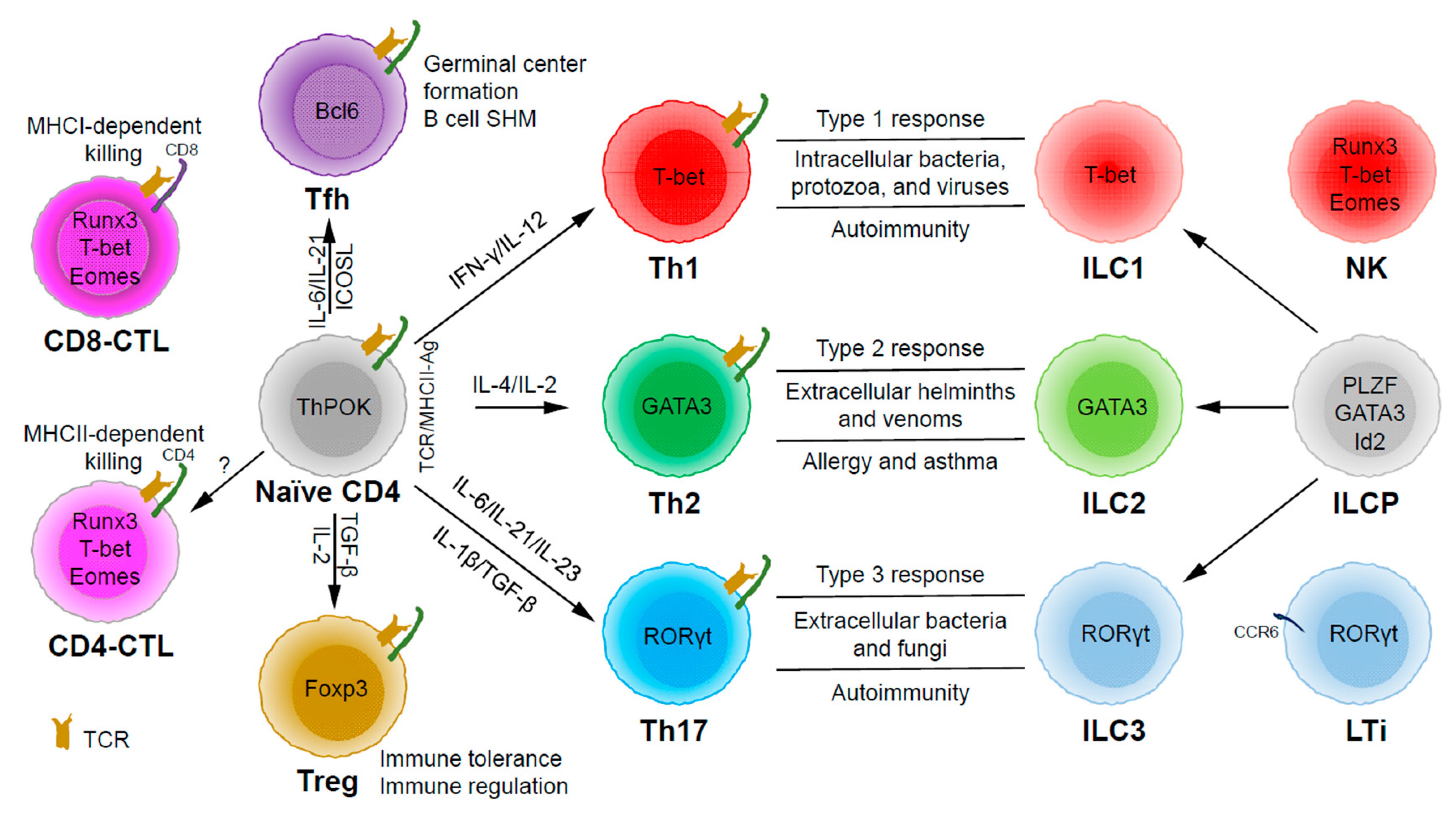
CD4 T Helper Cell Subsets and Related Human Immunological Disorders
Abstract
:1. Introduction
2. Th1 Cells and Related Diseases
2.1. Th1 Cells
2.2. Th1-Related Diseases
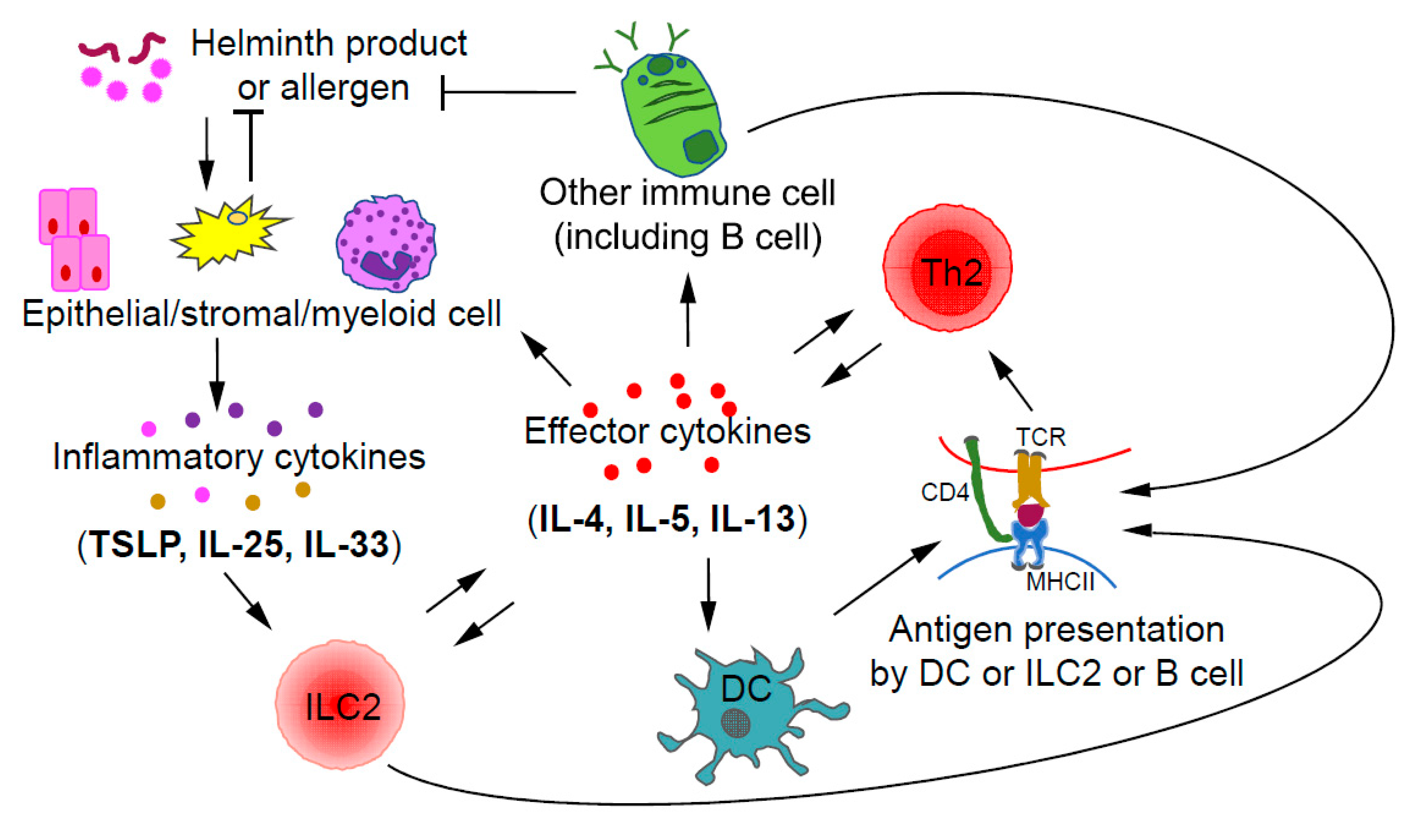
3. Th2 Cells and Related Diseases
3.1. Th2 Cells
3.2. Th2 Related Diseases
4. Th17 Cells and Related Diseases
4.1. Th17 Cells
4.2. CD4 T Cell Plasticity
4.3. Th17-Related Diseases
5. Treg Cells and Related Diseases
5.1. Treg Cells
5.2. Treg Related Diseases
6. Tfh Cells and Related Diseases
6.1. Tfh Cells
6.2. Tfh Subsets
6.3. Tfh Related Diseases
7. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Pennington, D.J.; Vermijlen, D.; Wise, E.L.; Clarke, S.L.; Tigelaar, R.E.; Hayday, A.C. The integration of conventional and unconventional T cells that characterizes cell-mediated responses. Adv. Immunol. 2005, 87, 27–59. [Google Scholar] [CrossRef] [PubMed]
- Girardi, S.R.A.M. Conventional and Unconventional T Cells. Clin. Basic Immunodermatol. 2008, 6, 85–104. [Google Scholar]
- Kumar, B.V.; Connors, T.J.; Farber, D.L. Human T Cell Development, Localization, and Function throughout Life. Immunity 2018, 48, 202–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruterbusch, M.; Pruner, K.B.; Shehata, L.; Pepper, M. In Vivo CD4(+) T Cell Differentiation and Function: Revisiting the Th1/Th2 Paradigm. Annu. Rev. Immunol. 2020, 38, 705–725. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yamane, H.; Paul, W.E. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 2010, 28, 445–489. [Google Scholar] [CrossRef] [Green Version]
- Annunziato, F.; Romagnani, C.; Romagnani, S. The 3 major types of innate and adaptive cell-mediated effector immunity. J. Allergy Clin. Immunol. 2015, 135, 626–635. [Google Scholar] [CrossRef]
- Fang, D.; Zhu, J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J. Exp. Med. 2017, 214, 1861–1876. [Google Scholar] [CrossRef]
- Chen, Y.; Kuchroo, V.K.; Inobe, J.; Hafler, D.A.; Weiner, H.L. Regulatory T cell clones induced by oral tolerance: Suppression of autoimmune encephalomyelitis. Science 1994, 265, 1237–1240. [Google Scholar] [CrossRef]
- Groux, H.; O’Garra, A.; Bigler, M.; Rouleau, M.; Antonenko, S.; de Vries, J.E.; Roncarolo, M.G. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature 1997, 389, 737–742. [Google Scholar] [CrossRef]
- Dardalhon, V.; Awasthi, A.; Kwon, H.; Galileos, G.; Gao, W.; Sobel, R.A.; Mitsdoerffer, M.; Strom, T.B.; Elyaman, W.; Ho, I.C.; et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat. Immunol. 2008, 9, 1347–1355. [Google Scholar] [CrossRef] [Green Version]
- Veldhoen, M.; Uyttenhove, C.; van Snick, J.; Helmby, H.; Westendorf, A.; Buer, J.; Martin, B.; Wilhelm, C.; Stockinger, B. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 2008, 9, 1341–1346. [Google Scholar] [CrossRef] [PubMed]
- Eyerich, S.; Eyerich, K.; Pennino, D.; Carbone, T.; Nasorri, F.; Pallotta, S.; Cianfarani, F.; Odorisio, T.; Traidl-Hoffmann, C.; Behrendt, H.; et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J. Clin. Investig. 2009, 119, 3573–3585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devarajan, P.; Jones, M.C.; Kugler-Umana, O.; Vong, A.M.; Xia, J.; Swain, S.L. Pathogen Recognition by CD4 Effectors Drives Key Effector and Most Memory Cell Generation against Respiratory Virus. Front. Immunol. 2018, 9, 596. [Google Scholar] [CrossRef]
- Juno, J.A.; van Bockel, D.; Kent, S.J.; Kelleher, A.D.; Zaunders, J.J.; Munier, C.M. Cytotoxic CD4 T Cells-Friend or Foe during Viral Infection? Front. Immunol. 2017, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, A.; Saito, T. CD4 CTL, a Cytotoxic Subset of CD4(+) T Cells, Their Differentiation and Function. Front. Immunol. 2017, 8, 194. [Google Scholar] [CrossRef] [Green Version]
- Eshima, K.; Chiba, S.; Suzuki, H.; Kokubo, K.; Kobayashi, H.; Iizuka, M.; Iwabuchi, K.; Shinohara, N. Ectopic expression of a T-box transcription factor, eomesodermin, renders CD4(+) Th cells cytotoxic by activating both perforin- and FasL-pathways. Immunol. Lett. 2012, 144, 7–15. [Google Scholar] [CrossRef]
- Reis, B.S.; Hoytema van Konijnenburg, D.P.; Grivennikov, S.I.; Mucida, D. Transcription factor T-bet regulates intraepithelial lymphocyte functional maturation. Immunity 2014, 41, 244–256. [Google Scholar] [CrossRef] [Green Version]
- Sledzinska, A.; Vila de Mucha, M.; Bergerhoff, K.; Hotblack, A.; Demane, D.F.; Ghorani, E.; Akarca, A.U.; Marzolini, M.A.V.; Solomon, I.; Vargas, F.A.; et al. Regulatory T Cells Restrain Interleukin-2- and Blimp-1-Dependent Acquisition of Cytotoxic Function by CD4(+) T Cells. Immunity 2020, 52, 151–166.e156. [Google Scholar] [CrossRef]
- Cheroutre, H.; Husain, M.M. CD4 CTL: Living up to the challenge. Semin. Immunol. 2013, 25, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Mucida, D.; Husain, M.M.; Muroi, S.; van Wijk, F.; Shinnakasu, R.; Naoe, Y.; Reis, B.S.; Huang, Y.; Lambolez, F.; Docherty, M.; et al. Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat. Immunol. 2013, 14, 281–289. [Google Scholar] [CrossRef]
- Reis, B.S.; Rogoz, A.; Costa-Pinto, F.A.; Taniuchi, I.; Mucida, D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nat. Immunol. 2013, 14, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Serroukh, Y.; Gu-Trantien, C.; Hooshiar Kashani, B.; Defrance, M.; Vu Manh, T.P.; Azouz, A.; Detavernier, A.; Hoyois, A.; Das, J.; Bizet, M.; et al. The transcription factors Runx3 and ThPOK cross-regulate acquisition of cytotoxic function by human Th1 lymphocytes. eLife 2018, 7, e30496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donnarumma, T.; Young, G.R.; Merkenschlager, J.; Eksmond, U.; Bongard, N.; Nutt, S.L.; Boyer, C.; Dittmer, U.; Le-Trilling, V.T.; Trilling, M.; et al. Opposing Development of Cytotoxic and Follicular Helper CD4 T Cells Controlled by the TCF-1-Bcl6 Nexus. Cell Rep. 2016, 17, 1571–1583. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, A.; Badr Mel, S.; Miyauchi, K.; Ishihara, C.; Onishi, R.; Guo, Z.; Sasaki, Y.; Ike, H.; Takumi, A.; Tsuji, N.M.; et al. CRTAM determines the CD4+ cytotoxic T lymphocyte lineage. J. Exp. Med. 2016, 213, 123–138. [Google Scholar] [CrossRef]
- Marshall, N.B.; Vong, A.M.; Devarajan, P.; Brauner, M.D.; Kuang, Y.; Nayar, R.; Schutten, E.A.; Castonguay, C.H.; Berg, L.J.; Nutt, S.L.; et al. NKG2C/E Marks the Unique Cytotoxic CD4 T Cell Subset, ThCTL, Generated by Influenza Infection. J. Immunol. 2017, 198, 1142–1155. [Google Scholar] [CrossRef] [Green Version]
- Meckiff, B.J.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Kusnadi, A.; Simon, H.; Eschweiler, S.; Grifoni, A.; Pelosi, E.; Weiskopf, D.; et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell 2020. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Kwek, S.S.; Raju, S.S.; Li, T.; McCarthy, E.; Chow, E.; Aran, D.; Ilano, A.; Pai, C.S.; Rancan, C.; et al. Intratumoral CD4(+) T Cells Mediate Anti-tumor Cytotoxicity in Human Bladder Cancer. Cell 2020, 181, 1612–1625. [Google Scholar] [CrossRef] [PubMed]
- Mattoo, H.; Stone, J.H.; Pillai, S. Clonally expanded cytotoxic CD4(+) T cells and the pathogenesis of IgG4-related disease. Autoimmunity 2017, 50, 19–24. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, K.; Kouno, T.; Ikawa, T.; Hayatsu, N.; Miyajima, Y.; Yabukami, H.; Terooatea, T.; Sasaki, T.; Suzuki, T.; Valentine, M.; et al. Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc. Natl. Acad. Sci. USA 2019, 116, 24242–24251. [Google Scholar] [CrossRef] [Green Version]
- Gurram, R.K.; Zhu, J. Orchestration between ILC2s and Th2 cells in shaping type 2 immune responses. Cell. Mol. Immunol. 2019, 16, 225–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockinger, B.; Omenetti, S. The dichotomous nature of T helper 17 cells. Nat. Rev. Immunol. 2017, 17, 535–544. [Google Scholar] [CrossRef]
