Relationship of 2D Affinity to T Cell Functional Outcomes
Abstract
:1. Introduction
2. The Affinity Measurements of TCR
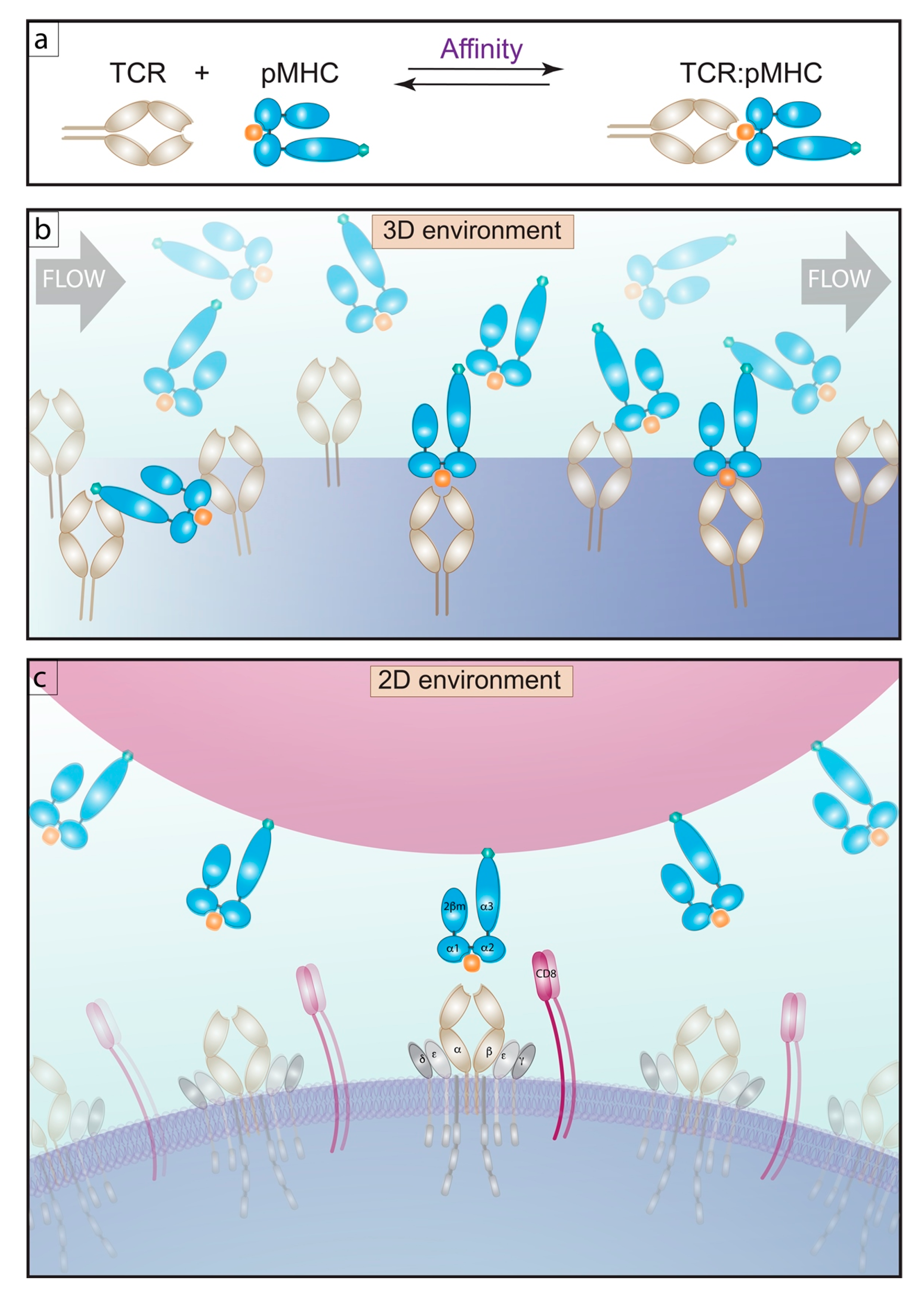
2.1. 3D Methods of Measuring Affinity
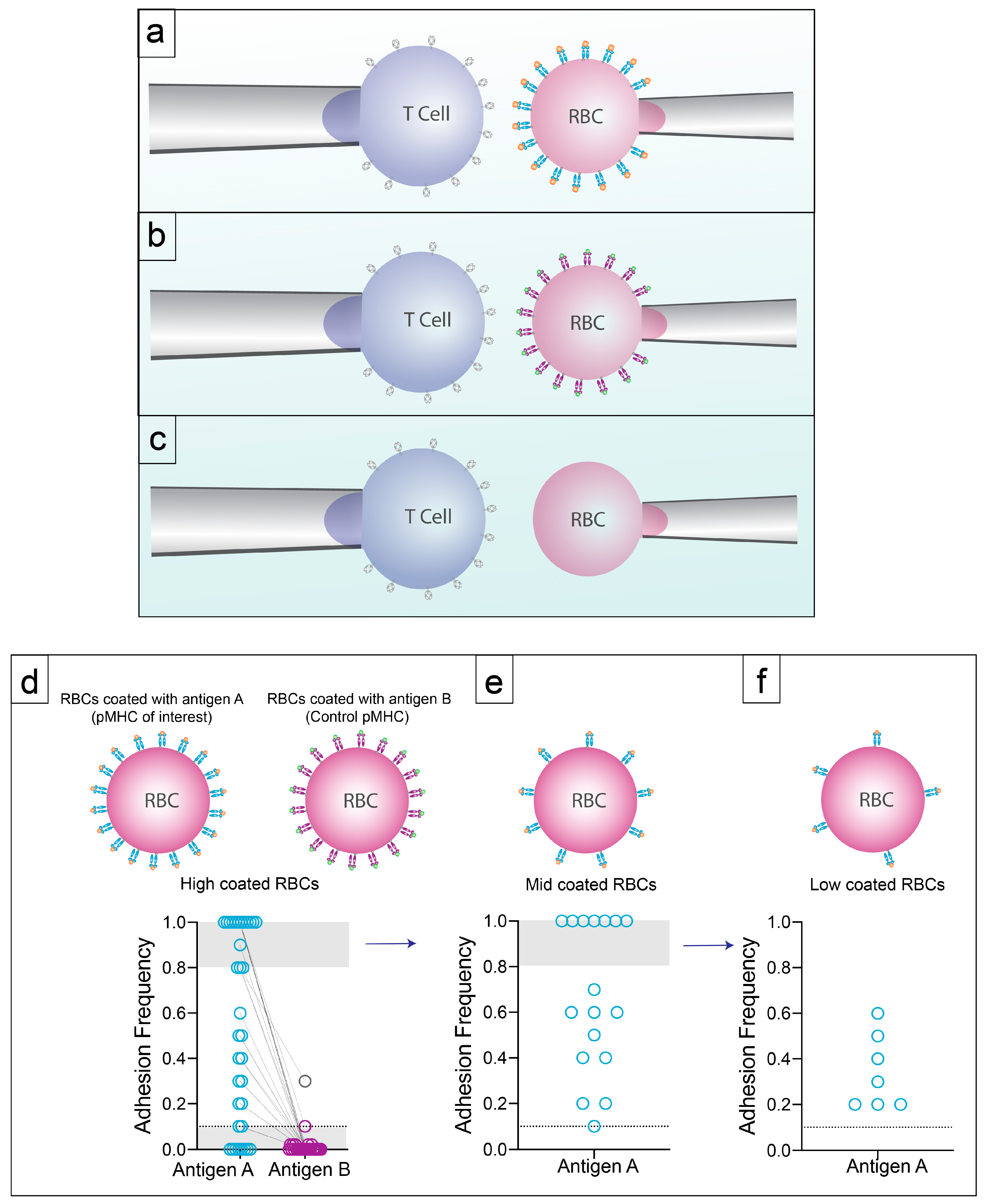
2.2. 2D Methods of Measuring Affinity
3. Are Lower Affinity T Cells Real?
3.1. Low Sensitivity Techniques
3.2. Specificity with High Sensitivity by 2D-Micropipette
4. Expansion and Frequency of Low Affinity CD4+ T Cells
5. The Affinity of TCR for pMHC Modulates TCR-Derived Signals
6. Factors Influencing Expansion of Lower Affinity T Cells in Addition to 2D Affinity of the TCR for pMHC
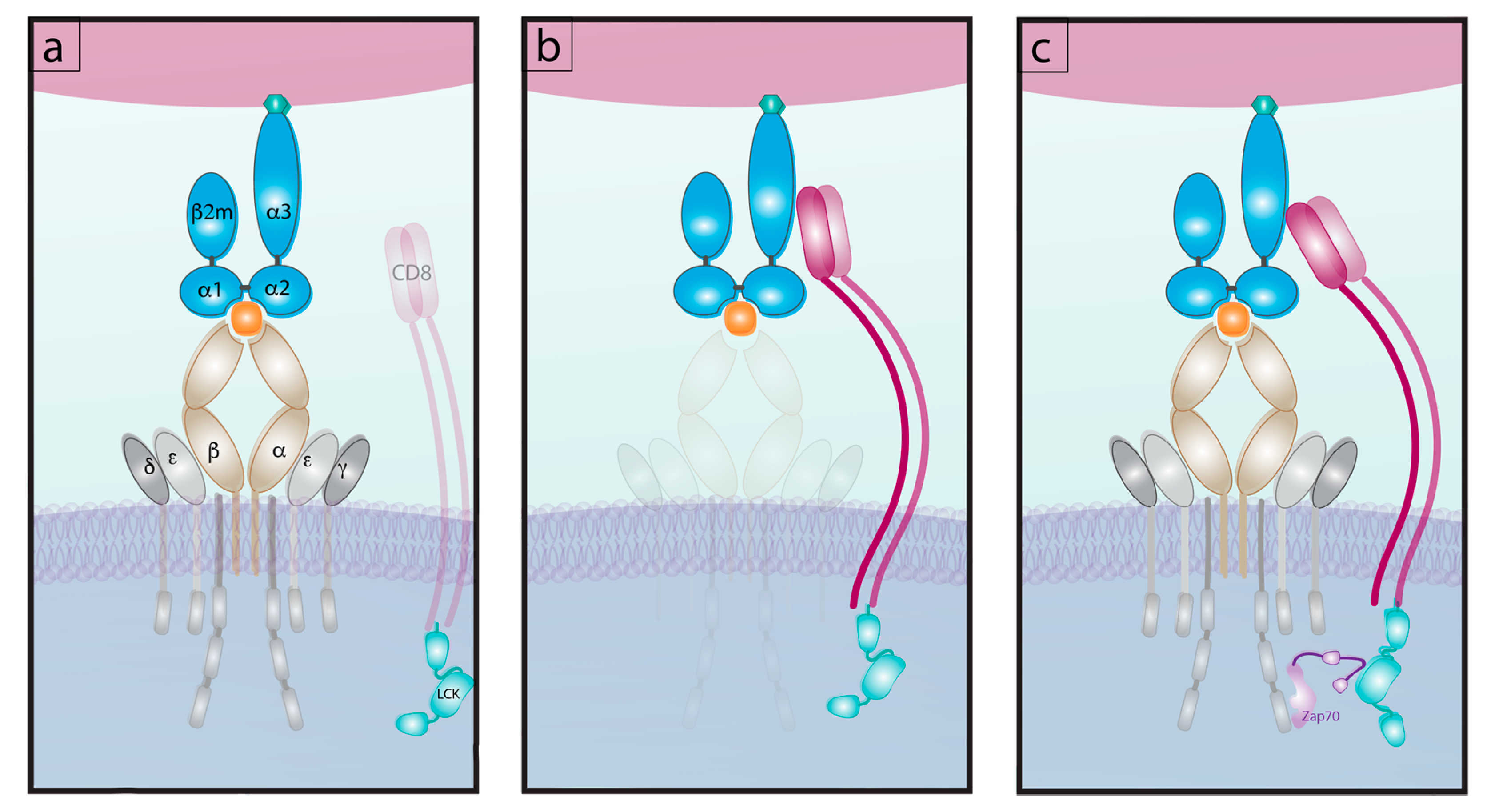
6.1. Differential Signaling from Co-Receptors
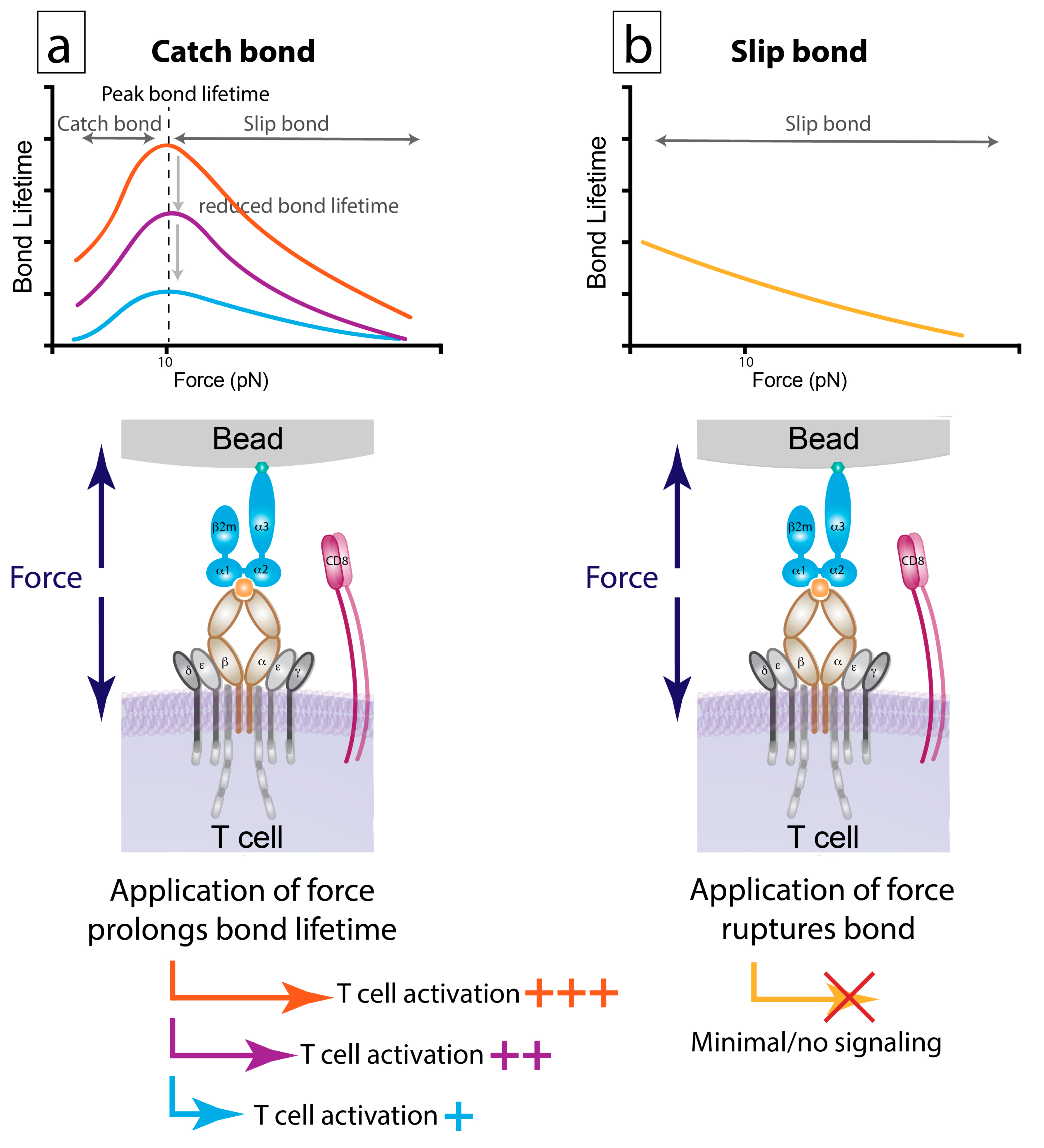
6.2. Bond Lifetime under Force: A Key Predictor of T Cell Function
7. Affinity Profile and CD4+ T Cell Effector Phenotypes
7.1. T Effector Cells (Teff)
7.2. T Follicular Helper Cells (Tfh)
7.3. Tregs
7.4. Memory T Cells
8. High and Low Affinity CD8+ T Cells
8.1. Effector CD8+ T Cells
8.2. Memory CD8+ T Cells
9. Conclusions
Funding
Conflicts of Interest
References
- Corse, E.; Gottschalk, R.A.; Allison, J.P. Strength of TCR-Peptide/MHC interactions and in vivo T cell responses. J. Immunol. 2011, 186, 5039–5045. [Google Scholar] [CrossRef] [PubMed]
- Van der Merwe, P.A.; Dushek, O. Mechanisms for T cell receptor triggering. Nature 2010, 11, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Krogsgaard, M.; Prado, N.; Adams, E.J.; He, X.L.; Chow, D.C.; Wilson, D.B.; Garcia, K.C.; Davis, M.M. Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation. Mol. Cell 2003, 12, 1367–1378. [Google Scholar] [CrossRef]
- Matsui, K.; Boniface, J.J.; Steffner, P.; Reay, P.A.; Davis, M.M. Kinetics of T-cell receptor binding to peptide/I-Ek complexes: Correlation of the dissociation rate with T-cell responsiveness. Proc. Natl. Acad. Sci. USA 1994, 91, 12862–12866. [Google Scholar] [CrossRef] [Green Version]
- Corr, M.; Slanetz, A.E.; Boyd, L.F.; Jelonek, M.T.; Khilko, S.; Al-ramadi, B.K. T cell receptor-MHC class I peptide interactions: Affinity, kinetics, and specificity. Science 1994, 265, 51–54. [Google Scholar] [CrossRef]
- Davis, M.M.; Krogsgaard, M.; Huppa, J.B.; Sumen, C.; Purbhoo, M.A.; Irvine, D.J.; Wu, L.C.; Ehrlich, L. Dynamics of cell surface molecules during T cell recognition. Annu. Rev. Biochem. 2003, 72, 717–742. [Google Scholar] [CrossRef]
- Irvine, D.; Purbhoo, M.; Krogsgaard, M.; Davis, M. Direct observation of ligand recognition by T cells. Nature 2002, 419, 845–849. [Google Scholar] [CrossRef] [PubMed]
- Purbhoo, M.A.; Irvine, D.J.; Huppa, J.B.; Davis, M.M. T cell killing does not require the formation of a stable mature immunological synapse. Nat. Immunol. 2004, 5, 524–530. [Google Scholar] [CrossRef]
- Harding, C.V.; Unanue, E.R. Quantitation of antigen-presenting cell MHC class II/peptide complexes necessary for T-cell stimulation. Nature 1990, 346, 574–576. [Google Scholar] [CrossRef]
- Germain, R.N.; Stefanová, I. The dynamics of T cell receptor signaling: Complex orchestration and the key roles of tempo and cooperation. Annu. Rev. Immunol. 1999, 17, 467–522. [Google Scholar] [CrossRef]
- Kersh, G.J.; Allen, P.M. Essential flexibility in the T cell recognition of antigen. Nature 1996, 380, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Jiang, N.; Ebert, P.J.R.; Kidd, B.A.; Müller, S.; Lund, P.J.; Juang, J.; Adachi, K.; Tse, T.; Birnbaum, M.E.; et al. Clonal deletion prunes but does not eliminate self-specific αβ CD8+ T lymphocytes. Immunity 2015, 42, 929–941. [Google Scholar] [CrossRef] [Green Version]
- Stritesky, G.L.; Jameson, S.C.; Hogquist, K.A. Selection of self-reactive T cells in the thymus. Annu. Rev. Immunol. 2012, 30, 95–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bielekova, B.; Sung, M.-H.; Kadom, N.; Simon, R.; McFarland, H.; Martin, R. Expansion and functional relevance of high-avidity myelin-specific CD4+ T cells in multiple sclerosis. J. Immunol. 2004, 172, 3893–3904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, K.C.; Radu, C.G.; Ho, J.; Ober, R.J.; Ward, E.S. Kinetics and thermodynamics of T cell receptor-autoantigen interactions in murine experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 2001, 98, 6818–6823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, J.D.; Chervin, A.S.; Kranz, D.M. T-cell receptor binding affinities and kinetics: Impact on T-cell activity and specificity. Immunology 2009, 126, 165–176. [Google Scholar] [CrossRef]
