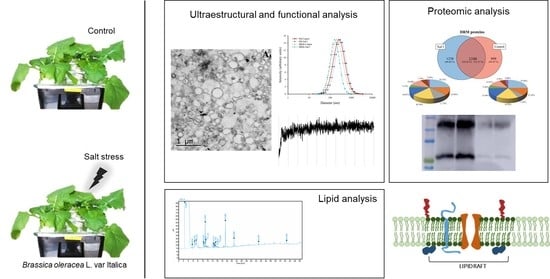
Detergent Resistant Membrane Domains in Broccoli Plasma Membrane Associated to the Response to Salinity Stress
Abstract
:1. Introduction
2. Results
2.1. Quantitative Analysis of Protein Amounts in DRM Isolation
2.2. Lipid Composition of Broccoli Root DRMs Compared to PM
2.3. Ultrastructural Analysis And Size of Broccoli Root DRMs Compared to PM
2.4. Vesicle Integrity and Functionality
2.5. PIP1 And PIP2 Aquaporin Quantification
2.6. Proteomic Analysis of DRMs from B. oleracea Root
3. Discussion
4. Materials and Methods
4.1. Plant Growth
4.2. Plasma Membrane Isolation and Enzyme Assay
4.3. Detergent-Resistant Membrane (DRM) Isolation
4.4. Isolation of Proteins Associated with Membrane Rafts and Plasma Membrane
4.5. Total Protein Quantification
4.6. Lipid Analysis
4.7. Transmission Electron Microscopy
4.8. Size of Membranes Vesicles
4.9. Stopped-Flow Light Scattering
4.10. Gel Electrophoresis and Immunoblotting
4.11. Proteomic Analysis
4.11.1. In-Gel Protein Digestion (Stacking Gel)
4.11.2. Liquid Chromatography and Mass Spectrometer Analysis (LC-ESI-MS/MS)
4.11.3. Proteomics Data Analysis and Sequence Search
4.12. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gronnier, J.; Gerbeau-Pissot, P.; Germain, V.; Mongrand, S.; Simon-Plas, F. Divide and Rule: Plant Plasma Membrane Organization. Trends Plant Sci. 2018, 23, 899–917. [Google Scholar] [CrossRef]
- Morel, J.; Claverol, S.; Mongrand, S.; Furt, F.; Fromentin, J.; Bessoule, J.-J.; Blein, J.-P.; Simon-Plas, F. Proteomics of Plant Detergent-resistant Membranes. Mol. Cell. Proteom. 2006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Q.; Liu, L.; Barkla, B.J. Membrane Lipid Remodeling in Response to Salinity. Int. J. Mol. Sci. 2019, 20, 4264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boursiac, Y.; Chen, S.; Luu, D.T.; Sorieul, M.; Van Den Dries, N.; Maurel, C. Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Plant Physiol. 2005, 139, 790–805. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Schraut, D.; Hartung, W.; Schäffner, A.R. Differential responses of maize MIP genes to salt stress and ABA. J. Exp. Bot. 2005, 56, 2971–2981. [Google Scholar] [CrossRef]
- Martínez-Ballesta, M.C.; Cabañero, F.J.; Maurel, C.; Olmos, E.; Carvajal, M. Nutritional Calcium as the regulator of the aquaporin activity in plants grown under salinity. Planta 2008, 228, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Maurel, C.; Verdoucq, L.; Luu, D.-T.; Santoni, V. Plant aquaporins: Membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 2008, 59, 595–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez Ballesta, M.C.; Carvajal, M. New challenges in plant aquaporin biotechnology. Plant Sci. 2014, 217, 71–77. [Google Scholar] [CrossRef]
- Muries, B.; Mohamed, F.; Carvajal, M.; Martínez-Ballesta, M.C. Identification and differential induction of the expression of aquaporins by salinity in broccoli plants. Mol. Biosyst. 2011, 7, 1322–1335. [Google Scholar] [CrossRef]
- Singer, S.J.; Nicolson, G.L. The fluid mosaic model of the structure of cell membranes. Science 1972, 175, 720–731. [Google Scholar] [CrossRef]
