Phospho-Mimetic Mutation at Ser602 Inactivates Human TRPA1 Channel
Abstract
:1. Introduction
2. Results
2.1. Phophomimetic Mutation S602D Abrogates Voltage-Dependent Activation of TRPA1
2.2. Salt-Bridge Formation is Not Involved in the Effects Caused by the S602D Mutation
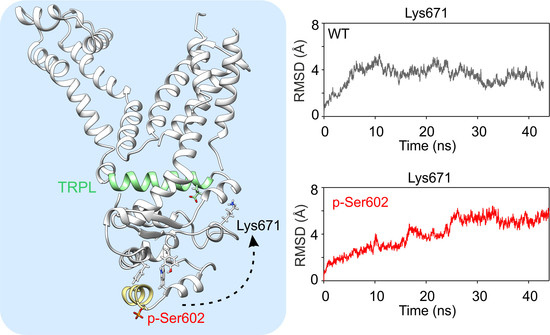
2.3. Molecular Dynamics Simulations of the Phosphorylation of TRPA1 at Ser602
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Constructs, and Transfection
4.2. Electrophysiology
4.3. Confocal Microscopy
4.4. Molecular Modeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TRPA1 | Transient receptor potential ankyrin subtype 1 |
| JT010 | 2-chloro-N-(4-(4-methoxyphenyl)thiazol-2-yl)-N-(3-methoxypropyl)-acetamide |
| AGC | Kinase family named after the protein kinase A, G, and C families |
| CMGC | Kinase family named after the initials of its subfamily members, including cyclin-dependent kinase, mitogen-activated protein kinase, glycogen synthase kinase and CDC-like kinase |
| AR16 | Ankyrin repeat 16 |
References
- Voolstra, O.; Huber, A. Post-Translational Modifications of TRP Channels. Cells 2014, 3, 258–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade, E.L.; Meotti, F.C.; Calixto, J.B. TRPA1 antagonists as potential analgesic drugs. Pharmacol. Ther. 2012, 133, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Story, G.M.; Peier, A.M.; Reeve, A.J.; Eid, S.R.; Mosbacher, J.; Hricik, T.R.; Earley, T.J.; Hergarden, A.C.; Andersson, D.A.; Hwang, S.W.; et al. ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell 2003, 112, 819–829. [Google Scholar] [CrossRef] [Green Version]
- Bautista, D.M.; Jordt, S.E.; Nikai, T.; Tsuruda, P.R.; Read, A.J.; Poblete, J.; Yamoah, E.N.; Basbaum, A.I.; Julius, D. TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell 2006, 124, 1269–1282. [Google Scholar] [CrossRef] [Green Version]
- Atoyan, R.; Shander, D.; Botchkareva, N.V. Non-neuronal expression of transient receptor potential type A1 (TRPA1) in human skin. J. Investig. Dermatol. 2009, 129, 2312–2315. [Google Scholar] [CrossRef] [Green Version]
- Earley, S.; Gonzales, A.L.; Crnich, R. Endothelium-dependent cerebral artery dilation mediated by TRPA1 and Ca2+-Activated K+ channels. Circ. Res. 2009, 104, 987–994. [Google Scholar] [CrossRef] [Green Version]
- Nozawa, K.; Kawabata-Shoda, E.; Doihara, H.; Kojima, R.; Okada, H.; Mochizuki, S.; Sano, Y.; Inamura, K.; Matsushime, H.; Koizumi, T.; et al. TRPA1 regulates gastrointestinal motility through serotonin release from enterochromaffin cells. Proc. Natl. Acad. Sci. USA 2009, 106, 3408–3413. [Google Scholar] [CrossRef] [Green Version]
- Okada, Y.; Reinach, P.S.; Shirai, K.; Kitano-Izutani, A.; Miyajima, M.; Yamanaka, O.; Sumioka, T.; Saika, S. Transient Receptor Potential Channels and Corneal Stromal Inflammation. Cornea 2015, 34 (Suppl. 11), S136–S141. [Google Scholar] [CrossRef]
- Nassini, R.; Pedretti, P.; Moretto, N.; Fusi, C.; Carnini, C.; Facchinetti, F.; Viscomi, A.R.; Pisano, A.R.; Stokesberry, S.; Brunmark, C.; et al. Transient receptor potential ankyrin 1 channel localized to non-neuronal airway cells promotes non-neurogenic inflammation. PLoS ONE 2012, 7, e42454. [Google Scholar] [CrossRef] [Green Version]