- Crotty, S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 2019, 50, 1132–1148. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, K.; Sugimoto-Ishige, A.; Harada, Y.; Adachi, Y.; Usami, Y.; Kaji, T.; Inoue, K.; Hasegawa, H.; Watanabe, T.; Hijikata, A.; et al. Protective neutralizing influenza antibody response in the absence of T follicular helper cells. Nat. Immunol. 2016, 17, 1447–1458. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Villar, M.; Hafler, D.A. Regulatory T cells in autoimmune disease. Nat. Immunol. 2018, 19, 665–673. [Google Scholar] [CrossRef]
- Shevach, E.M. CD4+ CD25+ suppressor T cells: More questions than answers. Nat. Rev. Immunol. 2002, 2, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E.; Artis, D.; Colonna, M.; Diefenbach, A.; Di Santo, J.P.; Eberl, G.; Koyasu, S.; Locksley, R.M.; McKenzie, A.N.J.; Mebius, R.E.; et al. Innate Lymphoid Cells: 10 Years On. Cell 2018, 174, 1054–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artis, D.; Spits, H. The biology of innate lymphoid cells. Nature 2015, 517, 293–301. [Google Scholar] [CrossRef]
- Cherrier, D.E.; Serafini, N.; Di Santo, J.P. Innate Lymphoid Cell Development: A T Cell Perspective. Immunity 2018, 48, 1091–1103. [Google Scholar] [CrossRef] [Green Version]
- Ebbo, M.; Crinier, A.; Vely, F.; Vivier, E. Innate lymphoid cells: Major players in inflammatory diseases. Nat. Rev. Immunol. 2017, 17, 665–678. [Google Scholar] [CrossRef]
- Zhong, C.; Zheng, M.; Zhu, J. Lymphoid tissue inducer-A divergent member of the ILC family. Cytokine Growth Factor Rev. 2018, 42, 5–12. [Google Scholar] [CrossRef]
- Sallusto, F. Heterogeneity of Human CD4(+) T Cells Against Microbes. Annu. Rev. Immunol. 2016, 34, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Hirahara, K.; Poholek, A.; Vahedi, G.; Laurence, A.; Kanno, Y.; Milner, J.D.; O’Shea, J.J. Mechanisms underlying helper T-cell plasticity: Implications for immune-mediated disease. J. Allergy Clin. Immunol. 2013, 131, 1276–1287. [Google Scholar] [CrossRef] [Green Version]
- Leonard, W.J.; Lin, J.X.; O’Shea, J.J. The gammac Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity 2019, 50, 832–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosmann, T.R.; Cherwinski, H.; Bond, M.W.; Giedlin, M.A.; Coffman, R.L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986, 136, 2348–2357. [Google Scholar]
- Zhu, J.; Jankovic, D.; Oler, A.J.; Wei, G.; Sharma, S.; Hu, G.; Guo, L.; Yagi, R.; Yamane, H.; Punkosdy, G.; et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity 2012, 37, 660–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanno, Y.; Vahedi, G.; Hirahara, K.; Singleton, K.; O’Shea, J.J. Transcriptional and epigenetic control of T helper cell specification: Molecular mechanisms underlying commitment and plasticity. Annu. Rev. Immunol. 2012, 30, 707–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullen, A.C.; Hutchins, A.S.; High, F.A.; Lee, H.W.; Sykes, K.J.; Chodosh, L.A.; Reiner, S.L. Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat. Immunol. 2002, 3, 652–658. [Google Scholar] [CrossRef]
- Djuretic, I.M.; Levanon, D.; Negreanu, V.; Groner, Y.; Rao, A.; Ansel, K.M. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 2007, 8, 145–153. [Google Scholar] [CrossRef]
- Yagi, R.; Junttila, I.S.; Wei, G.; Urban, J.F., Jr.; Zhao, K.; Paul, W.E.; Zhu, J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity 2010, 32, 507–517. [Google Scholar] [CrossRef] [Green Version]
- Grenningloh, R.; Kang, B.Y.; Ho, I.C. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J. Exp. Med. 2005, 201, 615–626. [Google Scholar] [CrossRef]
- Yu, F.; Sharma, S.; Jankovic, D.; Gurram, R.K.; Su, P.; Hu, G.; Li, R.; Rieder, S.; Zhao, K.; Sun, B.; et al. The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J. Exp. Med. 2018, 215, 1813–1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazarevic, V.; Chen, X.; Shim, J.H.; Hwang, E.S.; Jang, E.; Bolm, A.N.; Oukka, M.; Kuchroo, V.K.; Glimcher, L.H. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat. Immunol. 2011, 12, 96–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.K.; Turner, H.; Maynard, C.L.; Oliver, J.R.; Chen, D.; Elson, C.O.; Weaver, C.T. Late developmental plasticity in the T helper 17 lineage. Immunity 2009, 30, 92–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, G.; Wei, L.; Zhu, J.; Zang, C.; Hu-Li, J.; Yao, Z.; Cui, K.; Kanno, Y.; Roh, T.Y.; Watford, W.T.; et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 2009, 30, 155–167. [Google Scholar] [CrossRef] [Green Version]
- Bonecchi, R.; Bianchi, G.; Bordignon, P.P.; D’Ambrosio, D.; Lang, R.; Borsatti, A.; Sozzani, S.; Allavena, P.; Gray, P.A.; Mantovani, A.; et al. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 1998, 187, 129–134. [Google Scholar] [CrossRef]
- Klose, C.S.; Flach, M.; Mohle, L.; Rogell, L.; Hoyler, T.; Ebert, K.; Fabiunke, C.; Pfeifer, D.; Sexl, V.; Fonseca-Pereira, D.; et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 2014, 157, 340–356. [Google Scholar] [CrossRef] [Green Version]
- Weizman, O.E.; Adams, N.M.; Schuster, I.S.; Krishna, C.; Pritykin, Y.; Lau, C.; Degli-Esposti, M.A.; Leslie, C.S.; Sun, J.C.; O’Sullivan, T.E. ILC1 Confer Early Host Protection at Initial Sites of Viral Infection. Cell 2017, 171, 795–808. [Google Scholar] [CrossRef]
- Wei, G.; Abraham, B.J.; Yagi, R.; Jothi, R.; Cui, K.; Sharma, S.; Narlikar, L.; Northrup, D.L.; Tang, Q.; Paul, W.E.; et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 2011, 35, 299–311. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Tang, Q.; Sharma, S.; Yu, F.; Escobar, T.M.; Muljo, S.A.; Zhu, J.; Zhao, K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013, 14, 1190–1198. [Google Scholar] [CrossRef] [Green Version]
- Lund, R.J.; Loytomaki, M.; Naumanen, T.; Dixon, C.; Chen, Z.; Ahlfors, H.; Tuomela, S.; Tahvanainen, J.; Scheinin, J.; Henttinen, T.; et al. Genome-wide identification of novel genes involved in early Th1 and Th2 cell differentiation. J. Immunol. 2007, 178, 3648–3660. [Google Scholar] [CrossRef] [Green Version]
- Fang, D.; Cui, K.; Mao, K.; Hu, G.; Li, R.; Zheng, M.; Riteau, N.; Reiner, S.L.; Sher, A.; Zhao, K.; et al. Transient T-bet expression functionally specifies a distinct T follicular helper subset. J. Exp. Med. 2018, 215, 2705–2714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romagnani, S. The Th1/Th2 paradigm. Immunol. Today 1997, 18, 263–266. [Google Scholar] [CrossRef]
- Yang, R.; Mele, F.; Worley, L.; Langlais, D.; Rosain, J.; Benhsaien, I.; Elarabi, H.; Croft, C.A.; Doisne, J.M.; Zhang, P.; et al. Human T-bet governs innate and innate-like adaptive IFN-g immunity against mycobacteria. BioRxiv Prep. 2020. [Google Scholar] [CrossRef]
- Filipe-Santos, O.; Bustamante, J.; Chapgier, A.; Vogt, G.; de Beaucoudrey, L.; Feinberg, J.; Jouanguy, E.; Boisson-Dupuis, S.; Fieschi, C.; Picard, C.; et al. Inborn errors of IL-12/23- and IFN-gamma-mediated immunity: Molecular, cellular, and clinical features. Semin. Immunol. 2006, 18, 347–361. [Google Scholar] [CrossRef]
- Peng, D.; Kryczek, I.; Nagarsheth, N.; Zhao, L.; Wei, S.; Wang, W.; Sun, Y.; Zhao, E.; Vatan, L.; Szeliga, W.; et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015, 527, 249–253. [Google Scholar] [CrossRef] [Green Version]
- Paul, W.E.; Zhu, J. How are T(H)2-type immune responses initiated and amplified? Nat. Rev. Immunol. 2010, 10, 225–235. [Google Scholar] [CrossRef]
- Zhu, J.; Min, B.; Hu-Li, J.; Watson, C.J.; Grinberg, A.; Wang, Q.; Killeen, N.; Urban, J.F., Jr.; Guo, L.; Paul, W.E. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat. Immunol. 2004, 5, 1157–1165. [Google Scholar] [CrossRef]
- Zhu, J. Seventeen-Year Journey Working with a Master. Front. Immunol. 2018, 9, 960. [Google Scholar] [CrossRef] [Green Version]