- Kersh, G.J.; Kersh, E.N.; Fremont, D.H.; Allen, P.M. High- and low-potency ligands with similar affinities for the TCR: The importance of kinetic in TCR signaling. Immunity 1998, 9, 817–826. [Google Scholar] [CrossRef] [Green Version]
- Alam, S.M.; Davies, G.M.; Lin, C.M.; Zal, T.; Nasholds, W.; Jameson, S.C.; Hogquist, K.A.; Gascoigne, N.R.J.; Travers, P.J.; Jolla, L.; et al. Qualitative and quantitative differences in T cell receptor binding of agonist and antagonist ligands. Immunity 1999, 10, 227–237. [Google Scholar] [CrossRef] [Green Version]
- Rosette, C.; Werlen, G.; Daniels, M.A.; Holman, P.O.; Alam, S.M.; Travers, P.J.; Gascoigne, N.R.J.; Palmer, E.; Jameson, S.C. The Impact of duration versus extent of TCR occupancy on T cell activation: A revision of the kinetic proofreading model. Immunity 2001, 15, 59–70. [Google Scholar] [CrossRef] [Green Version]
- Hebeisen, M.; Schmidt, J.; Guillaume, P.; Baumgaertner, P.; Speiser, D.E.; Luescher, I.; Rufer, N. Identification of rare high-avidity, tumor-reactive CD8 þ T cells by monomeric TCR-ligand off-rates measurements on living cells. Cancer Res. 2015, 75, 1983–1991. [Google Scholar] [CrossRef] [Green Version]
- Schmid, D.A.; Irving, M.B.; Posevitz, V.; Hebeisen, M.; Posevitz-Fejfar, A.; Sarria, J.C.F.; Gomez-Eerland, R.; Thome, M.; Schumacher, T.N.M.; Romero, P.; et al. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. J. Immunol. 2010, 184, 4936–4946. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.J.; Narayanan, S.; Liu, B.; Birnbaum, M.E.; Kruse, A.C.; Bowerman, N.A.; Chen, W.; Levin, A.M.; Connolly, J.M.; Zhu, C.; et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity 2011, 35, 681–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, S.; Malecek, K.; Johnson, L.A.; Yu, Z.; Vega-Saenz de Miera, E.; Darvishian, F.; McGary, K.; Huang, K.; Boyer, J.; Corse, E.; et al. T-cell receptor affinity and avidity defines antitumor response and autoimmunity in T-cell immunotherapy. Proc. Natl. Acad. Sci. USA 2013, 110, 6973–6978. [Google Scholar] [CrossRef] [Green Version]
- Alam, S.M.; Travers, P.J.; Wung, J.L.; Nasholds, W.; Redpath, S.; Jameson, S.C.; Gascoigne, N.R.J. T-cell-receptor affinity and thymocyte positive selection. Nature 1996, 381, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Morgan, A.J.; Stewart-jones, G.; Shepherd, D.; Bossi, G.; Wooldridge, L.; Hutchinson, L.; Sewell, A.K.; Griffiths, G.M.; Van Der Merwe, A.; et al. Ca 2+ release from the endoplasmic reticulum of NY-ESO-1-specific T cells is modulated by the affinity of TCR and by the use of the CD8 coreceptor. J. Immunol. 2010, 184, 1829–1839. [Google Scholar] [CrossRef] [Green Version]
- Gras, S.; Chadderton, J.; Del Campo, C.M.; Quinn, K.M.; Rossjohn, J.; La Gruta, N.L.; Gras, S.; Chadderton, J.; Del Campo, C.M.; Farenc, C.; et al. Reversed T cell receptor docking on a major histocompatibility class I complex limits involvement in the immune response reversed T cell receptor docking on a major histocompatibility class I complex limits involvement in the immune response. Immunity 2016, 45, 749–760. [Google Scholar] [CrossRef] [Green Version]
- Holmberg, K.; Mariathasan, S.; Ohteki, T. TCR binding kinetics measured with MHC class I tetramers reveal a positive selecting peptide with relatively high affinity for TCR. J. Immunol. 2003, 171, 2427–2434. [Google Scholar] [CrossRef] [Green Version]
- Van der Merwe, P.A. The TCR triggering puzzle. Immunity 2001, 14, 665–668. [Google Scholar] [CrossRef] [Green Version]
- Lyons, D.S.; Lieberman, S.A.; Hampl, J.; Boniface, J.J.; Chien, Y.H.; Berg, L.J.; Davis, M.M. A TCR binds to antagonist ligands with lower affinities and faster dissociation rates than to agonists. Immunity 1996, 5, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Al-Ramadi, B.K.; Jelonek, M.T.; Boyd, L.F.; Margulies, D.H.; Bothwell, A.L. Lack of strict correlation of functional sensitization with the apparent affinity of MHC/peptide complexes for the TCR. J. Immunol. 1995, 155, 662–673. [Google Scholar]
