Cancer-Associated Fibroblasts Differentiated by Exosomes Isolated from Cancer Cells Promote Cancer Cell Invasion
Abstract
:1. Introduction
2. Results and Discussion
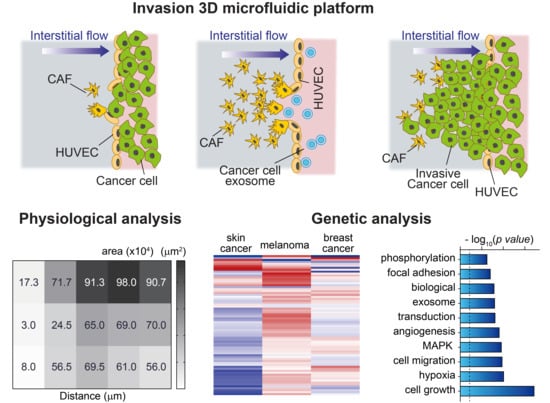
2.1. Physiological Interplay between CAFs and Cancer Cells Promotes Cancer Cell Invasion
2.2. Genetic Analysis of HUVEC Differentiation into CAF by Exosomes
2.3. Genetic Analysis of HUVEC Differentiation into CAF by Cancer Cells
2.4. Genetic Analysis of HUVEC Differentiation into CAF by Both Exosomes and Cancer Cells
3. Materials and Methods
3.1. Cultivation of Cancer Cells
3.2. Isolation of Cancer Cell-Derived Exosomes
3.3. Characterizations of Exosomes
3.4. Preparation of 3D Microfluidic Cancer Microenvironment
3.5. Culturing of HUVECs in Microfluidic Devices
3.6. Cultivation of Cancer-Associated Fibroblasts (CAFs) in the Microfluidic Device
3.7. Cultivation of Invasive Cancer Cells in Microfluidic Device
3.8. Immunostaining of Cells
3.9. Quantification of Invasive Cell Number, Invaded Area and Penetrated Distance of CAFs and Cancer Cells
3.10. RNA Extraction and NanoString nCounter Assay
3.11. Module Analysis of the Protein–Protein Interaction (PPI) Network
3.12. GeneOntology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Friedl, P.; Wolf, K. Tumour-cell invasion and migration: Diversity and escape mechanisms. Nat. Rev. Cancer 2003, 3, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Steeg, P.S. Targeting metastasis. Nat. Rev. Cancer 2016, 16, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Weidner, N.; Semple, J.P.; Welch, W.R.; Folkman, J. Tumor angiogenesis and metastasis—correlation in invasive breast carcinoma. N. Engl. J. Med. 1991, 324, 1–8. [Google Scholar] [CrossRef]
- Hunter, K.W.; Crawford, N.P.; Alsarraj, J. Mechanisms of metastasis. Breast Cancer Res. 2008, 10, S2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joyce, J.A.; Pollard, J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Zeisberg, E.M.; Tarnavski, O.; Zeisberg, M.; Dorfman, A.L.; McMullen, J.R.; Gustafsson, E.; Chandraker, A.; Yuan, X.; Pu, W.T.; Roberts, A.B. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 2007, 13, 952–961. [Google Scholar] [CrossRef]
- Truong, D.D.; Kratz, A.; Park, J.G.; Barrientos, E.S.; Saini, H.; Nguyen, T.; Pockaj, B.; Mouneimne, G.; LaBaer, J.; Nikkhah, M. A human organotypic microfluidic tumor model permits investigation of the interplay between patient-derived fibroblasts and breast cancer cells. Cancer Res. 2019, 79, 3139–3151. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, M.; De Ninno, A.; Mencattini, A.; Mermet-Meillon, F.; Fornabaio, G.; Evans, S.S.; Cossutta, M.; Khira, Y.; Han, W.; Sirven, P. Dissecting effects of anti-cancer drugs and cancer-associated fibroblasts by on-chip reconstitution of immunocompetent tumor microenvironments. Cell Rep. 2018, 25, 3884–3893 e3883. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Wang, L.-F.; Wang, R.-F. Role of cancer-associated fibroblasts in invasion and metastasis of gastric cancer. World J. Gastroenterol. 2015, 21, 9717–9726. [Google Scholar] [CrossRef]