- Wohlfert, E.A.; Grainger, J.R.; Bouladoux, N.; Konkel, J.E.; Oldenhove, G.; Ribeiro, C.H.; Hall, J.A.; Yagi, R.; Naik, S.; Bhairavabhotla, R.; et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J. Clin. Investig. 2011, 121, 4503–4515. [Google Scholar] [CrossRef]
- Fang, D.; Cui, K.; Hu, G.; Gurram, R.K.; Zhong, C.; Oler, A.J.; Yagi, R.; Zhao, M.; Sharma, S.; Liu, P.; et al. Bcl11b, a novel GATA3-interacting protein, suppresses Th1 while limiting Th2 cell differentiation. J. Exp. Med. 2018, 215, 1449–1462. [Google Scholar] [CrossRef]
- Kim, J.I.; Ho, I.C.; Grusby, M.J.; Glimcher, L.H. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity 1999, 10, 745–751. [Google Scholar] [CrossRef] [Green Version]
- Stritesky, G.L.; Muthukrishnan, R.; Sehra, S.; Goswami, R.; Pham, D.; Travers, J.; Nguyen, E.T.; Levy, D.E.; Kaplan, M.H. The transcription factor STAT3 is required for T helper 2 cell development. Immunity 2011, 34, 39–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amsen, D.; Blander, J.M.; Lee, G.R.; Tanigaki, K.; Honjo, T.; Flavell, R.A. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell 2004, 117, 515–526. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J. T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine 2015, 75, 14–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekiya, T.; Miyamasu, M.; Imanishi, M.; Yamada, H.; Nakajima, T.; Yamaguchi, M.; Fujisawa, T.; Pawankar, R.; Sano, Y.; Ohta, K.; et al. Inducible expression of a Th2-type CC chemokine thymus- and activation-regulated chemokine by human bronchial epithelial cells. J. Immunol. 2000, 165, 2205–2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panina-Bordignon, P.; Papi, A.; Mariani, M.; Di Lucia, P.; Casoni, G.; Bellettato, C.; Buonsanti, C.; Miotto, D.; Mapp, C.; Villa, A.; et al. The C-C chemokine receptors CCR4 and CCR8 identify airway T cells of allergen-challenged atopic asthmatics. J. Clin. Investig. 2001, 107, 1357–1364. [Google Scholar] [CrossRef] [Green Version]
- Knipfer, L.; Schulz-Kuhnt, A.; Kindermann, M.; Greif, V.; Symowski, C.; Voehringer, D.; Neurath, M.F.; Atreya, I.; Wirtz, S. A CCL1/CCR8-dependent feed-forward mechanism drives ILC2 functions in type 2-mediated inflammation. J. Exp. Med. 2019, 216, 2763–2777. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.J. TSLP in epithelial cell and dendritic cell cross talk. Adv. Immunol. 2009, 101, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Cosmi, L.; Annunziato, F.; Iwasaki, M.; Galli, G.; Manetti, R.; Maggi, E.; Nagata, K.; Romagnani, S. CRTH2 is the most reliable marker for the detection of circulating human type 2 Th and type 2 T cytotoxic cells in health and disease. Eur. J. Immunol. 2000, 30, 2972–2979. [Google Scholar] [CrossRef]
- Xu, D.; Chan, W.L.; Leung, B.P.; Huang, F.; Wheeler, R.; Piedrafita, D.; Robinson, J.H.; Liew, F.Y. Selective expression of a stable cell surface molecule on type 2 but not type 1 helper T cells. J. Exp. Med. 1998, 187, 787–794. [Google Scholar] [CrossRef]
- Zhong, C.; Zheng, M.; Cui, K.; Martins, A.J.; Hu, G.; Li, D.; Tessarollo, L.; Kozlov, S.; Keller, J.R.; Tsang, J.S.; et al. Differential Expression of the Transcription Factor GATA3 Specifies Lineage and Functions of Innate Lymphoid Cells. Immunity 2020, 52, 83–95.e84. [Google Scholar] [CrossRef]
- Halim, T.Y.; Steer, C.A.; Matha, L.; Gold, M.J.; Martinez-Gonzalez, I.; McNagny, K.M.; McKenzie, A.N.; Takei, F. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity 2014, 40, 425–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliphant, C.J.; Hwang, Y.Y.; Walker, J.A.; Salimi, M.; Wong, S.H.; Brewer, J.M.; Englezakis, A.; Barlow, J.L.; Hams, E.; Scanlon, S.T.; et al. MHCII-mediated dialog between group 2 innate lymphoid cells and CD4(+) T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity 2014, 41, 283–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Moltke, J.; Ji, M.; Liang, H.E.; Locksley, R.M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 2016, 529, 221–225. [Google Scholar] [CrossRef] [Green Version]
- Dewas, C.; Chen, X.; Honda, T.; Junttila, I.; Linton, J.; Udey, M.C.; Porcella, S.F.; Sturdevant, D.E.; Feigenbaum, L.; Koo, L.; et al. TSLP expression: Analysis with a ZsGreen TSLP reporter mouse. J. Immunol. 2015, 194, 1372–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Huang, Y.; Chen, X.; Hu-Li, J.; Urban, J.F., Jr.; Paul, W.E. Innate immunological function of TH2 cells in vivo. Nat. Immunol. 2015, 16, 1051–1059. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.E.; Reinhardt, R.L.; Bando, J.K.; Sullivan, B.M.; Ho, I.C.; Locksley, R.M. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat. Immunol. 2012, 13, 58–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peine, M.; Rausch, S.; Helmstetter, C.; Frohlich, A.; Hegazy, A.N.; Kuhl, A.A.; Grevelding, C.G.; Hofer, T.; Hartmann, S.; Lohning, M. Stable T-bet(+)GATA-3(+) Th1/Th2 hybrid cells arise in vivo, can develop directly from naive precursors, and limit immunopathologic inflammation. PLoS Biol. 2013, 11, e1001633. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.H.; Voo, K.S.; Liu, B.; Chen, C.Y.; Uygungil, B.; Spoede, W.; Bernstein, J.A.; Huston, D.P.; Liu, Y.J. A novel subset of CD4(+) T(H)2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. J. Exp. Med. 2010, 207, 2479–2491. [Google Scholar] [CrossRef]
- Irvin, C.; Zafar, I.; Good, J.; Rollins, D.; Christianson, C.; Gorska, M.M.; Martin, R.J.; Alam, R. Increased frequency of dual-positive TH2/TH17 cells in bronchoalveolar lavage fluid characterizes a population of patients with severe asthma. J. Allergy Clin. Immunol. 2014, 134, 1175–1186.e1177. [Google Scholar] [CrossRef] [Green Version]
- Hegazy, A.N.; Peine, M.; Helmstetter, C.; Panse, I.; Frohlich, A.; Bergthaler, A.; Flatz, L.; Pinschewer, D.D.; Radbruch, A.; Lohning, M. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity 2010, 32, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Zhong, C.; Northrup, D.L.; Yu, F.; Bouladoux, N.; Spencer, S.; Hu, G.; Barron, L.; Sharma, S.; Nakayama, T.; et al. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity 2014, 40, 378–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henriksson, J.; Chen, X.; Gomes, T.; Ullah, U.; Meyer, K.B.; Miragaia, R.; Duddy, G.; Pramanik, J.; Yusa, K.; Lahesmaa, R.; et al. Genome-wide CRISPR Screens in T Helper Cells Reveal Pervasive Crosstalk between Activation and Differentiation. Cell 2019, 176, 882–896.e818. [Google Scholar] [CrossRef]
- Pulendran, B.; Artis, D. New paradigms in type 2 immunity. Science 2012, 337, 431–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helm, R.M.; Burks, A.W. Mechanisms of food allergy. Curr. Opin. Immunol. 2000, 12, 647–653. [Google Scholar] [CrossRef]
- Palm, N.W.; Rosenstein, R.K.; Medzhitov, R. Allergic host defences. Nature 2012, 484, 465–472. [Google Scholar] [CrossRef]
- Thomas, W.R.; Hales, B.J. T and B cell responses to HDM allergens and antigens. Immunol. Res. 2007, 37, 187–199. [Google Scholar] [CrossRef]
- Mariani, V.; Gilles, S.; Jakob, T.; Thiel, M.; Mueller, M.J.; Ring, J.; Behrendt, H.; Traidl-Hoffmann, C. Immunomodulatory mediators from pollen enhance the migratory capacity of dendritic cells and license them for Th2 attraction. J. Immunol. 2007, 178, 7623–7631. [Google Scholar] [CrossRef] [Green Version]
- Coffman, R.L.; Sher, A.; Seder, R.A. Vaccine adjuvants: Putting innate immunity to work. Immunity 2010, 33, 492–503. [Google Scholar] [CrossRef] [Green Version]
- Ting, C.N.; Olson, M.C.; Barton, K.P.; Leiden, J.M. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature 1996, 384, 474–478. [Google Scholar] [CrossRef]