- Allison, K.A.; Sajti, E.; Collier, J.G.; Gosselin, D.; Troutman, T.D.; Stone, E.L.; Hedrick, S.M.; Glass, C.K. Affinity and dose of TCR engagement yield proportional enhancer and gene activity in CD4+ T cells. eLife 2016, 5, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, R.A.; Hathorn, M.M.; Beuneu, H.; Corse, E.; Dustin, M.L. Distinct in fl uences of peptide-MHC quality and quantity on in vivo T-cell responses. Proc. Natl. Acad. Sci. USA 2012, 109, 881–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Govern, C.C.; Paczosa, M.K.; Chakraborty, A.K.; Huseby, E.S. Fast on-rates allow short dwell time ligands to activate T cells. Proc. Natl. Acad. Sci. USA 2010, 107, 8724–8729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, S.; Maile, R.; Collins, E.J.; Frelinger, J.A. CD8+ T cell activation is governed by TCR-peptide/MHC affinity, not dissociation rate. J. Immunol. 2007, 179, 2952–2960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Zarnitsyna, V.I.; Liu, B.; Edwards, L.J.; Jiang, N.; Evavold, B.D.; Zhu, C. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 2010, 464, 932–936. [Google Scholar] [CrossRef] [Green Version]
- Kalergis, A.H.; Boucheron, N.; Doucey, M.A.; Palmieri, E.; Goyarts, E.C.; Vegh, Z.; Luescher, I.F.; Nathenson, S.G. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat. Immunol. 2001, 2, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Zehn, D.; Lee, S.Y.; Bevan, M.J. Complete but curtailed T-cell response to very low-affinity antigen. Nature 2009, 458, 211–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coombs, D.; Mora, J.E.; Palmieri, E.; Goldstein, B.; Gonza, P.A.; Nathenson, S.G.; Kalergis, A.M. T cell receptor binding kinetics required for T cell activation depend on the density of cognate ligand on the antigen-presenting cell. Proc. Natl. Acad. Sci. USA 2005, 102, 4824–4829. [Google Scholar] [CrossRef] [Green Version]
- Davis, M.M.; Boniface, J.J.; Reich, Z.; Lyons, D.; Hampl, J.; Arden, B.; Chien, Y. Ligand recognition by Aβ T cell receptors. Annu. Rev. Immunol. 1998, 16, 523–544. [Google Scholar] [CrossRef] [PubMed]
- Tolentino, T.P.; Wu, J.; Zarnitsyna, V.I.; Fang, Y.; Dustin, M.L.; Zhu, C. Measuring diffusion and binding kinetics by contact area FRAP. Biophys. J. 2008, 95, 920–930. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Fang, Y.; Zarnitsyna, V.I.; Tolentino, T.P.; Dustin, M.L.; Zhu, C. A coupled diffusion-kinetics model for analysis of contact-area FRAP experiment. Biophys. J. 2008, 95, 910–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huppa, J.B.; Axmann, M.; Mörtelmaier, M.A.; Lillemeier, B.F.; Newell, E.W.; Brameshuber, M.; Klein, L.O.; Schütz, G.J.; Davis, M.M. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 2010, 463, 963–967. [Google Scholar] [CrossRef] [Green Version]
- Edwards, L.J.; Zarnitsyna, V.I.; Hood, J.D.; Evavold, B.D.; Zhu, C. Insights into T cell recognition of antigen: Significance of two-dimensional kinetic parameters. Front. Immunol. 2012, 3, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanchfield, J.; Shorter, S.K.; Evavold, B.D. Monitoring the dynamics of T cell clonal diversity using recombinant peptide:MHC technology. Front. Immunol. 2013, 4, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.-Q.; Parker, P.; Ma, K.-Y.; He, C.; Shi, Q.; Cui, Z.; Williams, C.M.; Wendel, B.S.; Meriwether, A.I.; Salazar, M.A.; et al. Direct measurement of T cell receptor affinity and sequence from naive antiviral T cells. Sci. Transl. Med. 2016, 8, 341ra77. [Google Scholar] [CrossRef] [Green Version]
- Seo, Y.J.; Jothikumar, P.; Suthar, M.S.; Zhu, C.; Grakoui, A. Local cellular and cytokine cues in the spleen regulate in situ T cell receptor affinity, function, and fate of CD8+ T cells. Immunity 2016, 45, 988–998. [Google Scholar] [CrossRef] [Green Version]
- Chesla, S.E.; Selvaraj, P.; Zhu, C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys. J. 1998, 75, 1553–1572. [Google Scholar] [CrossRef] [Green Version]
- Harrison, D.L.; Fang, Y.; Huang, J. T-cell mechanobiology: Force sensation, potentiation, and translation. Front. Phys. 2019, 7, 1–18. [Google Scholar] [CrossRef]
- Huang, J.; Zeng, X.; Sigal, N.; Lund, P.J.; Su, L.F.; Huang, H.; Chien, Y.; Davis, M.M. Detection, phenotyping, and quantification of antigen-specific T cells using a peptide-MHC dodecamer. Proc. Natl. Acad. Sci. USA 2016, 113, E1890–E1897. [Google Scholar] [CrossRef] [Green Version]
- Batard, P.; Peterson, D.A.; Devêvre, E.; Guillaume, P.; Cerottini, J.; Rimoldi, D.; Speiser, D.E.; Winther, L.; Romero, P. Dextramers: New generation of fluorescent MHC class I/peptide multimers for visualization of antigen-specific CD8 + T cells. J. Immunol. Methods 2006, 310, 136–148. [Google Scholar] [CrossRef]