- Yu, M.; Cui, Y.; Zhang, X.; Li, R.; Lin, J. Organization and dynamics of functional plant membrane microdomains. Cell. Mol. Life Sci. 2020, 77, 75–287. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.G.W.; Jacobson, K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 2002, 296, 1821–1825. [Google Scholar] [CrossRef]
- Ferrer, A.; Altabella, T.; Arró, M.; Boronat, A. Emerging roles for conjugated sterols in plants. Prog. Lipid Res. 2017, 67, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, G.L. The Fluid—Mosaic Model of Membrane Structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1451–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaillais, Y.; Ott, T. The nanoscale organization of the plasma membrane and its importance in signaling: A proteolipid perspective. Plant Physiol. 2020, 182, 1682–1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, D.A.; Rose, J.K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 1992, 68, 533–544. [Google Scholar] [CrossRef]
- Minami, A.; Fujiwara, M.; Furuto, A.; Fukao, Y.; Yamashita, T.; Kamo, M.; Kawamura, Y.; Uemura, M. Alterations in detergent-resistant plasma membrane microdomains in Arabidopsis thaliana during cold acclimation. Plant Cell Physiol. 2009, 50, 341–359. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez-Carbonell, E.; Takahashi, D.; Lüthje, S.; González-Reyes, J.A.; Mongrand, S.; Contreras-Moreira, B.; Abadía, A.; Uemura, M.; Abadía, J.; López-Millán, A.F. A shotgun proteomic approach reveals that fe deficiency causes marked changes in the protein profiles of plasma membrane and detergent-resistant microdomain preparations from beta vulgaris roots. J. Proteome Res. 2016, 15, 2510–2524. [Google Scholar] [CrossRef] [Green Version]
- Macdonald, J.L.; Pike, L.J. A simplified method for the preparation of detergent-free lipid rafts. J. Lipid Res. 2005, 46, 1061–1067. [Google Scholar] [CrossRef] [Green Version]
- Smart, E.J.; Ying, Y.S.; Mineo, C.; Anderson, R.G.W. A detergent-free method for purifying caveolae membrane from tissue culture cells. Proc. Natl. Acad. Sci. USA 1995, 92, 10104–10108. [Google Scholar] [CrossRef] [Green Version]
- Peskan, T.; Westermann, M.; Oelmüller, R. Identification of low-density Triton X-100-insoluble plasma membrane microdomains in higher plants. Eur. J. Biochem. 2000, 267, 6989–6995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mongrand, S.; Morel, J.; Laroche, J.; Claverol, S.; Carde, J.P.; Hartmann, M.A.; Bonneu, M.; Simon-Plas, F.; Lessire, R.; Bessoule, J.J. Lipid rafts in higher plant cells: Purification and characterization of triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 2004, 279, 36277–36286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefebvre, B.; Furt, F.; Hartmann, M.-A.; Michaelson, L.V.; Carde, J.-P.; Sargueil-Boiron, F.; Rossignol, M.; Napier, J.A.; Cullimore, J.; Bessoule, J.-J.; et al. Characterization of Lipid Rafts from Medicago truncatula Root Plasma Membranes: A Proteomic Study Reveals the Presence of a Raft-Associated Redox System. Plant Physiol. 2007, 144, 402–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiwara, M.; Hamada, S.; Hiratsuka, M.; Fukao, Y.; Kawasaki, T.; Shimamoto, K. Proteome analysis of detergent-resistant membranes (DRMs) associated with osrac1-mediated innate immunity in rice. Plant Cell Physiol. 