- Van Esch, H.; Groenen, P.; Nesbit, M.A.; Schuffenhauer, S.; Lichtner, P.; Vanderlinden, G.; Harding, B.; Beetz, R.; Bilous, R.W.; Holdaway, I.; et al. GATA3 haplo-insufficiency causes human HDR syndrome. Nature 2000, 406, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Skapenko, A.; Leipe, J.; Niesner, U.; Devriendt, K.; Beetz, R.; Radbruch, A.; Kalden, J.R.; Lipsky, P.E.; Schulze-Koops, H. GATA-3 in human T cell helper type 2 development. J. Exp. Med. 2004, 199, 423–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, M.G.; Leiding, J.W.; Lyons, J.J.; Hsu, A.P.; Nelson, C.C.; Jones, N.; Fitzgerald, A.; Chien, W.W.; Workman, L.; Platts-Mills, T.A.; et al. GATA3 haploinsufficiency does not block allergic sensitization or atopic disease. J. Allergy Clin. Immunol. 2016, 137, 627–629.e622. [Google Scholar] [CrossRef] [Green Version]
- Klein Wolterink, R.G.; Serafini, N.; van Nimwegen, M.; Vosshenrich, C.A.; de Bruijn, M.J.; Fonseca Pereira, D.; Veiga Fernandes, H.; Hendriks, R.W.; Di Santo, J.P. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc. Natl. Acad. Sci. USA 2013, 110, 10240–10245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marrella, V.; Poliani, P.L.; Sobacchi, C.; Grassi, F.; Villa, A. Of Omenn and mice. Trends Immunol. 2008, 29, 133–140. [Google Scholar] [CrossRef]
- Villa, A.; Notarangelo, L.D. RAG gene defects at the verge of immunodeficiency and immune dysregulation. Immunol. Rev. 2019, 287, 73–90. [Google Scholar] [CrossRef]
- Milner, J.D.; Ward, J.M.; Keane-Myers, A.; Paul, W.E. Lymphopenic mice reconstituted with limited repertoire T cells develop severe, multiorgan, Th2-associated inflammatory disease. Proc. Natl. Acad. Sci. USA 2007, 104, 576–581. [Google Scholar] [CrossRef] [Green Version]
- Dolff, S.; Witzke, O.; Wilde, B. Th17 cells in renal inflammation and autoimmunity. Autoimmun. Rev. 2019, 18, 129–136. [Google Scholar] [CrossRef]
- Aggarwal, S.; Ghilardi, N.; Xie, M.H.; de Sauvage, F.J.; Gurney, A.L. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 2003, 278, 1910–1914. [Google Scholar] [CrossRef] [Green Version]
- Cua, D.J.; Sherlock, J.; Chen, Y.; Murphy, C.A.; Joyce, B.; Seymour, B.; Lucian, L.; To, W.; Kwan, S.; Churakova, T.; et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 2003, 421, 744–748. [Google Scholar] [CrossRef]
- Murphy, C.A.; Langrish, C.L.; Chen, Y.; Blumenschein, W.; McClanahan, T.; Kastelein, R.A.; Sedgwick, J.D.; Cua, D.J. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J. Exp. Med. 2003, 198, 1951–1957. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Li, Z.X.; Yang, X.O.; Chang, S.H.; Nurieva, R.; Wang, Y.H.; Wang, Y.; Hood, L.; Zhu, Z.; Tian, Q.; et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005, 6, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Bettelli, E.; Carrier, Y.J.; Gao, W.D.; Korn, T.; Strom, T.B.; Oukka, M.; Weiner, H.L.; Kuchroo, V.K. Reciprocal developmental pathways for the generation of pathogenic effector T(H)17 and regulatory T cells. Nature 2006, 441, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Harrington, L.E.; Hatton, R.D.; Mangan, P.R.; Turner, H.; Murphy, T.L.; Murphy, K.M.; Weaver, C.T. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005, 6, 1123–1132. [Google Scholar] [CrossRef]
- Gaublomme, J.T.; Yosef, N.; Lee, Y.; Gertner, R.S.; Yang, L.V.; Wu, C.; Pandolfi, P.P.; Mak, T.; Satija, R.; Shalek, A.K.; et al. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell 2015, 163, 1400–1412. [Google Scholar] [CrossRef] [Green Version]
- Ciofani, M.; Madar, A.; Galan, C.; Sellars, M.; Mace, K.; Pauli, F.; Agarwal, A.; Huang, W.; Parkurst, C.N.; Muratet, M.; et al. A Validated Regulatory Network for Th17 Cell Specification. Cell 2012, 151, 289–303. [Google Scholar] [CrossRef] [Green Version]
- Chung, Y.; Chang, S.H.; Martinez, G.J.; Yang, X.O.; Nurieva, R.; Kang, H.S.; Ma, L.; Watowich, S.S.; Jetten, A.M.; Tian, Q.; et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity 2009, 30, 576–587. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Wei, G.; Zhu, J.; Liao, W.; Leonard, W.J.; Zhao, K.; Paul, W. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc. Natl. Acad. Sci. USA 2009, 106, 13463–13468. [Google Scholar] [CrossRef] [Green Version]
- Dong, C. TH17 cells in development: An updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 2008, 8, 337–348. [Google Scholar] [CrossRef]
- Ghoreschi, K.; Laurence, A.; Yang, X.P.; Tato, C.M.; McGeachy, M.J.; Konkel, J.E.; Ramos, H.L.; Wei, L.; Davidson, T.S.; Bouladoux, N.; et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 2010, 467, 967–971. [Google Scholar] [CrossRef] [Green Version]
- Pawlak, M.; Ho, A.W.; Kuchroo, V.K. Cytokines and transcription factors in the differentiation of CD4+ T helper cell subsets and induction of tissue inflammation and autoimmunity. Curr. Opin. Immunol. 2020, 67, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Veldhoen, M.; Hocking, R.J.; Atkins, C.J.; Locksley, R.M.; Stockinger, B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 2006, 24, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, G.; Umemura, M. Interleukin-17 as an effector molecule of innate and acquired immunity against infections. Microbiol. Immunol. 2007, 51, 1139–1147. [Google Scholar] [CrossRef]
- Kim, C.H. Migration and function of Th17 cells. Inflamm. Allergy Drug Targets 2009, 8, 221–228. [Google Scholar] [CrossRef]
- Acosta-Rodriguez, E.V.; Rivino, L.; Geginat, J.; Jarrossay, D.; Gattorno, M.; Lanzavecchia, A.; Sallusto, F.; Napolitani, G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 2007, 8, 639–646. [Google Scholar] [CrossRef]
- Annunziato, F.; Cosmi, L.; Santarlasci, V.; Maggi, L.; Liotta, F.; Mazzinghi, B.; Parente, E.; Fili, L.; Ferri, S.; Frosali, F.; et al. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 2007, 204, 1849–1861. [Google Scholar] [CrossRef] [Green Version]
- Rankin, L.C.; Girard-Madoux, M.J.; Seillet, C.; Mielke, L.A.; Kerdiles, Y.; Fenis, A.; Wieduwild, E.; Putoczki, T.; Mondot, S.; Lantz, O.; et al. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat. Immunol. 2016, 17, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Sano, T.; Huang, W.; Hall, J.A.; Yang, Y.; Chen, A.; Gavzy, S.J.; Lee, J.Y.; Ziel, J.W.; Miraldi, E.R.; Domingos, A.I.; et al. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell 2015, 163, 381–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, K.; Baptista, A.P.; Tamoutounour, S.; Zhuang, L.; Bouladoux, N.; Martins, A.J.; Huang, Y.; Gerner, M.Y.; Belkaid, Y.; Germain, R.N. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 2018, 554, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Paul, W.E. Heterogeneity and plasticity of T helper cells. Cell Res. 2010, 20, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Awasthi, A.; Yosef, N.; Quintana, F.J.; Xiao, S.; Peters, A.; Wu, C.; Kleinewietfeld, M.; Kunder, S.; Hafler, D.A.; et al. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 2012, 13, 991–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Yosef, N.; Gaublomme, J.; Wu, C.; Lee, Y.; Clish, C.B.; Kaminski, J.; Xiao, S.; Meyer Zu Horste, G.; Pawlak, M.; et al. CD5L/AIM Regulates Lipid Biosynthesis and Restrains Th17 Cell Pathogenicity. Cell 2015, 163, 1413–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amir, M.; Chaudhari, S.; Wang, R.; Campbell, S.; Mosure, S.A.; Chopp, L.B.; Lu, Q.; Shang, J.; Pelletier, O.B.; He, Y.; et al. REV-ERBalpha Regulates TH17 Cell Development and Autoimmunity. Cell Rep. 2018, 25, 3733–3749.e3738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinewietfeld, M.; Manzel, A.; Titze, J.; Kvakan, H.; Yosef, N.; Linker, R.A.; Muller, D.N.; Hafler, D.A. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 2013, 496, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mahler, A.; Balogh, A.; Marko, L.; et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017, 551, 585–589. [Google Scholar] [CrossRef]