- Dolton, G.; Lissina, A.; Ladell, K.; Tungatt, K.; Jones, E.; Kronenberg-Versteeg, D.; Akpovwa, H.; Pentier, J.; Holland, C.; Godkin, A.; et al. Comparison of peptide-major histocompatibility complex tetramers and dextramers for the identification of antigen-specific T cells. Clin. Exp. Immunol. 2014, 177, 47–63. [Google Scholar] [CrossRef]
- Altman, A.J.D.; Moss, P.A.H.; Goulder, P.J.R.; Barouch, D.H.; Mcheyzer-williams, M.G.; Bell, J.I. Phenotypic analysis of antigen-specific T lymphocytes. Science 1996, 20–22. [Google Scholar] [CrossRef]
- Kim, C.; Wilson, T.; Fischer, K.F.; Williams, M.A. Sustained interactions between T cell receptors and antigens promote the differentiation of CD4+ memory T cells. Immunity 2013, 39, 508–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, F.; Kozono, H.; White, J.; Marrack, P.; Kappler, J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity 1998, 8, 675–682. [Google Scholar] [CrossRef] [Green Version]
- Wooldridge, L.; Wooldridge, L.; Cole, D.K.; Van Den Berg, H.A.; Price, D.A.; Sewell, A.K. Tricks with tetramers: How to get the most from multimeric peptide—MHC. Immunology 2009, 126, 147–164. [Google Scholar] [CrossRef] [PubMed]
- Wooldridge, L.; Van Den Berg, H.A.; Glick, M.; Gostick, E.; Laugel, B.; Hutchinson, S.L.; Milicic, A.; Brenchley, J.M.; Douek, D.C.; Price, D.A.; et al. Interaction between the CD8 coreceptor and major histocompatibility complex class I stabilizes T cell receptor-antigen complexes at the cell surface. J. Biol. Chem. 2005, 280, 27491–27501. [Google Scholar] [CrossRef] [Green Version]
- Holler, P.D.; Kranz, D.M. Quantitative analysis of the contribution of TCR/pepMHC affinity and CD8 to T cell activation. Immunity 2003, 18, 255–264. [Google Scholar] [CrossRef] [Green Version]
- Boniface, J.J.; Rabinowitz, J.D.; Wu, C.; Hampl, J.; Reich, Z.; Altman, J.D.; Kantor, R.M.; Beeson, C.; Mcconnell, H.M.; Davis, M.M. Initiation of signal transduction through the T cell receptor requires the multivalent engagement of peptide/MHC ligands. Immunity 1998, 9, 459–466. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Kern, P.; Chang, H.; Reinherz, E.L. T Cell receptor binding to a pMHCII ligand Is kinetically distinct from and independent of CD4 *. J. Biol. Chem. 2001, 276, 5659–5667. [Google Scholar] [CrossRef] [Green Version]
- Masopust, D.; Murali-krishna, K.; Ahmed, R. Quantitating the magnitude of the lymphocytic choriomeningitis virus-specific CD8 T-cell response: It is even bigger than we thought. J. Virol. 2007, 81, 2002–2011. [Google Scholar] [CrossRef] [Green Version]
- Stone, J.D.; Cochran, J.R.; Stern, L.J. T-cell activation by soluble MHC oligomers can be described by a two-parameter binding model. Biophys. J. 2001, 81, 2547–2557. [Google Scholar] [CrossRef] [Green Version]
- Rius, C.; Attaf, M.; Tungatt, K.; Bianchi, V.; Legut, M.; Bovay, A.; Donia, M.; thor Straten, P.; Peakman, M.; Svane, I.M.; et al. Peptide–MHC class I tetramers can fail to detect relevant functional T cell clonotypes and underestimate antigen-reactive T cell populations. J. Immunol. 2018, 200, 2263–2279. [Google Scholar] [CrossRef]
- Sabatino, J.J.; Huang, J.; Zhu, C.; Evavold, B.D. High prevalence of low affinity peptide–MHC II tetramer–negative effectors during polyclonal CD4+ T cell responses. J. Exp. Med. 2011, 208, 81–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kersh, A.E.; Edwards, L.J.; Evavold, B.D. Progression of relapsing-remitting demyelinating disease does not require increased TCR affinity or epitope spread. J. Immunol. 2014, 193, 4429–4438. [Google Scholar] [CrossRef] [Green Version]
- Krummey, S.M.; Morris, A.B.; Jacobs, J.R.; Evavold, B.D.; Kissick, H.T.; Ford, M.L. CD45RB status of CD8+ T cell memory defines T cell receptor affinity and persistence report CD45RB status of CD8+ T cell memory defines T cell receptor. Cell Rep. 2020, 30, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Minguet, S.; Ortı, A.R.; Schamel, W.W.A.; Risuen, R.M. A conformation- and avidity-based proofreading mechanism for the TCR—CD3 complex. Trends Immunol. 2006, 27, 176–182. [Google Scholar] [CrossRef]
- Dolton, G.; Tungatt, K.; Lloyd, A.; Theaker, S.M.; Trimby, A.; Christopher, J.; Donia, M.; Andrew, J.; Cole, D.K.; Thor, P.; et al. More tricks with tetramers: A practical guide to staining T cells with peptide—MHC multimers. Immunology 2015, 146, 11–22. [Google Scholar] [CrossRef]