- Pelon, F.; Bourachot, B.; Kieffer, Y.; Magagna, I.; Mermet-Meillon, F.; Bonnet, I.; Costa, A.; Givel, A.-M.; Attieh, Y.; Barbazan, J. Cancer-associated fibroblast heterogeneity in axillary lymph nodes drives metastases in breast cancer through complementary mechanisms. Nat. commu. 2020, 11, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.; Zhang, B.; Hu, Q.; Qin, Y.; Xu, W.; Liu, W.; Yu, X.; Xu, J. The impact of cancer-associated fibroblasts on major hallmarks of pancreatic cancer. Theranostics 2018, 8, 5072–5087. [Google Scholar] [CrossRef] [PubMed]
- Prakash, J. Cancer-associated fibroblasts: perspectives in cancer therapy. Trends Cancer 2016, 2, 277–279. [Google Scholar] [CrossRef]
- Erdogan, B.; Ao, M.; White, L.M.; Means, A.L.; Brewer, B.M.; Yang, L.; Washington, M.K.; Shi, C.; Franco, O.E.; Weaver, A.M. Cancer-associated fibroblasts promote directional cancer cell migration by aligning fibronectin. J. Cell Biol. 2017, 216, 3799–3816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saini, H.; Rahmani, K.; Veldhuizen, J.; Zare, A.; Allam, M.; Silva, C.; Kratz, A.; Truong, D.; Mouneimne, G.; LaBaer, J. The role of tumor-stroma interactions on desmoplasia and tumorigenicity within a microengineered 3D platform. Biomaterials 2020, 119975. [Google Scholar] [CrossRef]
- Clere, N.; Renault, S.; Corre, I. Endothelial-to-Mesenchymal Transition in Cancer. Front. Cell Dev. Biol. 2020, 8, 1–8. [Google Scholar] [CrossRef]
- Choi, K.J.; Nam, J.-K.; Kim, J.-H.; Choi, S.-H.; Lee, Y.-J. Endothelial-to-mesenchymal transition in anticancer therapy and normal tissue damage. Exp. Mol. Med. 2020, 52, 781–792. [Google Scholar] [CrossRef]
- Jiao, K.; Zhen, J.; Wu, M.; Teng, M.; Yang, K.; Zhou, Q.; Hu, C.; Zhou, M.; Li, Y.; Li, Z. 27-Hydroxycholesterol-induced EndMT acts via STAT3 signaling to promote breast cancer cell migration by altering the tumor microenvironment. Cancer Biol. Med. 2020, 17, 88–100. [Google Scholar]
- Motohara, T.; Masuda, K.; Morotti, M.; Zheng, Y.; El-Sahhar, S.; Chong, K.Y.; Wietek, N.; Alsaadi, A.; Karaminejadranjbar, M.; Hu, Z. An evolving story of the metastatic voyage of ovarian cancer cells: Cellular and molecular orchestration of the adipose-rich metastatic microenvironment. Oncogene 2019, 38, 2885–2898. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, G.J. Regulation of heterogeneous cancer-associated fibroblasts: The molecular pathology of activated signaling pathways. J. Exp. Clin. Cancer Res. 2020, 39, 1–15. [Google Scholar] [CrossRef]
- Piper, M.; Mueller, A.C.; Karam, S.D. The interplay between cancer associated fibroblasts and immune cells in the context of radiation therapy. Mol. Carcinog. 2020, 59, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Yeon, J.H.; Jeong, H.E.; Seo, H.; Cho, S.; Kim, K.; Na, D.; Chung, S.; Park, J.; Choi, N.; Kang, J.Y. Cancer-derived exosomes trigger endothelial to mesenchymal transition followed by the induction of cancer-associated fibroblasts. Acta Biomater. 2018, 76, 146–153. [Google Scholar] [CrossRef]
- Kim, H.; Lee, S.; Shin, E.; Seong, K.M.; Jin, Y.W.; Youn, H.; Youn, B. The Emerging Roles of Exosomes as EMT Regulators in Cancer. Cells 2020, 9, 861. [Google Scholar] [CrossRef] [PubMed]