- Bacher, P.; Hohnstein, T.; Beerbaum, E.; Rocker, M.; Blango, M.G.; Kaufmann, S.; Rohmel, J.; Eschenhagen, P.; Grehn, C.; Seidel, K.; et al. Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans. Cell 2019, 176, 1340–1355.e1315. [Google Scholar] [CrossRef] [Green Version]
- Britton, G.J.; Contijoch, E.J.; Mogno, I.; Vennaro, O.H.; Llewellyn, S.R.; Ng, R.; Li, Z.; Mortha, A.; Merad, M.; Das, A.; et al. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORgammat(+) Regulatory T Cells and Exacerbate Colitis in Mice. Immunity 2019, 50, 212–224.e214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, S.A.; Kim, M.; Kang, M.C.; Kong, J.S.; Kim, K.M.; Lee, S.; Hong, B.K.; Jeong, G.H.; Lee, J.; Shin, M.G.; et al. Placental growth factor regulates the generation of TH17 cells to link angiogenesis with autoimmunity. Nat. Immunol. 2019, 20, 1348–1359. [Google Scholar] [CrossRef]
- Harbour, S.N.; Maynard, C.L.; Zindl, C.L.; Schoeb, T.R.; Weaver, C.T. Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc. Natl. Acad. Sci. USA 2015, 112, 7061–7066. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.O.; Wolf, M.M.; Madden, M.Z.; Andrejeva, G.; Sugiura, A.; Contreras, D.C.; Maseda, D.; Liberti, M.V.; Paz, K.; Kishton, R.J.; et al. Distinct Regulation of Th17 and Th1 Cell Differentiation by Glutaminase-Dependent Metabolism. Cell 2018, 175, 1780–1795.e1719. [Google Scholar] [CrossRef] [Green Version]
- Kono, M.; Yoshida, N.; Maeda, K.; Tsokos, G.C. Transcriptional factor ICER promotes glutaminolysis and the generation of Th17 cells. Proc. Natl. Acad. Sci. USA 2018, 115, 2478–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, S.A.; Yang, W.; Yan, Z.; Qin, H.; Benveniste, E.N. CK2 Controls Th17 and Regulatory T Cell Differentiation Through Inhibition of FoxO1. J. Immunol. 2018, 201, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Karmaus, P.W.F.; Chen, X.; Lim, S.A.; Herrada, A.A.; Nguyen, T.M.; Xu, B.; Dhungana, Y.; Rankin, S.; Chen, W.; Rosencrance, C.; et al. Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 2019, 565, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Omenetti, S.; Bussi, C.; Metidji, A.; Iseppon, A.; Lee, S.; Tolaini, M.; Li, Y.; Kelly, G.; Chakravarty, P.; Shoaie, S.; et al. The Intestine Harbors Functionally Distinct Homeostatic Tissue-Resident and Inflammatory Th17 Cells. Immunity 2019, 51, 77–89.e76. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Godec, J.; Ben-Aissa, K.; Cui, K.; Zhao, K.; Pucsek, A.B.; Lee, Y.K.; Weaver, C.T.; Yagi, R.; Lazarevic, V. The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-gamma-producing T helper 17 cells. Immunity 2014, 40, 355–366. [Google Scholar] [CrossRef] [Green Version]
- Okada, S.; Markle, J.G.; Deenick, E.K.; Mele, F.; Averbuch, D.; Lagos, M.; Alzahrani, M.; Al-Muhsen, S.; Halwani, R.; Ma, C.S.; et al. IMMUNODEFICIENCIES. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science 2015, 349, 606–613. [Google Scholar] [CrossRef] [Green Version]
- Buckley, R.H. The hyper-IgE syndrome. Clin. Rev. Allergy Immunol. 2001, 20, 139–154. [Google Scholar]
- Holland, S.M.; DeLeo, F.R.; Elloumi, H.Z.; Hsu, A.P.; Uzel, G.; Brodsky, N.; Freeman, A.F.; Demidowich, A.; Davis, J.; Turner, M.L.; et al. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 2007, 357, 1608–1619. [Google Scholar]
- Minegishi, Y.; Saito, M.; Tsuchiya, S.; Tsuge, I.; Takada, H.; Hara, T.; Kawamura, N.; Ariga, T.; Pasic, S.; Stojkovic, O.; et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 2007, 448, 1058–1062. [Google Scholar] [PubMed]
- Deng, Y.; Li, T.; Xie, X.; Xia, D.; Ding, L.; Xiang, H.; Ma, J.J.; Li, W. Hyper IgE syndrome associated with novel and recurrent STAT3 mutations: Two case reports. Medicine 2019, 98, e14003. [Google Scholar] [CrossRef]
- Engelhardt, K.R.; McGhee, S.; Winkler, S.; Sassi, A.; Woellner, C.; Lopez-Herrera, G.; Chen, A.; Kim, H.S.; Lloret, M.G.; Schulze, I.; et al. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J. Allergy Clin. Immunol. 2009, 124, 1289–1302.e1284. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Davis, J.C.; Lamborn, I.T.; Freeman, A.F.; Jing, H.; Favreau, A.J.; Matthews, H.F.; Davis, J.; Turner, M.L.; Uzel, G.; et al. Combined immunodeficiency associated with DOCK8 mutations. N. Engl. J. Med. 2009, 361, 2046–2055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupuis, S.; Dargemont, C.; Fieschi, C.; Thomassin, N.; Rosenzweig, S.; Harris, J.; Holland, S.M.; Schreiber, R.D.; Casanova, J.L. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science 2001, 293, 300–303. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, S.; Jouanguy, E.; Al-Hajjar, S.; Fieschi, C.; Al-Mohsen, I.Z.; Al-Jumaah, S.; Yang, K.; Chapgier, A.; Eidenschenk, C.; Eid, P.; et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 2003, 33, 388–391. [Google Scholar] [CrossRef]
- Liu, L.; Okada, S.; Kong, X.F.; Kreins, A.Y.; Cypowyj, S.; Abhyankar, A.; Toubiana, J.; Itan, Y.; Audry, M.; Nitschke, P.; et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J. Exp. Med. 2011, 208, 1635–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van de Veerdonk, F.L.; Plantinga, T.S.; Hoischen, A.; Smeekens, S.P.; Joosten, L.A.; Gilissen, C.; Arts, P.; Rosentul, D.C.; Carmichael, A.J.; Smits-van der Graaf, C.A.; et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N. Engl. J. Med. 2011, 365, 54–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puel, A.; Cypowyj, S.; Bustamante, J.; Wright, J.F.; Liu, L.; Lim, H.K.; Migaud, M.; Israel, L.; Chrabieh, M.; Audry, M.; et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science 2011, 332, 65–68. [Google Scholar] [CrossRef] [Green Version]
- Ben-Sasson, S.Z.; Hu-Li, J.; Quiel, J.; Cauchetaux, S.; Ratner, M.; Shapira, I.; Dinarello, C.A.; Paul, W.E. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 7119–7124. [Google Scholar] [CrossRef] [Green Version]
- Masters, S.L.; Simon, A.; Aksentijevich, I.; Kastner, D.L. Horror autoinflammaticus: The molecular pathophysiology of autoinflammatory disease (*). Annu. Rev. Immunol. 2009, 27, 621–668. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.L.; Liang, S.; Li, J.; Napierata, L.; Brown, T.; Benoit, S.; Senices, M.; Gill, D.; Dunussi-Joannopoulos, K.; Collins, M.; et al. IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation. J. Clin. Investig. 2008, 118, 597–607. [Google Scholar] [CrossRef] [Green Version]
- Torres, T.; Faria, R. Ustekinumab: The “New Kid on the Block” in the Treatment of Psoriatic Arthritis. Drug Dev. Res. 2015, 76, 428–431. [Google Scholar] [CrossRef] [PubMed]
- McGeachy, M.J.; Cua, D.J.; Gaffen, S.L. The IL-17 Family of Cytokines in Health and Disease. Immunity 2019, 50, 892–906. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.R.; Littman, D.R. Small molecule inhibitors of RORgammat: Targeting Th17 cells and other applications. Eur. J. Immunol. 2012, 42, 2232–2237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakaguchi, S.; Sakaguchi, N.; Asano, M.; Itoh, M.; Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995, 155, 1151–1164. [Google Scholar]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef] [Green Version]
- Khattri, R.; Cox, T.; Yasayko, S.A.; Ramsdell, F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 2003, 4, 337–342. [Google Scholar] [CrossRef]
- Raffin, C.; Vo, L.T.; Bluestone, J.A. Treg cell-based therapies: Challenges and perspectives. Nat. Rev. Immunol. 2020, 20, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Shevach, E.M.; Thornton, A.M. tTregs, pTregs, and iTregs: Similarities and differences. Immunol. Rev. 2014, 259, 88–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanamori, M.; Nakatsukasa, H.; Okada, M.; Lu, Q.; Yoshimura, A. Induced Regulatory T Cells: Their Development, Stability, and Applications. Trends Immunol. 2016, 37, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Jin, W.; Hardegen, N.; Lei, K.J.; Li, L.; Marinos, N.; McGrady, G.; Wahl, S.M. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 2003, 198, 1875–1886. [Google Scholar] [CrossRef] [PubMed]