2009, 50, 1191–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, D.; Kawamura, Y.; Uemura, M. Detergent-resistant plasma membrane proteome to elucidate microdomain functions in plant cells. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, X.; Yang, Y.; Li, R.; He, Q.; Fang, X.; Luu, D.-T.; Maurel, C.; Lin, J. Single-Molecule Analysis of PIP2;1 Dynamics and Partitioning Reveals Multiple Modes of Arabidopsis Plasma Membrane Aquaporin Regulation. Plant Cell 2011, 23, 3780–3797. [Google Scholar] [CrossRef] [Green Version]
- Ravikumar, C. Therapeutic potential of Brassica oleracea (broccoli)—A review. Int. J. Drug Dev. Res. 2015, 7, 9–10. [Google Scholar]
- Martínez Ballesta, M.C.; Pérez-Sánchez, H.; Moreno, D.A.; Carvajal, M. Plant plasma membrane aquaporins in natural vesicles as potential stabilizers and carriers of glucosinolates. Colloids Surfaces B Biointerfaces 2016, 143, 318–326. [Google Scholar] [CrossRef]
- Martínez Ballesta, M.C.; García-Gomez, P.; Yepes-Molina, L.; Guarnizo, A.L.; Teruel, J.A.; Carvajal, M. Plasma membrane aquaporins mediates vesicle stability in broccoli. PLoS ONE 2018, 13, e0192422. [Google Scholar] [CrossRef] [Green Version]
- Rios, J.J.; Garcia-Ibañez, P.; Carvajal, M. The use of biovesicles to improve the efficiency of Zn foliar fertilization. Colloids Surfaces B Biointerfaces 2019, 173, 899–905. [Google Scholar] [CrossRef]
- Yepes-Molina, L.; Martínez-Ballesta, M.C.; Carvajal, M. Plant plasma membrane vesicles interaction with keratinocytes reveals their potential as carriers. J. Adv. Res. 2020, 23, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Bevan, M.; Bancroft, I.; Bent, E.; Love, K.; Goodman, H.; Dean, C.; Bergkamp, R.; Dirkse, W.; Van Staveren, M.; Stiekema, W.; et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 1998, 391, 485–488. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Pérez, L.; Martínez-Ballesta, M.C.; Maurel, C.; Carvajal, M. Changes in plasma membrane lipids, aquaporins and proton pump of broccoli roots, as an adaptation mechanism to salinity. Phytochemistry 2009, 70, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Kawamura, Y.; Yamashita, T.; Uemura, M. Detergent-resistant plasma membrane proteome in oat and rye: Similarities and dissimilarities between two monocotyledonous plants. J. Proteome Res. 2012, 11, 1654–1665. [Google Scholar] [CrossRef]
- Huang, D.; Sun, Y.; Ma, Z.; Ke, M.; Cui, Y.; Chen, Z.; Chen, C.; Ji, C.; Tran, T.M.; Yang, L.; et al. Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc. Natl. Acad. Sci. USA 2019, 116, 21274–21284. [Google Scholar] [CrossRef] [Green Version]
- Amtmann, A.; Bohnert, H.J.; Bressan, R.A. Abiotic stress and plant genome evolution. Search for new models. Plant Physiol. 2005, 138, 127–130. [Google Scholar] [CrossRef] [Green Version]
- Chalbi, N.; Martínez-Ballesta, M.C.; Youssef, N.B.; Carvajal, M. Intrinsic stability of Brassicaceae plasma membrane in relation to changes in proteins and lipids as a response to salinity. J. Plant Physiol. 2015, 175, 148–156. [Google Scholar] [CrossRef]