- Evans, E.; Leung, A.; Heinrich, V.; Zhu, C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc. Natl. Acad. Sci. USA 2004, 101, 11281–11286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andargachew, R.; Martinez, R.J.; Kolawole, E.M.; Evavold, B.D. CD4 T cell affinity diversity is equally maintained during acute and chronic infection. J. Immunol. 2018, 201, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Blanchfield, L.; Sabatino, J.J.; Lawrence, L.; Evavold, B.D. NFM cross-reactivity to MOG does not expand a critical threshold level of high-affinity T cells necessary for onset of demyelinating disease. J. Immunol. 2017, 199, 2680–2691. [Google Scholar] [CrossRef]
- Liu, B.; Hood, J.D.; Kolawole, E.M.; Woodruff, D.M.; Vignali, D.A.; Bettini, M.; Evavold, B.D. A hybrid insulin epitope maintains high 2D affinity for diabetogenic T cells in the periphery. Diabetes 2020, 69, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Jiang, N.; Zarnitsyna, V.I.; Evavold, B.D. Insights from in situ analysis of TCR-pMHC recognition: Response of an interaction network. Immunol. Rev. 2013, 251, 49–64. [Google Scholar] [CrossRef] [Green Version]
- Savage, P.A.; Boniface, J.J.; Davis, M.M. A kinetic basis for T cell receptor repertoire selection during an immune response. Immunity 1999, 10, 485–492. [Google Scholar] [CrossRef] [Green Version]
- Koehli, S.; Naeher, D.; Galati-Fournier, V.; Zehn, D.; Palmer, E. Optimal T-cell receptor affinity for inducing autoimmunity. Proc. Natl. Acad. Sci. USA 2014, 111, 17248–17253. [Google Scholar] [CrossRef] [Green Version]
- King, C.G.; Koehli, S.; Hausmann, B.; Schmaler, M.; Zehn, D.; Palmer, E. T cell affinity regulates asymmetric division, effector cell differentiation, and tissue pathology. Immunity 2012, 37, 709–720. [Google Scholar] [CrossRef] [Green Version]
- Krummey, S.M.; Martinez, R.J.; Andargachew, R.; Liu, D.; Wagener, M.; Kohlmeier, J.E.; Evavold, B.D.; Larsen, C.P.; Ford, M.L. Low-affinity memory CD8+ T cells mediate robust heterologous immunity. J. Immunol. 2016, 196, 2838–2846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laugel, B.; Van Den Berg, H.A.; Gostick, E.; Cole, D.K.; Wooldridge, L.; Boulter, J.; Milicic, A.; Price, D.A.; Sewell, A.K. Different T cell receptor affinity thresholds and CD8 coreceptor dependence govern cytotoxic T lymphocyte activation and tetramer binding properties. J. Biol. Chem. 2007, 282, 23799–23810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tungatt, K.; Bianchi, V.; Crowther, M.D.; Powell, W.E.; Schauenburg, A.J.; Trimby, A.; Donia, M.; Miles, J.J.; Holland, C.J.; Cole, D.K.; et al. Antibody stabilization of peptide–MHC multimers reveals functional T cells bearing extremely low-affinity TCRs. J. Immunol. 2015, 194, 463–474. [Google Scholar] [CrossRef]
- Gebe, J.A.; Falk, B.A.; Rock, K.A.; Kochik, S.A.; Heninger, A.K.; Reijonen, H.; Kwok, W.W.; Nepom, G.T. Low-avidity recognition by CD4+ T cells directed to self-antigens. Eur. J. Immunol. 2003, 33, 1409–1417. [Google Scholar] [CrossRef] [PubMed]
- Falta, M.T.; Fontenot, A.P.; Rosloniec, E.F.; Crawford, F.; Roark, C.L.; Bill, J.; Marrack, P.; Kappler, J.; Kotzin, B.L. Class II major histocompatibility complex—Peptide tetramer staining in relation to functional avidity and T cell receptor diversity in the mouse CD4+ T cell response to a rheumatoid arthritis—Associated antigen. Arthritis Rheum. 2005, 52, 1885–1896. [Google Scholar] [CrossRef]
- Hood, J.D.; Zarnitsyna, V.I.; Zhu, C.; Evavold, B.D. Regulatory and T effector cells have overlapping low to high ranges in TCR affinities for self during demyelinating disease. J. Immunol. 2015, 195, 4162–4170. [Google Scholar] [CrossRef] [PubMed]
- Bettini, M.; Scavuzzo, M.A.; Liu, B.; Kolawole, E.; Guo, L.; Evavold, B.D.; Borowiak, M.; Bettini, M.L. A critical insulin TCR contact residue selects high-affinity and pathogenic insulin-specific T cells. Diabetes 2020, 69, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.W.; Beisang, D.; Tubo, N.J.; Dileepan, T.; Wiesner, D.L.; Nielsen, K.; Wüthrich, M.; Klein, B.S.; Kotov, D.I.; Spanier, J.A.; et al. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity 2015, 42, 95–107. [Google Scholar] [CrossRef] [Green Version]