- Fantini, M.C.; Becker, C.; Monteleone, G.; Pallone, F.; Galle, P.R.; Neurath, M.F. Cutting edge: TGF-beta induces a regulatory phenotype in CD4+CD25- T cells through Foxp3 induction and down-regulation of Smad7. J. Immunol. 2004, 172, 5149–5153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, S.; Zhang, N.; Yopp, A.C.; Chen, D.; Mao, M.; Chen, D.; Zhang, H.; Ding, Y.; Bromberg, J.S. TGF-beta induces Foxp3 + T-regulatory cells from CD4 + CD25—Precursors. Am. J. Transpl. 2004, 4, 1614–1627. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.G.; Wang, J.H.; Gray, J.D.; Soucier, H.; Horwitz, D.A. Natural and induced CD4+CD25+ cells educate CD4+CD25- cells to develop suppressive activity: The role of IL-2, TGF-beta, and IL-10. J. Immunol. 2004, 172, 5213–5221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiering, C.; Krausgruber, T.; Chomka, A.; Frohlich, A.; Adelmann, K.; Wohlfert, E.A.; Pott, J.; Griseri, T.; Bollrath, J.; Hegazy, A.N.; et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 2014, 513, 564–568. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.M.; Hall, J.A.; Blank, R.B.; Bouladoux, N.; Oukka, M.; Mora, J.R.; Belkaid, Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 2007, 204, 1775–1785. [Google Scholar] [CrossRef] [Green Version]
- Coombes, J.L.; Siddiqui, K.R.; Arancibia-Carcamo, C.V.; Hall, J.; Sun, C.M.; Belkaid, Y.; Powrie, F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 2007, 204, 1757–1764. [Google Scholar] [CrossRef]
- Lathrop, S.K.; Bloom, S.M.; Rao, S.M.; Nutsch, K.; Lio, C.W.; Santacruz, N.; Peterson, D.A.; Stappenbeck, T.S.; Hsieh, C.S. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011, 478, 250–254. [Google Scholar] [CrossRef]
- Thornton, A.M.; Korty, P.E.; Tran, D.Q.; Wohlfert, E.A.; Murray, P.E.; Belkaid, Y.; Shevach, E.M. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 2010, 184, 3433–3441. [Google Scholar] [CrossRef] [Green Version]
- Yadav, M.; Louvet, C.; Davini, D.; Gardner, J.M.; Martinez-Llordella, M.; Bailey-Bucktrout, S.; Anthony, B.A.; Sverdrup, F.M.; Head, R.; Kuster, D.J.; et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J. Exp. Med. 2012, 209, 1713–1722. [Google Scholar] [CrossRef] [Green Version]
- Floess, S.; Freyer, J.; Siewert, C.; Baron, U.; Olek, S.; Polansky, J.; Schlawe, K.; Chang, H.D.; Bopp, T.; Schmitt, E.; et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007, 5, e38. [Google Scholar] [CrossRef] [PubMed]
- Polansky, J.K.; Kretschmer, K.; Freyer, J.; Floess, S.; Garbe, A.; Baron, U.; Olek, S.; Hamann, A.; von Boehmer, H.; Huehn, J. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 2008, 38, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Ohnmacht, C.; Park, J.H.; Cording, S.; Wing, J.B.; Atarashi, K.; Obata, Y.; Gaboriau-Routhiau, V.; Marques, R.; Dulauroy, S.; Fedoseeva, M.; et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 2015, 349, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Sefik, E.; Geva-Zatorsky, N.; Oh, S.; Konnikova, L.; Zemmour, D.; McGuire, A.M.; Burzyn, D.; Ortiz-Lopez, A.; Lobera, M.; Yang, J.; et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 2015, 349, 993–997. [Google Scholar] [CrossRef] [Green Version]
- Takimoto, T.; Wakabayashi, Y.; Sekiya, T.; Inoue, N.; Morita, R.; Ichiyama, K.; Takahashi, R.; Asakawa, M.; Muto, G.; Mori, T.; et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 2010, 185, 842–855. [Google Scholar] [CrossRef] [Green Version]
- Zorn, E.; Nelson, E.A.; Mohseni, M.; Porcheray, F.; Kim, H.; Litsa, D.; Bellucci, R.; Raderschall, E.; Canning, C.; Soiffer, R.J.; et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood 2006, 108, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- Burchill, M.A.; Yang, J.; Vogtenhuber, C.; Blazar, B.R.; Farrar, M.A. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007, 178, 280–290. [Google Scholar] [CrossRef] [Green Version]
- Rudra, D.; Egawa, T.; Chong, M.M.; Treuting, P.; Littman, D.R.; Rudensky, A.Y. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 2009, 10, 1170–1177. [Google Scholar] [CrossRef]
- Sekiya, T.; Kashiwagi, I.; Inoue, N.; Morita, R.; Hori, S.; Waldmann, H.; Rudensky, A.Y.; Ichinose, H.; Metzger, D.; Chambon, P.; et al. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2011, 2, 269. [Google Scholar] [CrossRef] [Green Version]
- Kerdiles, Y.M.; Stone, E.L.; Beisner, D.R.; McGargill, M.A.; Ch’en, I.L.; Stockmann, C.; Katayama, C.D.; Hedrick, S.M. Foxo transcription factors control regulatory T cell development and function. Immunity 2010, 33, 890–904. [Google Scholar] [CrossRef] [Green Version]
- Kitagawa, Y.; Ohkura, N.; Kidani, Y.; Vandenbon, A.; Hirota, K.; Kawakami, R.; Yasuda, K.; Motooka, D.; Nakamura, S.; Kondo, M.; et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 2017, 18, 173–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Getnet, D.; Grosso, J.F.; Goldberg, M.V.; Harris, T.J.; Yen, H.R.; Bruno, T.C.; Durham, N.M.; Hipkiss, E.L.; Pyle, K.J.; Wada, S.; et al. A role for the transcription factor Helios in human CD4(+)CD25(+) regulatory T cells. Mol. Immunol. 2010, 47, 1595–1600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shevach, E.M. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity 2009, 30, 636–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spolski, R.; Li, P.; Leonard, W.J. Biology and regulation of IL-2: From molecular mechanisms to human therapy. Nat. Rev. Immunol. 2018, 18, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gerner, M.Y.; Van Panhuys, N.; Levine, A.G.; Rudensky, A.Y.; Germain, R.N. Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature 2015, 528, 225–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onishi, Y.; Fehervari, Z.; Yamaguchi, T.; Sakaguchi, S. Foxp3+ natural regulatory T cells preferentially form aggregates on dendritic cells in vitro and actively inhibit their maturation. Proc. Natl. Acad. Sci. USA 2008, 105, 10113–10118. [Google Scholar] [CrossRef] [Green Version]
- Grossman, W.J.; Verbsky, J.W.; Barchet, W.; Colonna, M.; Atkinson, J.P.; Ley, T.J. Human T regulatory cells can use the perforin pathway to cause autologous target cell death. Immunity 2004, 21, 589–601. [Google Scholar] [CrossRef] [Green Version]
- Akkaya, B.; Oya, Y.; Akkaya, M.; Al Souz, J.; Holstein, A.H.; Kamenyeva, O.; Kabat, J.; Matsumura, R.; Dorward, D.W.; Glass, D.D.; et al. Regulatory T cells mediate specific suppression by depleting peptide-MHC class II from dendritic cells. Nat. Immunol. 2019, 20, 218–231. [Google Scholar] [CrossRef]
- Bennett, C.L.; Christie, J.; Ramsdell, F.; Brunkow, M.E.; Ferguson, P.J.; Whitesell, L.; Kelly, T.E.; Saulsbury, F.T.; Chance, P.F.; Ochs, H.D. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001, 27, 20–21. [Google Scholar] [CrossRef]
- Brunkow, M.E.; Jeffery, E.W.; Hjerrild, K.A.; Paeper, B.; Clark, L.B.; Yasayko, S.A.; Wilkinson, J.E.; Galas, D.; Ziegler, S.F.; Ramsdell, F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001, 27, 68–73. [Google Scholar] [CrossRef]
- Wildin, R.S.; Ramsdell, F.; Peake, J.; Faravelli, F.; Casanova, J.L.; Buist, N.; Levy-Lahad, E.; Mazzella, M.; Goulet, O.; Perroni, L.; et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 2001, 27, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Van Gool, F.; Nguyen, M.L.T.; Mumbach, M.R.; Satpathy, A.T.; Rosenthal, W.L.; Giacometti, S.; Le, D.T.; Liu, W.; Brusko, T.M.; Anderson, M.S.; et al. A Mutation in the Transcription Factor Foxp3 Drives T Helper 2 Effector Function in Regulatory T Cells. Immunity 2019, 50, 362–377.e366. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.H.; Hu, H.; Jin, J.; Puebla-Osorio, N.; Xiao, Y.; Gilbert, B.E.; Brink, R.; Ullrich, S.E.; Sun, S.C. TRAF3 regulates the effector function of regulatory T cells and humoral immune responses. J. Exp. Med. 2014, 211, 137–151. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Huynh, A.; Whitcher, G.; Chang, J.; Maltzman, J.S.; Turka, L.A. An obligate cell-intrinsic function for CD28 in Tregs. J. Clin. Investig. 2013, 123, 580–593. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, M.; Miyazaki, K.; Chen, S.; Itoi, M.; Miller, M.; Lu, L.F.; Varki, N.; Chang, A.N.; Broide, D.H.; Murre, C. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nat. Immunol. 2014, 15, 767–776. [Google Scholar] [CrossRef]