- Valitova, J.N.; Sulkarnayeva, A.G.; Minibayeva, F.V. Plant sterols: Diversity, biosynthesis, and physiological functions. Biochemistry 2016, 81, 819–834. [Google Scholar] [CrossRef]
- Staneva, G.; Seigneuret, M.; Koumanov, K.; Trugnan, G.; Angelova, M.I. Detergents induce raft-like domains budding and fission from giant unilamellar heterogeneous vesicles: A direct microscopy observation. Chem. Phys. Lipids 2005, 136, 55–66. [Google Scholar] [CrossRef]
- Grosjean, K.; Der, C.; Robert, F.; Thomas, D.; Mongrand, S.; Simon-Plas, F.; Gerbeau-Pissot, P. Interactions between lipids and proteins are critical for organization of plasma membrane-ordered domains in tobacco BY-2 cells. J. Exp. Bot. 2018, 69, 3545–3557. [Google Scholar] [CrossRef] [PubMed]
- Bieberich, E. Sphingolipids and lipid rafts: Novel concepts and methods of analysis. Chem. Phys. Lipids 2018, 216, 114–131. [Google Scholar] [CrossRef]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef] [Green Version]
- Sandelius, A.S.; Penel, C.; Auderset, G.; Brightman, A.; Millard, M.; Morré, D.J. Isolation of Highly Purified Fractions of Plasma Membrane and Tonoplast from the Same Homogenate of Soybean Hypocotyls by Free-Flow Electrophoresis. Plant Physiol. 1986, 81, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Jen, A.; Warley, A.; Lawrence, M.J.; Quinn, P.J.; Morris, R.J. Isolation at physiological temperature of detergent-resistant membranes with properties expected of lipid rafts: The influence of buffer composition. Biochem. J. 2009, 417, 525–533. [Google Scholar] [CrossRef] [Green Version]
- Dubois, L.; Löf, L.; Larsson, A.; Hultenby, K.; Waldenstrom, A.; Hamali-Moghaddam, M.; Ronquist, G.; Ronquist, K.G. Human erythrocyte-derived nanovesicles can readily be loaded with doxorubicin and act as anticancer agents. Cancer Res. Front. 2018, 4, 13–26. [Google Scholar] [CrossRef]
- van Gestel, R.A.; Brouwers, J.F.; Ultee, A.; Helms, J.B.; Gadella, B.M. Ultrastructure and lipid composition of detergent-resistant membranes derived from mammalian sperm and two types of epithelial cells. Cell Tissue Res. 2016, 363, 129–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef] [Green Version]
- Shogomori, H.; Brown, D.A. Use of detergents to study membrane rafts: The good, the bad, and the ugly. Biol. Chem. 2003, 384, 1259–1263. [Google Scholar] [CrossRef] [PubMed]
- Murase, K.; Fujiwara, T.; Umemura, Y.; Suzuki, K.; Iino, R.; Yamashita, H.; Saito, M.; Murakoshi, H.; Ritchie, K.; Kusumi, A. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys. J. 2004, 88, 4075–4093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, J.; Briggs, M.M.; McIntosh, T.J. Water permeability of aquaporin-4 channel depends on bilayer composition, thickness, and elasticity. Biophys. J. 2012, 103, 1899–1908. [Google Scholar] [CrossRef] [Green Version]
- Belugin, B.V.; Zhestkova, I.M.; Piotrovskii, M.S.; Lapshin, N.K.; Trofimova, M.S. PIP1 aquaporins, sterols, and osmotic water permeability of plasma membranes from etiolated pea seedlings. Biochem. Suppl. Ser. A Membr. Cell Biol. 2017, 11, 168–176. [Google Scholar] [CrossRef]
- Kai, L.; Kaldenhoff, R. A refined model of water and CO2 membrane diffusion: Effects and contribution of sterols and proteins. Sci. Rep. 2014, 4, 6665. [Google Scholar] [CrossRef] [Green Version]