- Bussard, K.M.; Mutkus, L.; Stumpf, K.; Gomez-Manzano, C.; Marini, F.C. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016, 18, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Othman, N.; Jamal, R.; Abu, N. Cancer-derived exosomes as effectors of key inflammation-related players. Front. Immunol. 2019, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Chen, M.; Jiang, R.; Guo, Y.; Wu, M.; Zhang, X. Exosome-related tumor microenvironment. J. Cancer 2018, 9, 3084–3092. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, S.; Yoshioka, Y.; Prieto-Vila, M.; Ochiya, T. Involvement of extracellular vesicles in vascular-related functions in cancer progression and metastasis. Int. J. Mol. Sci. 2019, 20, 2584. [Google Scholar] [CrossRef] [Green Version]
- Conigliaro, A.; Cicchini, C. Exosome-mediated signaling in epithelial to mesenchymal transition and tumor progression. J. Clin. Med. 2019, 8, 26. [Google Scholar] [CrossRef] [Green Version]
- Richards, K.E.; Zeleniak, A.E.; Fishel, M.L.; Wu, J.; Littlepage, L.E.; Hill, R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene 2017, 36, 1770. [Google Scholar] [CrossRef] [Green Version]
- Ruivo, C.F.; Adem, B.; Silva, M.; Melo, S.A. The biology of cancer exosomes: Insights and new perspectives. Cancer Res. 2017, 77, 6480–6488. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Luo, J.-d.; Jiang, H.; Duan, D.D. Tumor exosomes: A double-edged sword in cancer therapy. Acta Pharmacol. Sin. 2018, 39, 534–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Zhong, J.; Zhong, B.; Huang, J.; Jiang, L.; Jiang, Y.; Yuan, J.; Sun, J.; Dai, L.; Yang, C. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020, 476, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Deng, W.; Klinke II, D.J. Exosomes: Improved methods to characterize their morphology, RNA content, and surface protein biomarkers. Analyst 2015, 140, 6631–6642. [Google Scholar] [CrossRef] [Green Version]
- Ning, X.; Zhang, H.; Wang, C.; Song, X. Exosomes released by gastric cancer cells induce transition of pericytes into cancer-associated fibroblasts. Med. Sci. Monit. 2018, 24, 2350–2359. [Google Scholar] [CrossRef]
- Erdogan, B.; Webb, D.J. Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochem. Soc. Trans. 2017, 45, 229–236. [Google Scholar] [CrossRef] [Green Version]
- Saiki, I.; Murataxd, J.; Yoneda, J.; Kobayashi, H.; Azuma, I. Influence of fibroblasts on the invasion and migration of highly or weakly metastatic B16 melanoma cells. Int. J. Cancer 1994, 56, 867–873. [Google Scholar] [CrossRef]
- Nicolson, G.L.; Dulski, K.; Basson, C.; Welch, D.R. Preferential organ attachment and invasion in vitro by B16 melanoma cells selected for differing metastatic colonization and invasive properties. Invasion Metastasis 1985, 5, 144–158. [Google Scholar]
- Didona, D.; Paolino, G.; Bottoni, U.; Cantisani, C. Non melanoma skin cancer pathogenesis overview. Biomedicines 2018, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Apalla, Z.; Nashan, D.; Weller, R.B.; Castellsague, X. Skin cancer: Epidemiology, disease burden, pathophysiology, diagnosis, and therapeutic approaches. Dermatol. Ther. 2017, 7, 5–19. [Google Scholar] [CrossRef] [Green Version]