- Chang, J.H.; Xiao, Y.; Hu, H.; Jin, J.; Yu, J.; Zhou, X.; Wu, X.; Johnson, H.M.; Akira, S.; Pasparakis, M.; et al. Ubc13 maintains the suppressive function of regulatory T cells and prevents their conversion into effector-like T cells. Nat. Immunol. 2012, 13, 481–490. [Google Scholar] [CrossRef]
- Layman, A.A.K.; Deng, G.; O’Leary, C.E.; Tadros, S.; Thomas, R.M.; Dybas, J.M.; Moser, E.K.; Wells, A.D.; Doliba, N.M.; Oliver, P.M. Ndfip1 restricts mTORC1 signalling and glycolysis in regulatory T cells to prevent autoinflammatory disease. Nat. Commun. 2017, 8, 15677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, H.; Grinberg-Bleyer, Y.; Liao, W.; Maloney, D.; Wang, P.; Wu, Z.; Wang, J.; Bhatt, D.M.; Heise, N.; Schmid, R.M.; et al. An NF-kappaB Transcription-Factor-Dependent Lineage-Specific Transcriptional Program Promotes Regulatory T Cell Identity and Function. Immunity 2017, 47, 450–465.e455. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, M.; Lopez-Ocasio, M.; Metidji, A.; Rieder, S.A.; Shevach, E.M.; Thornton, A.M. Helios Controls a Limited Subset of Regulatory T Cell Functions. J. Immunol. 2016, 196, 144–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpenter, A.C.; Wohlfert, E.; Chopp, L.B.; Vacchio, M.S.; Nie, J.; Zhao, Y.; Shetty, J.; Xiao, Q.; Deng, C.; Tran, B.; et al. Control of Regulatory T Cell Differentiation by the Transcription Factors Thpok and LRF. J. Immunol. 2017, 199, 1716–1728. [Google Scholar] [CrossRef]
- DuPage, M.; Chopra, G.; Quiros, J.; Rosenthal, W.L.; Morar, M.M.; Holohan, D.; Zhang, R.; Turka, L.; Marson, A.; Bluestone, J.A. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity 2015, 42, 227–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roychoudhuri, R.; Hirahara, K.; Mousavi, K.; Clever, D.; Klebanoff, C.A.; Bonelli, M.; Sciume, G.; Zare, H.; Vahedi, G.; Dema, B.; et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 2013, 498, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Gasper, D.J.; Lee, S.H.; Plisch, E.H.; Svaren, J.; Suresh, M. Bach2 regulates homeostasis of Foxp3+ regulatory T cells and protects against fatal lung disease in mice. J. Immunol. 2014, 192, 985–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, M.; Tanaka, Y.; Kuwabara, T.; Naito, T.; Kohwi-Shigematsu, T.; Watanabe, A. SATB1 Plays a Critical Role in Establishment of Immune Tolerance. J. Immunol. 2016, 196, 563–572. [Google Scholar] [CrossRef] [Green Version]
- Cretney, E.; Xin, A.; Shi, W.; Minnich, M.; Masson, F.; Miasari, M.; Belz, G.T.; Smyth, G.K.; Busslinger, M.; Nutt, S.L.; et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 2011, 12, 304–311. [Google Scholar] [CrossRef]
- Yang, K.; Blanco, D.B.; Neale, G.; Vogel, P.; Avila, J.; Clish, C.B.; Wu, C.; Shrestha, S.; Rankin, S.; Long, L.; et al. Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature 2017, 548, 602–606. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Rao, S.; Tsai, L.M.; Lee, S.K.; He, Y.; Sutcliffe, E.L.; Srivastava, M.; Linterman, M.; Zheng, L.; Simpson, N.; et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 2009, 31, 457–468. [Google Scholar] [CrossRef]
- Nurieva, R.I.; Chung, Y.; Martinez, G.J.; Yang, X.O.; Tanaka, S.; Matskevitch, T.D.; Wang, Y.H.; Dong, C. Bcl6 mediates the development of T follicular helper cells. Science 2009, 325, 1001–1005. [Google Scholar] [CrossRef] [Green Version]
- Johnston, R.J.; Poholek, A.C.; DiToro, D.; Yusuf, I.; Eto, D.; Barnett, B.; Dent, A.L.; Craft, J.; Crotty, S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 2009, 325, 1006–1010. [Google Scholar] [CrossRef] [Green Version]
- Vinuesa, C.G.; Linterman, M.A.; Yu, D.; MacLennan, I.C. Follicular Helper T Cells. Annu. Rev. Immunol. 2016, 34, 335–368. [Google Scholar] [CrossRef]
- Roco, J.A.; Mesin, L.; Binder, S.C.; Nefzger, C.; Gonzalez-Figueroa, P.; Canete, P.F.; Ellyard, J.; Shen, Q.; Robert, P.A.; Cappello, J.; et al. Class-Switch Recombination Occurs Infrequently in Germinal Centers. Immunity 2019, 51, 337–350.e337. [Google Scholar] [CrossRef]
- Nurieva, R.I.; Chung, Y.; Hwang, D.; Yang, X.O.; Kang, H.S.; Ma, L.; Wang, Y.H.; Watowich, S.S.; Jetten, A.M.; Tian, Q.; et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 2008, 29, 138–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, K.T.; Kanno, Y.; Cannons, J.L.; Handon, R.; Bible, P.; Elkahloun, A.G.; Anderson, S.M.; Wei, L.; Sun, H.; O’Shea, J.J.; et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity 2011, 35, 622–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fazilleau, N.; McHeyzer-Williams, L.J.; Rosen, H.; McHeyzer-Williams, M.G. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat. Immunol. 2009, 10, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Spolski, R.; Leonard, W.J. Interleukin-21: A double-edged sword with therapeutic potential. Nat. Rev. Drug Discov. 2014, 13, 379–395. [Google Scholar] [CrossRef]
- DiToro, D.; Winstead, C.J.; Pham, D.; Witte, S.; Andargachew, R.; Singer, J.R.; Wilson, C.G.; Zindl, C.L.; Luther, R.J.; Silberger, D.J.; et al. Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science 2018, 361, eaao2933. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S. T follicular helper cell differentiation, function, and roles in disease. Immunity 2014, 41, 529–542. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.S.; Deenick, E.K.; Batten, M.; Tangye, S.G. The origins, function, and regulation of T follicular helper cells. J. Exp. Med. 2012, 209, 1241–1253. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Chen, X.; Zhong, B.; Wang, A.; Wang, X.; Chu, F.; Nurieva, R.I.; Yan, X.; Chen, P.; van der Flier, L.G.; et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature 2014, 507, 513–518. [Google Scholar] [CrossRef]
- Xiao, N.M.; Eto, D.; Elly, C.; Peng, G.Y.; Crotty, S.; Liu, Y.C. The E3 ubiquitin ligase Itch is required for the differentiation of follicular helper T cells. Nat. Immunol. 2014, 15, 657–666. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Zhao, Y.; Zou, L.; Zhang, D.; Aki, D.; Liu, Y.C. The E3 ligase VHL promotes follicular helper T cell differentiation via glycolytic-epigenetic control. J. Exp. Med. 2019, 216, 1664–1681. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Zhu, L.; Lu, Z.; Chen, H.; Fan, L.; Xue, Q.; Shi, J.; Li, M.; Li, H.; Gong, J.; et al. IL-37 Represses the Autoimmunity in Myasthenia Gravis via Directly Targeting Follicular Th and B Cells. J. Immunol. 2020, 204, 1736–1745. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Raybuck, A.L.; Blagih, J.; Kemboi, E.; Haase, V.H.; Jones, R.G.; Boothby, M.R. Hypoxia-inducible factors in CD4(+) T cells promote metabolism, switch cytokine secretion, and T cell help in humoral immunity. Proc. Natl. Acad. Sci. USA 2019, 116, 8975–8984. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Yang, Q.; Cao, Y.J.; Yuan, W.M.; Lei, A.H.; Zhou, P.; Zhou, W.; Liu, Y.D.; Shi, M.H.; Yang, Q.; et al. Cyclooxygenase-1 Regulates the Development of Follicular Th Cells via Prostaglandin E2. J. Immunol. 2019, 203, 864–872. [Google Scholar] [CrossRef]