- Santoni, V.; Vinh, J.; Pflieger, D.; Sommerer, N.; Maurel, C. A proteomic study reveals novel insights into the diversity of aquaporin forms expressed in the plasma membrane of plant roots. Biochem. J 2003, 373, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Santoni, V.; Kieffer, S.; Desclaux, D.; Masson, F.; Rabilloud, T. Membrane proteomics: Use of additive main effects with multiplicative interaction model to classify plasma membrane proteins according to their solubility and electrophoretic properties. Electrophoresis 2000, 21, 3329–3344. [Google Scholar] [CrossRef]
- Santoni, V.; Molloy, M.; Rabilloud, T. Membrane proteins and proteomics: Un amour impossible? Electrophoresis 2000, 21, 1054–1070. [Google Scholar] [CrossRef]
- Hu, S.; Musante, L.; Tataruch, D.; Xu, X.; Kretz, O.; Henry, M.; Meleady, P.; Luo, H.; Zou, H.; Jiang, Y.; et al. Purification and Identification of Membrane Proteins from Urinary Extracellular Vesicles using Triton X-114 Phase Partitioning. J. Proteome Res. 2018, 17, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Casado-Vela, J.; Muries, B.; Carvajal, M.; Iloro, I.; Elortza, F.; Martínez-Ballesta, M.C. Analysis of root plasma membrane aquaporins from brassica oleracea: Post-translational modifications, de novo sequencing and detection of isoforms by high resolution mass spectrometry. J. Proteome Res. 2010, 9, 3479–3494. [Google Scholar] [CrossRef] [PubMed]
- Suga, S.; Komatsu, S.; Maeshima, M. Aquaporin isoforms responsive to salt and water stresses and phytohormones in radish seedlings. Plant Cell Physiol. 2002, 43, 1229–1237. [Google Scholar] [CrossRef] [Green Version]
- Martínez Ballesta, M.C.; Muries, B.; Moreno, D.Á.; Dominguez-Perles, R.; García-Viguera, C.; Carvajal, M. Involvement of a glucosinolate (sinigrin) in the regulation of water transport in Brassica oleracea grown under salt stress. Physiol. Plant. 2014, 150, 145–160. [Google Scholar] [CrossRef]
- Belugin, B.V.; Zhestkova, I.M.; Trofimova, M.S. Affinity of PIP-aquaporins to sterol-enriched domains in plasma membrane of the cells of etiolated pea seedlings. Biochem. Suppl. Ser. A Membr. Cell Biol. 2011, 5, 56–63. [Google Scholar] [CrossRef]
- Alexandersson, E.; Saalbach, G.; Larsson, C.; Kjellbom, P. Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol. 2004, 45, 1543–1556. [Google Scholar] [CrossRef] [Green Version]
- Davies, E.; Fillingham, B.D.; Oto, Y.; Abe, S. Evidence for the existence of cytoskeleton-bound polysomes in plants. Cell Biol. Int. Rep. 1991, 15, 973–981. [Google Scholar] [CrossRef]
- Pang, Q.; Chen, S.; Dai, S.; Chen, Y.; Wang, Y.; Yan, X. Comparative proteomics of salt tolerance in arabidopsis thaliana and thellungiella halophila. J. Proteome Res. 2010, 9, 2584–2599. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Dietz, K.J. Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Front. Plant Sci. 2016, 7, 548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rausell, A.; Kanhonou, R.; Yenush, L.; Serrano, R.; Ros, R. The translation initiation factor elF1A is an important determinant in the tolerance to NaCl stress in yeast and plants. Plant J. 2003, 34. [Google Scholar] [CrossRef] [Green Version]