- Martinez, J.-C.; Otley, C.C. The management of melanoma and nonmelanoma skin cancer: A review for the primary care physician. Mayo Clin. Proc. 2001, 76, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Sonnenschein, C.; Soto, A.M. Cancer metastases: So close and so far. J. Natl. Cancer Inst. 2015, 107, 1253–1265. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Wang, L.; Liu, S.; Wang, Y.; Liu, Y.; Wang, M.; Nakshatri, H.; Li, B.-Y.; Yokota, H. Attraction and Compaction of Migratory Breast Cancer Cells by Bone Matrix Proteins through Tumor-Osteocyte Interactions. Sci. Rep. 2018, 8, 5420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasari, S.; Fang, Y.; Mitra, A.K. Cancer associated fibroblasts: Naughty neighbors that drive ovarian cancer progression. Cancers 2018, 10, 406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-mediated metastasis: Communication from a distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Cesano, A. nCounter® PanCancer immune profiling panel (NanoString technologies, Inc., Seattle, WA). J. Immunother. Cancer 2015, 3, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suh, S.-S.; Yoo, J.Y.; Cui, R.; Kaur, B.; Huebner, K.; Lee, T.-K.; Aqeilan, R.I.; Croce, C.M. FHIT suppresses epithelial-mesenchymal transition (EMT) and metastasis in lung cancer through modulation of microRNAs. PLoS Genet 2014, 10, e1004652. [Google Scholar] [CrossRef] [PubMed]
- Shintani, Y.; Fujiwara, A.; Kimura, T.; Kawamura, T.; Funaki, S.; Minami, M.; Okumura, M. IL-6 secreted from cancer-associated fibroblasts mediates chemoresistance in NSCLC by increasing epithelial-mesenchymal transition signaling. J. Thorac. Oncol. 2016, 11, 1482–1492. [Google Scholar] [CrossRef] [Green Version]
- Goulet, C.R.; Champagne, A.; Bernard, G.; Vandal, D.; Chabaud, S.; Pouliot, F.; Bolduc, S. Cancer-associated fibroblasts induce epithelial–mesenchymal transition of bladder cancer cells through paracrine IL-6 signalling. BMC Cancer 2019, 19, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Stivarou, T.; Patsavoudi, E. Extracellular molecules involved in cancer cell invasion. Cancers 2015, 7, 238–265. [Google Scholar] [CrossRef] [Green Version]
- Ciavarella, S.; Laurenzana, A.; De Summa, S.; Pilato, B.; Chilla, A.; Lacalamita, R.; Minoia, C.; Margheri, F.; Iacobazzi, A.; Rana, A. u-PAR expression in cancer associated fibroblast: New acquisitions in multiple myeloma progression. BMC Cancer 2017, 17, 215. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Song, E. Turning foes to friends: Targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 2019, 18, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Daley, W.P.; Peters, S.B.; Larsen, M. Extracellular matrix dynamics in development and regenerative medicine. J. Cell Sci. 2008, 121, 255–264. [Google Scholar] [CrossRef] [Green Version]
- Kumari, S.; Malla, R. New insight on the role of plasminogen receptor in cancer progression. Cancer Growth Metastasis 2015, 8, 35–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quemener, C.; Gabison, E.E.; Naïmi, B.; Lescaille, G.; Bougatef, F.; Podgorniak, M.P.; Labarchède, G.; Lebbé, C.; Calvo, F.; Menashi, S. Extracellular matrix metalloproteinase inducer up-regulates the urokinase-type plasminogen activator system promoting tumor cell invasion. Cancer Res. 2007, 67, 9–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langhammer, S.; Scheerer, J. Breaking the crosstalk of the cellular tumorigenic network: Hypothesis for addressing resistances to targeted therapies in advanced NSCLC. Oncotarget 2017, 8, 43555–43570. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Hwang, Y.S. Cancer-associated fibroblast stimulates cancer cell invasion in an interleukin-1 receptor (IL-1R)-dependent manner. Oncol. Lett. 2019, 18, 4645–4650. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, S.; Tiffen, J.; Hersey, P. Histone modifications, modifiers and readers in melanoma resistance to targeted and immune therapy. Cancers 2015, 7, 1959–1982. [Google Scholar] [CrossRef]