- Vacchio, M.S.; Ciucci, T.; Gao, Y.; Watanabe, M.; Balmaceno-Criss, M.; McGinty, M.T.; Huang, A.; Xiao, Q.; McConkey, C.; Zhao, Y.; et al. A Thpok-Directed Transcriptional Circuitry Promotes Bcl6 and Maf Expression to Orchestrate T Follicular Helper Differentiation. Immunity 2019, 51, 465–478.e466. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhao, X.; Wang, X.; Feng, H.; Gou, M.; Jin, W.; Wang, X.; Liu, X.; Dong, C. The Transcription Factor Tox2 Drives T Follicular Helper Cell Development via Regulating Chromatin Accessibility. Immunity 2019, 51, 826–839.e825. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Dutta, P.R.; Cerasoli, D.M.; Kelsoe, G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. V. Affinity maturation develops in two stages of clonal selection. J. Exp. Med. 1998, 187, 885–895. [Google Scholar] [CrossRef]
- Han, S.; Hathcock, K.; Zheng, B.; Kepler, T.B.; Hodes, R.; Kelsoe, G. Cellular interaction in germinal centers. Roles of CD40 ligand and B7-2 in established germinal centers. J. Immunol. 1995, 155, 556–567. [Google Scholar] [PubMed]
- Zotos, D.; Coquet, J.M.; Zhang, Y.; Light, A.; D’Costa, K.; Kallies, A.; Corcoran, L.M.; Godfrey, D.I.; Toellner, K.M.; Smyth, M.J.; et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J. Exp. Med. 2010, 207, 365–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linterman, M.A.; Beaton, L.; Yu, D.; Ramiscal, R.R.; Srivastava, M.; Hogan, J.J.; Verma, N.K.; Smyth, M.J.; Rigby, R.J.; Vinuesa, C.G. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J. Exp. Med. 2010, 207, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Van Besouw, N.M.; Mendoza Rojas, A.; Baan, C.C. The role of follicular T helper cells in the humoral alloimmune response after clinical organ transplantation. HLA 2019, 94, 407–414. [Google Scholar] [CrossRef] [Green Version]
- Gowthaman, U.; Chen, J.S.; Zhang, B.; Flynn, W.F.; Lu, Y.; Song, W.; Joseph, J.; Gertie, J.A.; Xu, L.; Collet, M.A.; et al. Identification of a T follicular helper cell subset that drives anaphylactic IgE. Science 2019, 365, eaaw6433. [Google Scholar] [CrossRef] [PubMed]
- Simpson, N.; Gatenby, P.A.; Wilson, A.; Malik, S.; Fulcher, D.A.; Tangye, S.G.; Manku, H.; Vyse, T.J.; Roncador, G.; Huttley, G.A.; et al. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 2010, 62, 234–244. [Google Scholar] [CrossRef]
- Faliti, C.E.; Gualtierotti, R.; Rottoli, E.; Gerosa, M.; Perruzza, L.; Romagnani, A.; Pellegrini, G.; De Ponte Conti, B.; Rossi, R.L.; Idzko, M.; et al. P2X7 receptor restrains pathogenic Tfh cell generation in systemic lupus erythematosus. J. Exp. Med. 2019, 216, 317–336. [Google Scholar] [CrossRef]
- Psianou, K.; Panagoulias, I.; Papanastasiou, A.D.; de Lastic, A.L.; Rodi, M.; Spantidea, P.I.; Degn, S.E.; Georgiou, P.; Mouzaki, A. Clinical and immunological parameters of Sjogren’s syndrome. Autoimmun. Rev. 2018, 17, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.; Schmitt, N.; Bentebibel, S.E.; Ranganathan, R.; Bourdery, L.; Zurawski, G.; Foucat, E.; Dullaers, M.; Oh, S.; Sabzghabaei, N.; et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 2011, 34, 108–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, J.; Wei, Y.; Fonseca, V.R.; Graca, L.; Yu, D. T follicular helper cells and T follicular regulatory cells in rheumatic diseases. Nat. Rev. Rheumatol. 2019, 15, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.S.; Avery, D.T.; Chan, A.; Batten, M.; Bustamante, J.; Boisson-Dupuis, S.; Arkwright, P.D.; Kreins, A.Y.; Averbuch, D.; Engelhard, D.; et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood 2012, 119, 3997–4008. [Google Scholar] [CrossRef]
- Lee, W.I.; Torgerson, T.R.; Schumacher, M.J.; Yel, L.; Zhu, Q.; Ochs, H.D. Molecular analysis of a large cohort of patients with the hyper immunoglobulin M (IgM) syndrome. Blood 2005, 105, 1881–1890. [Google Scholar] [CrossRef]
- Du, X.; Tang, W.; Chen, X.; Zeng, T.; Wang, Y.; Chen, Z.; Xu, T.; Zhou, L.; Tang, X.; An, Y.; et al. Clinical, genetic and immunological characteristics of 40 Chinese patients with CD40 ligand deficiency. Scand. J. Immunol. 2019, 90, e12798. [Google Scholar] [CrossRef]
- Bossaller, L.; Burger, J.; Draeger, R.; Grimbacher, B.; Knoth, R.; Plebani, A.; Durandy, A.; Baumann, U.; Schlesier, M.; Welcher, A.A.; et al. ICOS deficiency is associated with a severe reduction of CXCR5+CD4 germinal center Th cells. J. Immunol. 2006, 177, 4927–4932. [Google Scholar] [CrossRef] [Green Version]
- Strutt, T.M.; McKinstry, K.K.; Dibble, J.P.; Winchell, C.; Kuang, Y.; Curtis, J.D.; Huston, G.; Dutton, R.W.; Swain, S.L. Memory CD4+ T cells induce innate responses independently of pathogen. Nat. Med. 2010, 16, 558–564. [Google Scholar] [CrossRef] [Green Version]
- Kawabe, T.; Jankovic, D.; Kawabe, S.; Huang, Y.; Lee, P.H.; Yamane, H.; Zhu, J.; Sher, A.; Germain, R.N.; Paul, W.E. Memory-phenotype CD4(+) T cells spontaneously generated under steady-state conditions exert innate TH1-like effector function. Sci. Immunol. 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Sharma, S.; Edwards, J.; Feigenbaum, L.; Zhu, J. Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat. Immunol. 2015, 16, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Chong, M.M.; Littman, D.R. Plasticity of CD4+ T cell lineage differentiation. Immunity 2009, 30, 646–655. [Google Scholar] [CrossRef] [Green Version]
- Sakaguchi, S.; Vignali, D.A.; Rudensky, A.Y.; Niec, R.E.; Waldmann, H. The plasticity and stability of regulatory T cells. Nat. Rev. Immunol. 2013, 13, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Stubbington, M.J.T.; Rozenblatt-Rosen, O.; Regev, A.; Teichmann, S.A. Single-cell transcriptomics to explore the immune system in health and disease. Science 2017, 358, 58–63. [Google Scholar] [CrossRef] [Green Version]
- Ledford, H. Quest to use CRISPR against disease gains ground. Nature 2020, 577, 156. [Google Scholar] [CrossRef]
- Renaud, J.P.; Chari, A.; Ciferri, C.; Liu, W.T.; Remigy, H.W.; Stark, H.; Wiesmann, C. Cryo-EM in drug discovery: Achievements, limitations and prospects. Nat. Rev. Drug Discov. 2018, 17, 471–492. [Google Scholar] [CrossRef] [PubMed]
- Bendall, S.C.; Simonds, E.F.; Qiu, P.; El-ad, D.A.; Krutzik, P.O.; Finck, R.; Bruggner, R.V.; Melamed, R.; Trejo, A.; Ornatsky, O.I.; et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011, 332, 687–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Germain, R.N.; Robey, E.A.; Cahalan, M.D. A decade of imaging cellular motility and interaction dynamics in the immune system. Science 2012, 336, 1676–1681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, H.; Kastenmuller, W.; Germain, R.N. Spatiotemporal basis of innate and adaptive immunity in secondary lymphoid tissue. Annu. Rev. Cell Dev. Biol. 2014, 30, 141–167. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; van Panhuys, N.; Kastenmuller, W.; Germain, R.N. The future of immunoimaging—Deeper, bigger, more precise, and definitively more colorful. Eur. J. Immunol. 2013, 43, 1407–1412. [Google Scholar] [CrossRef]
- Schreiner, D.; King, C.G. CD4+ Memory T Cells at Home in the Tissue: Mechanisms for Health and Disease. Front. Immunol. 2018, 9, 2394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jameson, S.C.; Masopust, D. Understanding Subset Diversity in T Cell Memory. Immunity 2018, 48, 214–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Zhu, J. CD4 T Helper Cell Subsets and Related Human Immunological Disorders. Int. J. Mol. Sci. 2020, 21, 8011. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218011
Zhu X, Zhu J. CD4 T Helper Cell Subsets and Related Human Immunological Disorders. International Journal of Molecular Sciences. 2020; 21(21):8011. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218011
Chicago/Turabian StyleZhu, Xiaoliang, and Jinfang Zhu. 2020. "CD4 T Helper Cell Subsets and Related Human Immunological Disorders" International Journal of Molecular Sciences 21, no. 21: 8011. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218011