- Martinez, R.J.; Andargachew, R.; Martinez, H.A.; Evavold, B.D. Low-affinity CD4 T cells are major responders in the primary immune response. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Rosenthal, K.M.; Edwards, L.J.; Sabatino, J.J.; Hood, J.D.; Wasserman, H.A.; Zhu, C.; Evavold, B.D. Low 2-dimensional CD4 T cell receptor affinity for myelin sets in motion delayed response kinetics. PLoS ONE 2012, 7, e32562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schubert, D.A.; Gordo, S.; Sabatino, J.J.; Vardhana, S.; Gagnon, E.; Sethi, D.K.; Seth, N.P.; Choudhuri, K.; Reijonen, H.; Nepom, G.T.; et al. Self-reactive human CD4 T cell clones form unusual immunological synapses. J. Exp. Med. 2012, 209, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Bettini, M.; Blanchfield, L.; Castellaw, A.; Zhang, Q.; Nakayama, M.; Smeltzer, M.P.; Zhang, H.; Hogquist, K.A.; Evavold, B.D.; Vignali, D.A.A. TCR affinity and tolerance mechanisms converge to shape T cell diabetogenic potential. J. Immunol. 2014, 193, 571–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Chen, W.; Evavold, B.D.; Zhu, C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 2014, 157, 357–368. [Google Scholar] [CrossRef] [Green Version]
- Pryshchep, S.; Zarnitsyna, V.I.; Hong, J.; Evavold, B.D.; Zhu, C. Accumulation of serial forces on TCR and CD8 frequently applied by agonist antigenic peptides embedded in MHC molecules triggers calcium in T cells. J. Immunol. 2014, 193, 68–76. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Blanchfield, L.; Ma, V.P.-Y.; Andargachew, R.; Galior, K.; Liu, Z.; Evavold, B.; Salaita, K. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc. Natl. Acad. Sci. USA 2016, 113, 5610–5615. [Google Scholar] [CrossRef] [Green Version]
- Artyomov, M.N.; Lis, M.; Devadas, S.; Davis, M.M.; Chakraborty, A.K. CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc. Natl. Acad. Sci. USA 2010, 107, 16916–16921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Edwards, L.J.; Evavold, B.D.; Zhu, C. Kinetics of MHC-CD8 interaction at the T cell membrane. J. Immunol. 2007, 179, 7653–7662. [Google Scholar] [CrossRef] [Green Version]
- Kolawole, E.M.; Andargachew, R.; Liu, B.; Jacobs, J.R.; Evavold, B.D. 2D kinetic analysis of TCR and CD8 coreceptor for LCMV GP33 epitopes. Front. Immunol. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Jiang, N.; Huang, J.; Edwards, L.J.; Liu, B.; Zhang, Y.; Beal, C.D.; Evavold, B.D.; Zhu, C. Two-stage cooperative T cell receptor-peptide major histocompatibility complex-CD8 trimolecular interactions amplify antigen discrimination. Immunity 2011, 34, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Aleksic, M.; Dushek, O.; Zhang, H.; Shenderov, E.; Chen, J.; Cerundolo, V.; Coombs, D.; Van Der Merwe, P.A. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity 2010, 32, 163–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klammt, C.; Novotná, L.; Li, D.T.; Wolf, M.; Blount, A.; Zhang, K.; Fitchett, J.R.; Lillemeier, B.F. T cell receptor dwell times control the kinase activity of Zap70. Nat. Immunol. 2015, 16, 961–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadinski, B.D.; Blevins, S.J.; Spidale, N.A.; Duke, B.R.; Huseby, P.G.; Stern, L.J.; Huseby, E.S. A temporal thymic selection switch and ligand binding kinetics constrain neonatal Foxp3+ Treg cell development. Nat. Immunol. 2019, 20, 1046–1058. [Google Scholar] [CrossRef]
- Huse, M. Mechanical forces in the immune system. Nat. Rev. Immunol. 2017, 17, 679–690. [Google Scholar] [CrossRef]
- Depoil, D.; Dustin, M.L. Force and affinity in ligand discrimination by the TCR. Trends Immunol. 2014, 35, 597–603. [Google Scholar] [CrossRef]
- Cost, A.L.; Ringer, P.; Chrostek-Grashoff, A.; Grashoff, C. How to measure molecular forces in cells: A guide to evaluating genetically-encoded FRET-based tension sensors. Cell. Mol. Bioeng. 2015, 8, 96–105. [Google Scholar] [CrossRef] [Green Version]
- Hu, K.H.; Butte, M.J. T cell activation requires force generation. J. Cell Biol. 2016, 213, 535–542. [Google Scholar] [CrossRef]
- Hosseini, B.H.; Louban, I.; Djandji, D.; Wabnitz, G.H.; Janosch, D.; Bulbuc, N.; Samstag, Y.; GUnzer, M.; Spatz, J.P.; Hammerling, G.J. Immune synapse formation determines interaction forces between T cells and antigen-presenting cells measured by atomic force microscopy. Proc. Natl. Acad. Sci. USA 2009, 106, 17852–17857. [Google Scholar] [CrossRef] [Green Version]