- Mukhopadhyay, I.; Gomes, P.; Aranake, S.; Shetty, M.; Karnik, P.; Damle, M.; Kuruganti, S.; Thorat, S.; Khairatkar-Joshi, N. Expression of functional TRPA1 receptor on human lung fibroblast and epithelial cells. J. Recept. Signal Transduct. 2011, 31, 350–358. [Google Scholar] [CrossRef]
- Corey, D.P.; Garcia-Anoveros, J.; Holt, J.R.; Kwan, K.Y.; Lin, S.Y.; Vollrath, M.A.; Amalfitano, A.; Cheung, E.L.; Derfler, B.H.; Duggan, A.; et al. TRPA1 is a candidate for the mechanosensitive transduction channel of vertebrate hair cells. Nature 2004, 432, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Shigetomi, E.; Tong, X.; Kwan, K.Y.; Corey, D.P.; Khakh, B.S. TRPA1 channels regulate astrocyte resting calcium and inhibitory synapse efficacy through GAT-3. Nat. Neurosci. 2011, 15, 70–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamilton, N.B.; Kolodziejczyk, K.; Kougioumtzidou, E.; Attwell, D. Proton-gated Ca(2+)-permeable TRP channels damage myelin in conditions mimicking ischaemia. Nature 2016, 529, 523–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Logu, F.; Nassini, R.; Materazzi, S.; Carvalho Goncalves, M.; Nosi, D.; Rossi Degl’Innocenti, D.; Marone, I.M.; Ferreira, J.; Li Puma, S.; Benemei, S.; et al. Schwann cell TRPA1 mediates neuroinflammation that sustains macrophage-dependent neuropathic pain in mice. Nat. Commun. 2017, 8, 1887. [Google Scholar] [CrossRef]
- El Karim, I.A.; Linden, G.J.; Curtis, T.M.; About, I.; McGahon, M.K.; Irwin, C.R.; Lundy, F.T. Human odontoblasts express functional thermo-sensitive TRP channels: Implications for dentin sensitivity. Pain 2011, 152, 2211–2223. [Google Scholar] [CrossRef]
- Kochukov, M.Y.; McNearney, T.A.; Fu, Y.; Westlund, K.N. Thermosensitive TRP ion channels mediate cytosolic calcium response in human synoviocytes. Am. J. Physiol. Cell Physiol. 2006, 291, C424–C432. [Google Scholar] [CrossRef]
- Nilius, B.; Appendino, G.; Owsianik, G. The transient receptor potential channel TRPA1: From gene to pathophysiology. Pflug. Arch. 2012, 464, 425–458. [Google Scholar] [CrossRef]
- Talavera, K.; Startek, J.B.; Alvarez-Collazo, J.; Boonen, B.; Alpizar, Y.A.; Sanchez, A.; Naert, R.; Nilius, B. Mammalian transient receptor potential TRPA1 channels: From structure to disease. Physiol. Rev. 2019, 100, 725–803. [Google Scholar] [CrossRef]
- Viana, F. TRPA1 channels: Molecular sentinels of cellular stress and tissue damage. J. Physiol. 2016, 594, 4151–4169. [Google Scholar] [CrossRef] [Green Version]
- Zygmunt, P.M.; Hogestatt, E.D. TRPA1. Handb. Exp. Pharmacol. 2014, 222, 583–630. [Google Scholar] [CrossRef]
- Taylor-Clark, T.E.; Undem, B.J.; Macglashan, D.W., Jr.; Ghatta, S.; Carr, M.J.; McAlexander, M.A. Prostaglandin-induced activation of nociceptive neurons via direct interaction with transient receptor potential A1 (TRPA1). Mol. Pharmacol. 2008, 73, 274–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz-Orengo, L.; Dhaka, A.; Heuermann, R.J.; Young, T.J.; Montana, M.C.; Cavanaugh, E.J.; Kim, D.; Story, G.M. Cutaneous nociception evoked by 15-delta PGJ2 via activation of ion channel TRPA1. Mol. Pain 2008, 4, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redmond, W.J.; Gu, L.; Camo, M.; McIntyre, P.; Connor, M. Ligand determinants of fatty acid activation of the pronociceptive ion channel TRPA1. PeerJ 2014, 2, e248. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Dai, Y.; Fukuoka, T.; Yamanaka, H.; Kobayashi, K.; Obata, K.; Cui, X.; Tominaga, M.; Noguchi, K. Phospholipase C and protein kinase A mediate bradykinin sensitization of TRPA1: A molecular mechanism of inflammatory pain. Brain 2008, 131, 1241–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brackley, A.D.; Gomez, R.; Guerrero, K.A.; Akopian, A.N.; Glucksman, M.J.; Du, J.; Carlton, S.M.; Jeske, N.A. A-Kinase Anchoring Protein 79/150 Scaffolds Transient Receptor Potential A 1 Phosphorylation and Sensitization by Metabotropic Glutamate Receptor Activation. Sci. Rep. 2017, 7, 1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meents, J.E.; Fischer, M.J.; McNaughton, P.A. Sensitization of TRPA1 by Protein Kinase A. PLoS ONE 2017, 12, e0170097. [Google Scholar] [CrossRef] [PubMed]
- Anand, U.; Otto, W.R.; Facer, P.; Zebda, N.; Selmer, I.; Gunthorpe, M.J.; Chessell, I.P.; Sinisi, M.; Birch, R.; Anand, P. TRPA1 receptor localisation in the human peripheral nervous system and functional studies in cultured human and rat sensory neurons. Neurosci. Lett. 2008, 438, 221–227. [Google Scholar] [CrossRef]
- Obata, K.; Katsura, H.; Mizushima, T.; Yamanaka, H.; Kobayashi, K.; Dai, Y.; Fukuoka, T.; Tokunaga, A.; Tominaga, M.; Noguchi, K. TRPA1 induced in sensory neurons contributes to cold hyperalgesia after inflammation and nerve injury. J. Clin. Investig. 2005, 115, 2393–2401. [Google Scholar] [CrossRef] [Green Version]
- Sulak, M.A.; Ghosh, M.; Sinharoy, P.; Andrei, S.R.; Damron, D.S. Modulation of TRPA1 channel activity by Cdk5 in sensory neurons. Channels 2018, 12, 65–75. [Google Scholar] [CrossRef]
- Hall, B.E.; Prochazkova, M.; Sapio, M.R.; Minetos, P.; Kurochkina, N.; Binukumar, B.K.; Amin, N.D.; Terse, A.; Joseph, J.; Raithel, S.J.; et al. Phosphorylation of the Transient Receptor Potential Ankyrin 1 by Cyclin-dependent Kinase 5 affects Chemo-nociception. Sci. Rep. 2018, 8, 1177. [Google Scholar] [CrossRef]
- Dai, Y.; Wang, S.; Tominaga, M.; Yamamoto, S.; Fukuoka, T.; Higashi, T.; Kobayashi, K.; Obata, K.; Yamanaka, H.; Noguchi, K. Sensitization of TRPA1 by PAR2 contributes to the sensation of inflammatory pain. J. Clin. Investig. 2007, 117, 1979–1987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Li, L.; McNaughton, P.A. Proinflammatory mediators modulate the heat-activated ion channel TRPV1 via the scaffolding protein AKAP79/150. Neuron 2008, 59, 450–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimova, L.; Barvikova, K.; Macikova, L.; Vyklicka, L.; Sinica, V.; Barvik, I.; Vlachova, V. Proximal C-Terminus Serves as a Signaling Hub for TRPA1 Channel Regulation via Its Interacting Molecules and Supramolecular Complexes. Front. Physiol. 2020, 11, 189. [Google Scholar] [CrossRef] [PubMed]
- Miyano, K.; Shiraishi, S.; Minami, K.; Sudo, Y.; Suzuki, M.; Yokoyama, T.; Terawaki, K.; Nonaka, M.; Murata, H.; Higami, Y.; et al. Carboplatin Enhances the Activity of Human Transient Receptor Potential Ankyrin 1 through the Cyclic AMP-Protein Kinase A-A-Kinase Anchoring Protein (AKAP) Pathways. Int. J. Mol. Sci. 2019, 20, 3271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Kobayashi, K.; Kogure, Y.; Yamanaka, H.; Yamamoto, S.; Yagi, H.; Noguchi, K.; Dai, Y. Negative Regulation of TRPA1 by AMPK in Primary Sensory Neurons as a Potential Mechanism of Painful Diabetic Neuropathy. Diabetes 2018, 67, 98–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, K.; Sadofsky, L.R.; Morice, A.H. Genetic variants affecting human TRPA1 or TRPM8 structure can be classified in vitro as ‘well expressed’, ‘poorly expressed’ or ‘salvageable’. Biosci. Rep. 2015, 35, e00255. [Google Scholar] [CrossRef]
