing type 2 diabetes mellitus in a high cardiovascular risk population: A nested case-control study. J. Clin. Endocrinol. Metab. 2013, 98, 4524–4531. [Google Scholar] [CrossRef] [PubMed]
- Okuno, S.; Ishimura, E.; Tsuboniwa, N.; Norimine, K.; Yamakawa, K.; Shoji, S.; Mori, K.; Nishizawa, Y.; Inaba, M. Significant inverse relationship between serum undercarboxylated osteocalcin and glycemic control in maintenance hemodialysis patients. Osteoporos. Int. 2013, 24, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Takashi, Y.; Koga, M.; Matsuzawa, Y.; Saito, J.; Omura, M.; Nishikawa, T. Undercarboxylated osteocalcin can predict insulin secretion ability in type 2 diabetes. J. Diabetes Investig. 2017, 8, 471–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shea, M.K.; Gundberg, C.M.; Meigs, J.B.; E Dallal, G.; Saltzman, E.; Yoshida, M.; Jacques, P.F.; Booth, S.L. Gamma-carboxylation of osteocalcin and insulin resistance in older men and women. Am. J. Clin. Nutr. 2009, 90, 1230–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, K.; Emoto, M.; Motoyama, K.; Lee, E.; Yamada, S.; Morioka, T.; Imanishi, Y.; Shoji, T.; Inaba, M. Undercarboxylated osteocalcin does not correlate with insulin resistance as assessed by euglycemic hyperinsulinemic clamp technique in patients with type 2 diabetes mellitus. Diabetol. Metab. Syndr. 2012, 4, 53. [Google Scholar] [CrossRef] [Green Version]
- Giudici, K.V.P.; Fisberg, R.M.; Marchioni, D.M.L.; Peters, B.S.E.; Martini, L.A. Crosstalk Between Bone and Fat Tissue: Associations Between Vitamin D, Osteocalcin, Adipokines, and Markers of Glucose Metabolism Among Adolescents. J. Am. Coll. Nutr. 2017, 36, 273–280. [Google Scholar] [CrossRef]
- De Bruin, M.L.; Westberg-Rasmussen, S.; Lykkeboe, S.; Handberg, A.; Hartmann, B.; Holst, J.J.; Hermansen, K.; Vestergaard, P.; Gregersen, S. Glucose Tolerance Tests and Osteocalcin Responses in Healthy People. Front. Endocrinol. 2018, 9, 356. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Ivaska, K.K.; Alén, M.; Wang, Q.; Törmäkangas, T.; Xu, L.; Wiklund, P.; Mikkola, T.M.; Pekkala, S.; Tian, H.; et al. Serum osteocalcin is not associated with glucose but is inversely associated with leptin across generations of nondiabetic women. J. Clin. Endocrinol. Metab. 2012, 97, 4106–4114. [Google Scholar] [CrossRef] [Green Version]
- Prats-Puig, A.; Osiniri, I.; Soriano-Rodríguez, P.; Carreras-Badosa, G.; Buñuel-Álvarez, J.C.; Vila-Pablos, C.; De Zegher, F.; Ibáñez, L.; Bassols, J.; López-Bermejo, A. Undercarboxylated osteocalcin relates to cardiovascular risk markers in offspring of families with metabolic syndrome. Atherosclerosis 2014, 233, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Enriquez, I.T.B.-G.S.; Ballesteros-Gonzalez, I.T.; N-Bernal, S.P.-G.; Pascoe-Gonzalez, S.; Rivera-Leon, E.A.; Bastidas-Ramirez, B.E.; Rivas-Carrillo, J.D.; Alcala-Zermeno, J.L.; Armendariz-Borunda, J.; Llamas-Covarrubias, I.M.; et al. Serum levels of undercarboxylated osteocalcin are related to cardiovascular risk factors in patients with type 2 diabetes mellitus and healthy subjects. World J. Diabetes 2017, 8, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Riquelme-Gallego, B.; García-Molina, L.; Cano-Ibáñez, N.; Sánchez-Delgado, G.; Andújar-Vera, F.; García-Fontana, C.; González-Salvatierra, S.; García-Recio, E.; Martínez-Ruiz, V.; Bueno-Cavanillas, A.; et al. Circulating Undercarboxylated Osteocalcin as Estimator of Cardiovascular and Type 2 Diabetes Risk in Metabolic Syndrome Patients. Sci. Rep. 2020, 10, 1840. [Google Scholar] [CrossRef] [PubMed]
- Pi, M.; Wu, Y.; Quarles, L.D. GPRC6A mediates responses to osteocalcin in beta-cells in vitro and pancreas in vivo. J. Bone Miner. Res. 2011, 26, 1680–1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smajilovic, S.; Clemmensen, C.; Johansen, L.D.; Wellendorph, P.; Holst, J.J.; Thams, P.G.; Ogo, E.; Bräuner-Osborne, H. The L-alpha-amino acid receptor GPRC6A is expressed in the islets of Langerhans but is not involved in L-arginine-induced insulin release. Amino Acids 2013, 44, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Pi, M.; Wu, Y.; Lenchik, N.I.; Gerling, I.; Quarles, L.D. GPRC6A mediates the effects of L-arginine on insulin secretion in mouse pancreatic islets. Endocrinology 2012, 153, 4608–4615. [Google Scholar] [CrossRef] [PubMed]