- Puech, P.H.; Nevoltris, D.; Robert, P.; Limozin, L.; Boyer, C.; Bongrand, P. Force measurements of TCR/pMHC recognition at T cell surface. PLoS ONE 2011, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Das, D.K.; Feng, Y.; Mallis, R.J.; Li, X.; Keskin, D.B.; Hussey, R.E.; Brady, S.K.; Wang, J.-H.; Wagner, G.; Reinherz, E.L.; et al. Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc. Natl. Acad. Sci. USA 2015, 112, 1517–1522. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Persaud, S.P.; Horvath, S.; Allen, P.M.; Evavold, B.D.; Zhu, C. Force-regulated in situ TCR–peptide-bound MHC class II kinetics determine functions of CD4+ T cells. J. Immunol. 2015, 195, 3557–3564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Brazin, K.N.; Kobayashi, E.; Mallis, R.J.; Reinherz, E.L.; Lang, M.J. Mechanosensing drives acuity of αβ T-cell recognition. Proc. Natl. Acad. Sci. USA 2017, 114, E8204–E8213. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Ge, C.; Jothikumar, P.; Yuan, Z.; Liu, B.; Bai, K.; Li, K.; Rittase, W.; Shinzawa, M.; Zhang, Y.; et al. A TCR mechanotransduction signaling loop induces negative selection in the thymus. Nat. Immunol. 2018, 19, 1379–1392. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Chen, W.; Natarajan, K.; Li, Z.; Margulies, D.H.; Zhu, C. The cellular environment regulates in situ kinetics of T-cell receptor interaction with peptide major histocompatibility complex. Eur. J. Immunol. 2015, 45, 2099–2110. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, T.; Liu, B.; Zhu, C.; Chen, W. Mechano-regulation of peptide-MHC class I conformations determines TCR antigen article mechano-regulation of peptide-MHC class I conformations determines. Mol. Cell 2019, 73, 1015–1027. [Google Scholar] [CrossRef] [Green Version]
- Sibener, L.V.; Fernandes, R.A.; Kolawole, E.M.; Carbone, C.B.; Liu, F.; McAffee, D.; Birnbaum, M.E.; Yang, X.; Su, L.F.; Yu, W.; et al. Isolation of a structural mechanism for uncoupling T cell receptor signaling from peptide-MHC binding. Cell 2018, 174, 672–687.e27. [Google Scholar] [CrossRef]
- Iezzi, G.; Karjalainen, K.; Lanzavecchia, A. The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity 1998, 8, 89–95. [Google Scholar] [CrossRef] [Green Version]
- Tuosto, L.; Acuto, O. CD28 affects the earliest signaling events generated by TCR engagement. Eur. J. Immunol. 1998, 28, 2131–2142. [Google Scholar] [CrossRef]
- Garcia, K.C.; Scott, C.A.; Brunmark, A.; Carbone, F.R.; Peterson, P.A.; Wilson, I.A.; Teyton, L. CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature 1996, 384, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Carbone, C.B.; Kern, N.; Fernandes, R.A.; Hui, E.; Su, X.; Garcia, K.C.; Vale, R.D. In vitro reconstitution of T cell receptor-mediated segregation of the CD45 phosphatase. Proc. Natl. Acad. Sci. USA 2017, 114, E9338–E9345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniels, M.A.; Teixeiro, E. TCR signaling in T cell memory. Front. Immunol. 2015, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.T.; Takeuchi, K.; Sun, Z.Y.J.; Touma, M.; Castro, C.E.; Fahmy, A.; Lang, M.J.; Wagner, G.; Reinherz, E.L. The αβ T cell receptor is an anisotropic mechanosensor. J. Biol. Chem. 2009, 284, 31028–31037. [Google Scholar] [CrossRef] [Green Version]
- Gourley, T.S.; Wherry, E.J.; Masopust, D.; Ahmed, R. Generation and maintenance of immunological memory. Semin. Immunol. 2004, 16, 323–333. [Google Scholar] [CrossRef]
- Bettelli, E.; Pagany, M.; Weiner, H.L.; Linington, C.; Sobel, R.A.; Kuchroo, V.K. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J. Exp. Med. 2003, 197, 1073–1081. [Google Scholar] [CrossRef] [Green Version]
- Alli, R.; Nguyen, P.; Geiger, T. Retrogenic modeling of experimental allergic encephalomyelitis associates T-cell frequency but not T-cell receptor functional affinity with pathogenicity. J. Immunol. 2008, 181, 136–145. [Google Scholar] [CrossRef]
- Yu, D.; Rao, S.; Tsai, L.M.; Lee, S.K.; He, Y.; Sutcliffe, E.L.; Srivastava, M.; Linterman, M.; Zheng, L.; Simpson, N.; et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 2009, 31, 457–468. [Google Scholar] [CrossRef]
- Nurieva, R.I.; Chung, Y.; Martinez, G.J.; Yang, X.O.; Tanaka, S.; Matskevitch, T.D.; Wang, Y.; Dong, C. Bcl6 mediates the development of T follicular helper cells. Science 2009, 325, 1001–1005. [Google Scholar] [CrossRef] [Green Version]
- Differentiation, H.C.; Action, C.; Cell-dependent, A.; Palmer, E.M.; Seventer, C. A Van Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 2009, 325, 1006–1010. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.S.; Kageyama, R.; Eto, D.; Escobar, T.C.; Johnston, R.J.; Monticelli, L.; Lao, C.; Crotty, S. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 2011, 34, 932–946. [Google Scholar] [CrossRef] [Green Version]