- Shahollari, B.; Peskan-Berghöfer, T.; Oelmüller, R. Receptor kinases with leucine-rich repeats are enriched in Triton X-100 insoluble plasma membrane microdomains from plants. Physiol. Plant. 2004, 122, 397–403. [Google Scholar] [CrossRef]
- Borner, G.H.H.; Sherrier, D.J.; Weimar, T.; Michaelson, L.V.; Hawkins, N.D.; MacAskill, A.; Napier, J.A.; Beale, M.H.; Lilley, K.S.; Dupree, P. Analysis of detergent-resistant membranes in arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 2005, 137, 104–116. [Google Scholar] [CrossRef] [Green Version]
- Krügel, U.; He, H.X.; Gier, K.; Reins, J.; Chincinska, I.; Grimm, B.; Schulze, W.X.; Kühn, C. The potato sucrose transporter StSUT1 interacts with a DRM-associated protein disulfide isomerase. Mol. Plant 2012, 5, 43–62. [Google Scholar] [CrossRef] [Green Version]
- Jang, J.Y.; Kim, D.G.; Kim, Y.O.; Kim, J.S.; Kang, H. An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol. Biol. 2004, 54, 713–725. [Google Scholar] [CrossRef]
- Pou, A.; Jeanguenin, L.; Milhiet, T.; Batoko, H.; Chaumont, F.; Hachez, C. Salinity-mediated transcriptional and post-translational regulation of the Arabidopsis aquaporin PIP2;7. Plant Mol. Biol. 2016, 92, 731–744. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Ji, W.; Bai, X.; Cai, H.; Zhu, D.; Sun, X.L.; Chen, L.J.; Zhu, Y.M. A novel Glycine soja tonoplast intrinsic protein gene responds to abiotic stress and depresses salt and dehydration tolerance in transgenic Arabidopsis thaliana. J. Plant Physiol. 2011, 168, 1241–1248. [Google Scholar] [CrossRef] [PubMed]
- Larsson, C.; Widell, S.; Kjellbom, P. Preparation of high-purity plasma membranes. Methods Enzymol. 1987, 148, 558–568. [Google Scholar] [CrossRef]
- Widell, S.; Larsson, C. A Critical Evaluation of Markers Used in Plasma Membrane Purification. In The Plant Plasma Membrane; Springer: Berlin, Heidelberg/Germany, 1990; pp. 16–43. [Google Scholar]
- O’Neill, S.; Bennett, A.; Spanswick, R. Characterization of a NO3-Sensitive H+-ATPase from Corn Roots. Plant Physiol. 1983, 72, 837–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundborg, T.; Widell, S.; Larsson, C. Distribution of ATPases in wheat root membranes separated by phase partition. Physiol. Plant. 1981, 52, 89–95. [Google Scholar] [CrossRef]
- Hodges, T.K.; Leonard, R. Purification of a Plasma Membrane-Bound Adenosine Triphosphatase from Plant Roots. Methods Enzym. 1974, 36, 392–406. [Google Scholar]
- Mas, A.; Navarro-Pedreño, J.; Cooke, D.T.; James, C.S. Characterization and lipid composition of the plasma membrane in grape leaves. Phytochemistry 1994, 35, 1249–1253. [Google Scholar] [CrossRef]
- Barrajon-Catalan, E.; Menendez-Gutierrez, M.P.; Falco, A.; Carrato, A.; Saceda, M.; Micol, V. Selective death of human breast cancer cells by lytic immunoliposomes: Correlation with their HER2 expression level. Cancer Lett. 2010, 290, 192–203. [Google Scholar] [CrossRef]
- Maurel, C.; Tacnet, F.; Güclü, J.; Guern, J.; Ripoche, P. Purified vesicles of tobacco cell vacuolar and plasma membranes exhibit dramatically different water permeability and water channel activity. Proc. Natl. Acad. Sci. USA 1997, 94, 7103–7108. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, 506–515. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]