- Kadkova, A.; Synytsya, V.; Krusek, J.; Zimova, L.; Vlachova, V. Molecular basis of TRPA1 regulation in nociceptive neurons. A review. Physiol. Res. 2017, 66, 425–439. [Google Scholar] [CrossRef]
- Meents, J.E.; Ciotu, C.I.; Fischer, M.J.M. TRPA1: A molecular view. J. Neurophysiol. 2019, 121, 427–443. [Google Scholar] [CrossRef]
- Zhao, J.; Lin King, J.V.; Paulsen, C.E.; Cheng, Y.; Julius, D. Irritant-evoked activation and calcium modulation of the TRPA1 receptor. Nature 2020, 585, 141–145. [Google Scholar] [CrossRef]
- Suo, Y.; Wang, Z.; Zubcevic, L.; Hsu, A.L.; He, Q.; Borgnia, M.J.; Ji, R.R.; Lee, S.Y. Structural insights into Electrophile Irritant Sensing by the human TRPA1 channel. Neuron 2020, 105, 882–894. [Google Scholar] [CrossRef]
- Savage, S.R.; Zhang, B. Using phosphoproteomics data to understand cellular signaling: A comprehensive guide to bioinformatics resources. Clin. Proteom. 2020, 17, 27. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, C.E.; Armache, J.P.; Gao, Y.; Cheng, Y.; Julius, D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 2015, 520, 511–517. [Google Scholar] [CrossRef] [Green Version]
- Sura, L.; Zima, V.; Marsakova, L.; Hynkova, A.; Barvik, I.; Vlachova, V. C-terminal Acidic Cluster Is Involved in Ca2+-induced Regulation of Human Transient Receptor Potential Ankyrin 1 Channel. J. Biol. Chem. 2012, 287, 18067–18077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hynkova, A.; Marsakova, L.; Vaskova, J.; Vlachova, V. N-terminal tetrapeptide T/SPLH motifs contribute to multimodal activation of human TRPA1 channel. Sci. Rep. 2016, 6, 28700. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T. Why nature chose phosphate to modify proteins. Philos. Trans. R. Soc. B-Biol. Sci. 2012, 367, 2513–2516. [Google Scholar] [CrossRef]
- Dephoure, N.; Gould, K.L.; Gygi, S.P.; Kellogg, D.R. Mapping and analysis of phosphorylation sites: A quick guide for cell biologists. Mol. Biol. Cell 2013, 24, 535–542. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Kornhauser, J.M.; Latham, V.; Murray, B.; Nandhikonda, V.; Nord, A.; Skrzypek, E.; Wheeler, T.; Zhang, B.; Gnad, F. 15 years of PhosphoSitePlus(R): Integrating post-translationally modified sites, disease variants and isoforms. Nucleic Acids Res. 2019, 47, D433–D441. [Google Scholar] [CrossRef] [Green Version]
- Needham, E.J.; Parker, B.L.; Burykin, T.; James, D.E.; Humphrey, S.J. Illuminating the dark phosphoproteome. Sci. Signal. 2019, 12, eaau8645. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Xu, H.; Lin, S.; Deng, W.; Zhou, J.; Zhang, Y.; Shi, Y.; Peng, D.; Xue, Y. GPS 5.0: An Update on the Prediction of Kinase-specific Phosphorylation Sites in Proteins. Genom. Proteom. Bioinform. 2020, 18, 72–80. [Google Scholar] [CrossRef]