- Fazilleau, N.; Mcheyzer-williams, L.J.; Rosen, H.; Mcheyzer, M.G. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat. Immunol. 2009, 10, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Ditoro, D.; Winstead, C.; Pham, D.; Witte, S.; Andargachew, R.; Singer, J.R.; Wilson, C.G.; Zindl, C.L.; Luther, R.J.; Silberger, D.J.; et al. Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science 2018, 361. [Google Scholar] [CrossRef]
- Ren, H.M.; Kolawole, E.M.; Ren, M.; Jin, G.; Netherby-Winslow, C.S.; Wade, Q.; Shwetank; Rahman, Z.S.M.; Evavold, B.D.; Lukacher, A.E. IL-21 from high-affinity CD4 T cells drives differentiation of brain-resident CD8 T cells during persistent viral infection. Sci. Immunol. 2020, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Moran, A.E.; Hogquist, K. T-cell receptor affinity in thymic development. Immunology 2011, 135, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.; Zheng, Y.; Liang, Y.; Fontenot, J.D.; Rudensky, A.Y. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat. Immunol. 2006, 7, 401–410. [Google Scholar] [CrossRef]
- Pacholczyk, R.; Kern, J. The T-cell receptor repertoire of regulatory T cells. Immunology 2008, 125, 450–458. [Google Scholar] [CrossRef]
- Sprouse, M.L.; Bettini, M.L.; Bettini, M.; Sprouse, M.L.; Scavuzzo, M.A.; Blum, S.; Shevchenko, I.; Lee, T.; Makedonas, G.; Borowiak, M.; et al. High self-reactivity drives T-bet and potentiates Treg function in tissue-specific autoimmunity Find the latest version: High self-reactivity drives T-bet and potentiates treg function in tissue-specific autoimmunity. JCI Insight 2018, 3, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Frost, E.L.; Kersh, A.E.; Evavold, B.D.; Lukacher, A.E. Cutting edge: Resident memory CD8 T cells express high-affinity TCRs. J. Immunol. 2015, 195, 3520–3524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, L.F.; Kidd, B.A.; Han, A.; Kotzin, J.J.; Davis, M.M. Virus-specific CD4+ memory-phenotype T cells are abundant in unexposed adults. Immunity 2013, 38, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020, 94, eabd3871. [Google Scholar] [CrossRef] [PubMed]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Crotty, S. Pre-existing immunity to SARS-CoV-2: The knowns and unknowns. Nat. Rev. Immunol. 2020, 20, 457–458. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef]
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.A.; Endeman, H.; van den Akker, J.P.C.; Molenkamp, R.; Koopmans, M.P.G.; van Gorp, E.C.M.; et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5, 1–14. [Google Scholar] [CrossRef]
- Cole, D.K.; Bulek, A.M.; Dolton, G.; Schauenberg, A.J.; Szomolay, B.; Rittase, W.; Trimby, A.; Jothikumar, P.; Fuller, A.; Skowera, A.; et al. Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. J. Clin. Investig. 2016, 126, 2191–2204. [Google Scholar] [CrossRef] [Green Version]
- Moore, T.; Wagner, C.R.; Scurti, G.M.; Hutchens, K.A.; Godellas, C.; Clark, A.L.; Kolawole, E.M.; Lance, M.; Singh, N.K.; Huyke, F.A.; et al. Clinical and immunologic evaluation of three metastatic melanoma patients treated with autologous melanoma-reactive TCR-transduced T cells. Cancer Immunol. Immunother. 2018, 67, 311–325. [Google Scholar] [CrossRef]
- Knudson, K.M.; Goplen, N.P.; Cunningham, C.A.; Daniels, M.A.; Teixeiro, E. Low-affinity T cells are programmed to maintain normal primary responses but are impaired in their recall to low-affinity ligands. Cell Rep. 2013, 4, 554–565. [Google Scholar] [CrossRef] [Green Version]
- Sanecka, A.; Yoshida, N.; Kolawole, E.M.; Patel, H.; Evavold, B.D.; Frickel, E.-M. T cell receptor–major histocompatibility complex interaction strength defines trafficking and CD103+ memory status of CD8 T cells in the brain. Front. Immunol. 2018, 9, 1–15. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolawole, E.M.; Lamb, T.J.; Evavold, B.D. Relationship of 2D Affinity to T Cell Functional Outcomes. Int. J. Mol. Sci. 2020, 21, 7969. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21217969
Kolawole EM, Lamb TJ, Evavold BD. Relationship of 2D Affinity to T Cell Functional Outcomes. International Journal of Molecular Sciences. 2020; 21(21):7969. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21217969
Chicago/Turabian StyleKolawole, Elizabeth M., Tracey J. Lamb, and Brian D. Evavold. 2020. "Relationship of 2D Affinity to T Cell Functional Outcomes" International Journal of Molecular Sciences 21, no. 21: 7969. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21217969