| Fraction | Treatment | % Fatty Acids | |||||
|---|---|---|---|---|---|---|---|
| Palmitoleic (C16:1) | Oleic (C18:1) | Linoleic (C18:2) | Linolenic (C18:3) | DBI | RUFA | ||
| Control | PM | 56.14 ± 8.18 a | 10.99 ± 0.04 ab | 29.02 ± 1.64 a | 1.96 ± 0.36 b | 129.40 ± 2.96 a | 1.77 ± 0.50 b |
| DRMs | 68.62 ± 10.46 a | 5.23 ± 1.92 b | 14.61 ± 4.46 ab | 11.53 ± 4.08 a | 136.28 ± 13.61 a | 5.01 ± 0.14 a | |
| NaCl | PM | 60.91 ± 6.43 a | 7.91 ± 3.24 b | 20.46 ± 5.54 a | 11.87 ± 2.67 a | 137.66 ± 2.22 a | 4.94 ± 1.27 a |
| DRMs | 53.62 ± 10.07 a | 34.77 ± 9.57 a | 5.77 ± 0.30 b | 7.21 ± 1.13 a | 117.4 ± 0.70 a | 0.38 ± 0.09 c | |
| UniProt Accession | Description | MW (Da) | Score | PSMs | Peptides | Coverage |
|---|---|---|---|---|---|---|
| Control | ||||||
| A0A178VIZ0 | PIP1-1 | 30,897 | 72 | 2 | 1 | 21 |
| A0A078IDF7 | PIP1-1 | 30,852 | 206 | 4 | 3 | 29.7 |
| A0A0D3C6T1 | PIP1-3 | 30,825 | 135 | 2 | 2 | 29.7 |
| A0A0D3DZM3 | PIP1-4 | 30,769 | 194 | 2 | 2 | 29.7 |
| F2X5K3 | PIP1-6 | 16,451 | 198 | 2 | 2 | 21.3 |
| A0A0D3B5C8 | PIP2-1 | 30,208 | 736 | 18 | 9 | 39.6 |
| O80369 | PIP2-1 | 30,670 | 380 | 8 | 6 | 39.7 |
| A0A0D3BRA6 | PIP2-2 | 30,512 | 137 | 3 | 2 | 39.3 |
| A0A0D3BRA5 | PIP2-2 | 30,495 | 108 | 2 | 2 | 39.3 |
| A0A0D3B2E7 | PIP2-4 | 30,293 | 466 | 7 | 6 | 37.9 |
| A0A078F7W0 | PIP2-4 | 27,697 | 151 | 2 | 2 | 35.1 |
| A0A0D3C4M6 | TIP1-1 | 25,635 | 59 | 3 | 2 | 6.8 |
| A0A078JDN1 | TIP2-2 | 25,148 | 106 | 2 | 2 | 7.2 |
| NaCl | ||||||
| A0A178VIZ0 | PIP1-1 | 30,897 | 61 | 2 | 1 | 21 |
| R0HDM0 | PIP1-1 | 30,779 | 273 | 4 | 4 | 29 |
| A0A078G0R2 | PIP1-2 | 30,792 | 961 | 19 | 13 | 35 |
| A0A078HJQ7 | PIP1-2 | 30,736 | 177 | 2 | 2 | 35 |
| A0A0D3BRA6 | PIP2-1 | 30,512 | 74 | 2 | 1 | 34 |
| O80369 | PIP2-1 | 30,670 | 293 | 6 | 4 | 29 |
| M4CLX8 | PIP2-2 | 25,497 | 59 | 2 | 2 | 27 |
| A0A0D3B2E7 | PIP2-4 | 30,293 | 472 | 7 | 7 | 38 |
| A0A078IUF9 | PIP2-7 | 29,949 | 91 | 3 | 2 | 30.2 |
| A0A0D3C4M6 | TIP1-1 | 25,635 | 130 | 3 | 3 | 7 |
| A0A078GXB0 | TIP2-1 | 24,983 | 205 | 3 | 3 | 7.3 |
| A0A078J7S3 | TIP2-3 | 25,348 | 165 | 6 | 2 | 7.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yepes-Molina, L.; Carvajal, M.; Martínez-Ballesta, M.C. Detergent Resistant Membrane Domains in Broccoli Plasma Membrane Associated to the Response to Salinity Stress. Int. J. Mol. Sci. 2020, 21, 7694. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21207694
Yepes-Molina L, Carvajal M, Martínez-Ballesta MC. Detergent Resistant Membrane Domains in Broccoli Plasma Membrane Associated to the Response to Salinity Stress. International Journal of Molecular Sciences. 2020; 21(20):7694. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21207694
Chicago/Turabian StyleYepes-Molina, Lucía, Micaela Carvajal, and Maria Carmen Martínez-Ballesta. 2020. "Detergent Resistant Membrane Domains in Broccoli Plasma Membrane Associated to the Response to Salinity Stress" International Journal of Molecular Sciences 21, no. 20: 7694. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21207694