- Pi, M.; Wu, Y.; I Lenchik, N.; Gerling, I.; Quarles, L.D. Evidence for Osteocalcin Binding and Activation of GPRC6A in beta-Cells. Endocrinology 2016, 157, 1866–1880. [Google Scholar] [CrossRef] [Green Version]
- Rueda, P.; Harley, E.; Lü, Y.; Stewart, G.D.; Fabb, S.; Diepenhorst, N.; Cremers, B.; Rouillon, M.-H.; Wehrle, I.; Géant, A.; et al. Murine GPRC6A Mediates Cellular Responses to L-Amino Acids, but Not Osteocalcin Variants. PLoS ONE 2016, 11, e0146846. [Google Scholar] [CrossRef]
- Wei, J.; Hanna, T.; Suda, N.; Karsenty, G.; Ducy, P. Osteocalcin promotes beta-cell proliferation during development and adulthood through Gprc6a. Diabetes 2014, 63, 1021–1031. [Google Scholar] [CrossRef] [Green Version]
- Pi, M.; Chen, L.; Huang, M.-Z.; Zhu, W.; Ringhofer, B.; Luo, J.; Christenson, L.; Li, B.; Zhang, J.; Jackson, P.D.; et al. GPRC6A null mice exhibit osteopenia, feminization and metabolic syndrome. PLoS ONE 2008, 3, e3858. [Google Scholar] [CrossRef]
- Wellendorph, P.; Johansen, L.D.; Jensen, A.A.; Casanova, E.; Gassmann, M.; Deprez, P.; Clément-Lacroix, P.; Bettler, B.; Bräuner-Osborne, H. No evidence for a bone phenotype in GPRC6A knockout mice under normal physiological conditions. J. Mol. Endocrinol. 2009, 42, 215–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorgensen, C.V.; Gasparini, S.J.; Tu, J.; Zhou, H.; Seibel, M.J.; Brauner-Osborne, H. Metabolic and skeletal homeostasis are maintained in full locus GPRC6A knockout mice. Sci. Rep. 2019, 9, 5995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mera, P.; Laue, K.; Wei, J.; Berger, J.M.; Karsenty, G. Osteocalcin is necessary and sufficient to maintain muscle mass in older mice. Mol. Metab. 2016, 5, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Schulz, L.C.; Palmisano, B.; Singh, P.; Berger, J.M.; Yadav, V.K.; Mera, P.; Ellingsgaard, H.; Hidalgo, J.; Brüning, J.C.; et al. Muscle-derived interleukin 6 increases exercise capacity by signaling in osteoblasts. J. Clin. Investig. 2020, 130, 2888–2902. [Google Scholar] [CrossRef] [Green Version]
- Khrimian, L.; Obri, A.; Ramos-Brossier, M.; Rousseaud, A.; Moriceau, S.; Nicot, A.-S.; Mera, P.; Kosmidis, S.; Karnavas, T.; Saudou, F.; et al. Gpr158 mediates osteocalcin’s regulation of cognition. J. Exp. Med. 2017, 214, 2859–2873. [Google Scholar] [CrossRef]
- Ferron, M.; McKee, M.D.; Levine, R.L.; Ducy, P.; Karsenty, G. Intermittent injections of osteocalcin improve glucose metabolism and prevent type 2 diabetes in mice. Bone 2012, 50, 568–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Karsenty’s Group | Williams’ Group | Our Group | ||
|---|---|---|---|---|
| Method | Deletion of Bglap and Bglap2 Using ES Cells | Deletion of Bglap and Bglap2 by CRISPR/Cas9 | Deletion of Bglap and Bglap2 Using ES Cells | |
| genetic background | 129Sv;C57BL/6J | C57BL/6J | C57BL/6J;C3H | C57BL/6N |
| cortical bone | ↑↑ | → | → | → |
| trabecular bone | ↑↑ | ↑ | → | → |
| bone formation | ↑ | ↑ | nd | → |
| bone resorption | ↑ | → | nd | → |
| ovariectomy | bone resorption Ocn−/− > wild-type | nd | nd | similar to wild-type |
| cortical BMD | nd | → or ↓ | → | → |
| trabecular BMD | nd | → | → | → |
| crystallinity | ↓ | ↑ | → | → |
| mineral:matrix ratio | → | → | → | → |
| carbonate:phos-phate ratio | → | ↓ | ↑ | → |
| collagen maturity | nd | nd | ↑ | → |
| size or shape of BAp | nd | thin, plate-like | nd | Size → |
| bone strength | ↑ | ↑ or ↓ | → | ↓ |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komori, T. Functions of Osteocalcin in Bone, Pancreas, Testis, and Muscle. Int. J. Mol. Sci. 2020, 21, 7513. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21207513
Komori T. Functions of Osteocalcin in Bone, Pancreas, Testis, and Muscle. International Journal of Molecular Sciences. 2020; 21(20):7513. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21207513
Chicago/Turabian StyleKomori, Toshihisa. 2020. "Functions of Osteocalcin in Bone, Pancreas, Testis, and Muscle" International Journal of Molecular Sciences 21, no. 20: 7513. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21207513