- Patrick, R.; Kobe, B.; Le Cao, K.A.; Boden, M. PhosphoPICK-SNP: Quantifying the effect of amino acid variants on protein phosphorylation. Bioinformatics 2017, 33, 1773–1781. [Google Scholar] [CrossRef] [Green Version]
- Blom, N.; Sicheritz-Ponten, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Horn, H.; Schoof, E.M.; Kim, J.; Robin, X.; Miller, M.L.; Diella, F.; Palma, A.; Cesareni, G.; Jensen, L.J.; Linding, R. KinomeXplorer: An integrated platform for kinome biology studies. Nat. Methods 2014, 11, 603–604. [Google Scholar] [CrossRef]
- Li, T.; Li, F.; Zhang, X. Prediction of kinase-specific phosphorylation sites with sequence features by a log-odds ratio approach. Proteins 2008, 70, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Thelen, J.J.; Dunker, A.K.; Xu, D. Musite, a tool for global prediction of general and kinase-specific phosphorylation sites. Mol. Cell. Proteom. MCP 2010, 9, 2586–2600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearlman, S.M.; Serber, Z.; Ferrell, J.E., Jr. A mechanism for the evolution of phosphorylation sites. Cell 2011, 147, 934–946. [Google Scholar] [CrossRef] [Green Version]
- Gnad, F.; Ren, S.; Cox, J.; Olsen, J.V.; Macek, B.; Oroshi, M.; Mann, M. PHOSIDA (phosphorylation site database): Management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007, 8, R250. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Beglov, D.; Roux, B. Finite Representation of an Infinite Bulk System-Solvent Boundary Potential for Computer-Simulations. J. Chem. Phys. 1994, 100, 9050–9063. [Google Scholar] [CrossRef] [Green Version]
- Schlenkrich, M.; Brickmann, J.; MacKerell, A.D., Jr.; Karplus, M. An Empirical Potential Energy Function for Phospholipids: Criteria for Parameter Optimization and Applications. In Biological Membranes: A Molecular Perspective from Computation and Experiment; Merz, K., Jr., Roux, B., Eds.; Birkhauser Boston: Cambridge, MA, USA, 1996; pp. 31–81. [Google Scholar]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]




| Predicted Protein Kinase | Prediction Server |
|---|---|
| Protein kinase A | GPS 5.0 [49], PhosphoPICK [50] |
| Protein kinase C | GPS 5.0, NetPhos [51], NetPhorest, NetworKIN [52] |
| Protein kinase D1 | GPS 5.0 [49], PhosphoPICK [50] |
| Glycogen synthase kinase-3 | GPS 5.0 [49] |
| AKT2 kinase | GPS 5.0 [49] |
| Rho-associated protein kinase | PhoScan [53] |
| Mitogen-activated protein (MAP) kinase 3 | Musite [54] |
| MAP kinase-activated protein kinase 5 | GPS 5.0 [49] |
| ULK1 protein kinase | GPS 5.0 [49] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barvikova, K.; Barvik, I.; Sinica, V.; Zimova, L.; Vlachova, V. Phospho-Mimetic Mutation at Ser602 Inactivates Human TRPA1 Channel. Int. J. Mol. Sci. 2020, 21, 7995. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21217995
Barvikova K, Barvik I, Sinica V, Zimova L, Vlachova V. Phospho-Mimetic Mutation at Ser602 Inactivates Human TRPA1 Channel. International Journal of Molecular Sciences. 2020; 21(21):7995. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21217995
Chicago/Turabian StyleBarvikova, Kristyna, Ivan Barvik, Viktor Sinica, Lucie Zimova, and Viktorie Vlachova. 2020. "Phospho-Mimetic Mutation at Ser602 Inactivates Human TRPA1 Channel" International Journal of Molecular Sciences 21, no. 21: 7995. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21217995