- Yoshida, G.J.; Azuma, A.; Miura, Y.; Orimo, A. Activated fibroblast program orchestrates tumor initiation and progression; molecular mechanisms and the associated therapeutic strategies. Int. J. Mol. Sci. 2019, 20, 2256. [Google Scholar] [CrossRef] [Green Version]
- Hirata, E.; Girotti, M.R.; Viros, A.; Hooper, S.; Spencer-Dene, B.; Matsuda, M.; Larkin, J.; Marais, R.; Sahai, E. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin β1/FAK signaling. Cancer Cell 2015, 27, 574–588. [Google Scholar] [CrossRef] [Green Version]
- Chan, X.; Singh, A.; Osman, N.; Piva, T. Role played by signalling pathways in overcoming BRAF inhibitor resistance in melanoma. Int. J. Mol. Sci. 2017, 18, 1527. [Google Scholar] [CrossRef]
- Rosell, R.; Karachaliou, N.; Morales-Espinosa, D.; Costa, C.; Molina, M.A.; Sansano, I.; Gasco, A.; Viteri, S.; Massuti, B.; Wei, J. Adaptive resistance to targeted therapies in cancer. Transl. Lung Cancer Res. 2013, 2, 152–159. [Google Scholar]
- Mainiero, F.; Murgia, C.; Wary, K.K.; Curatola, A.M.; Pepe, A.; Blumemberg, M.; Westwick, J.K.; Der, C.J.; Giancotti, F.G. The coupling of α6β4 integrin to Ras–MAP kinase pathways mediated by Shc controls keratinocyte proliferation. EMBO J. 1997, 16, 2365–2375. [Google Scholar] [CrossRef] [Green Version]
- Culig, Z. Cytokine disbalance in common human cancers. BBA Mol. Cell. Res. 2011, 1813, 308–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirotković-Skerlev, M.; Kulić, A.; Bradić, L.; Čačev, T. Protumor effects of proinflammatory mediators in breast cancer. Period. Biol. 2012, 114, 489–496. [Google Scholar]
- Benedicto, A.; Romayor, I.; Arteta, B. Role of liver ICAM-1 in metastasis. Oncol. Lett. 2017, 14, 3883–3892. [Google Scholar] [CrossRef] [Green Version]
- Gialeli, C.; Viola, M.; Barbouri, D.; Kletsas, D.; Passi, A.; Karamanos, N. Dynamic interplay between breast cancer cells and normal endothelium mediates the expression of matrix macromolecules, proteasome activity and functional properties of endothelial cells. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 2549–2559. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Sohn, Y.J.; Lee, R.; Yoo, H.J.; Kang, J.Y.; Choi, N.; Na, D.; Yeon, J.H. Cancer-Associated Fibroblasts Differentiated by Exosomes Isolated from Cancer Cells Promote Cancer Cell Invasion. Int. J. Mol. Sci. 2020, 21, 8153. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218153
Kim K, Sohn YJ, Lee R, Yoo HJ, Kang JY, Choi N, Na D, Yeon JH. Cancer-Associated Fibroblasts Differentiated by Exosomes Isolated from Cancer Cells Promote Cancer Cell Invasion. International Journal of Molecular Sciences. 2020; 21(21):8153. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218153
Chicago/Turabian StyleKim, Kimin, Yeh Joo Sohn, Ruri Lee, Hye Ju Yoo, Ji Yoon Kang, Nakwon Choi, Dokyun Na, and Ju Hun Yeon. 2020. "Cancer-Associated Fibroblasts Differentiated by Exosomes Isolated from Cancer Cells Promote Cancer Cell Invasion" International Journal of Molecular Sciences 21, no. 21: 8153. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218153