Molecular Interaction of Protein-Pigment C-Phycocyanin with Bovine Serum Albumin in a Gomphosis Structure Inhibiting Amyloid Formation
Abstract
:1. Introduction
2. Results
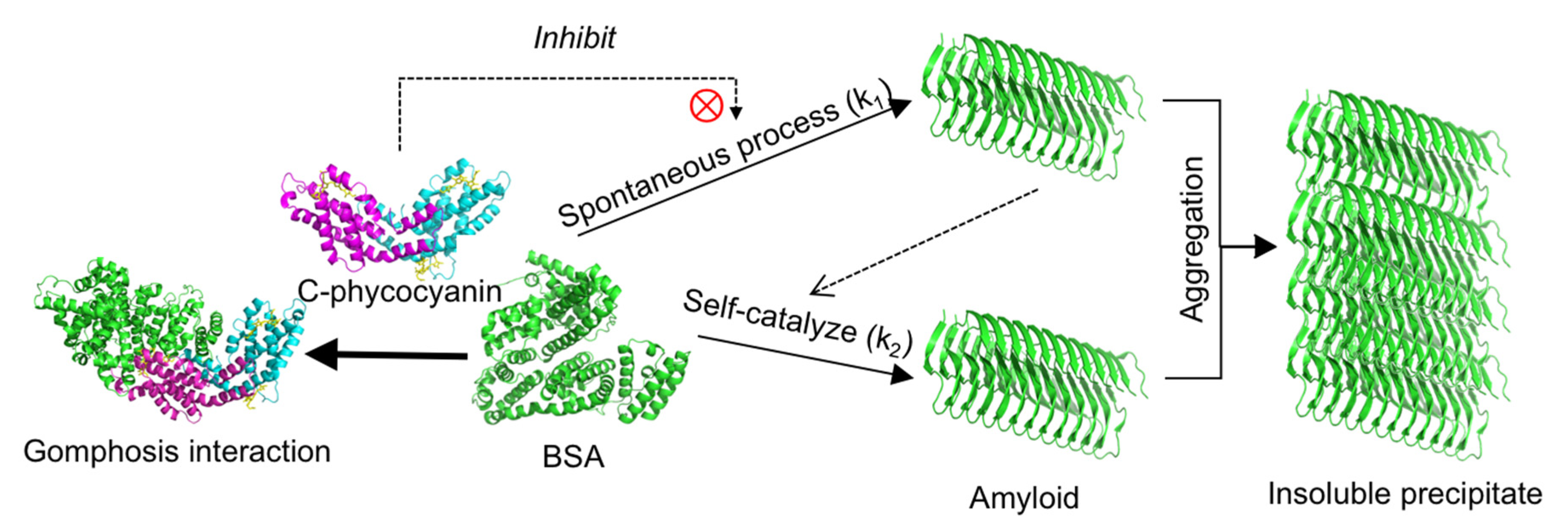
2.1. Inhibitory Effect of C-phycocyanin on Formation of Amyloid Fibrils
2.2. Fluorescence Intensity and Kinetic Fitting Results
2.3. CD Spectra
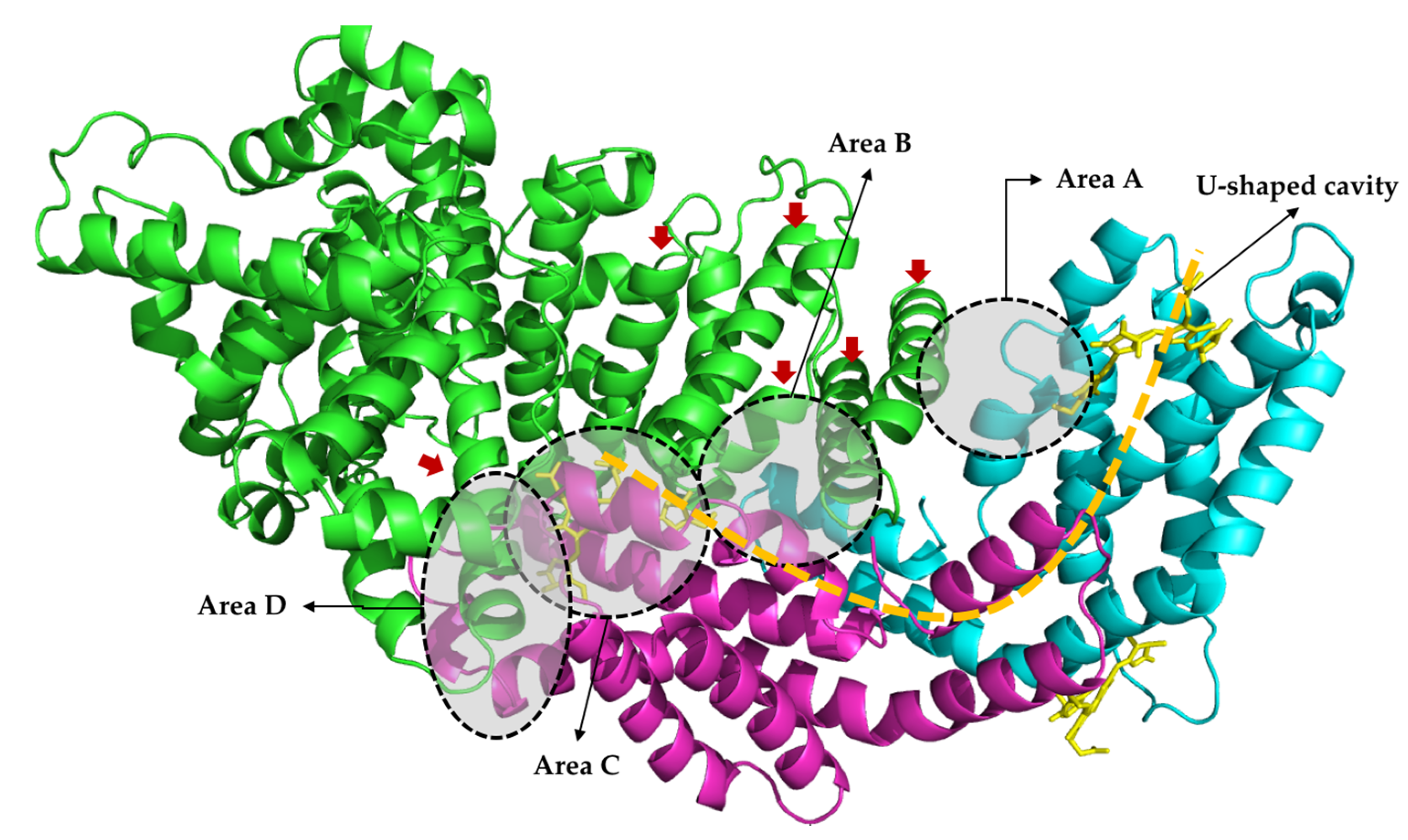
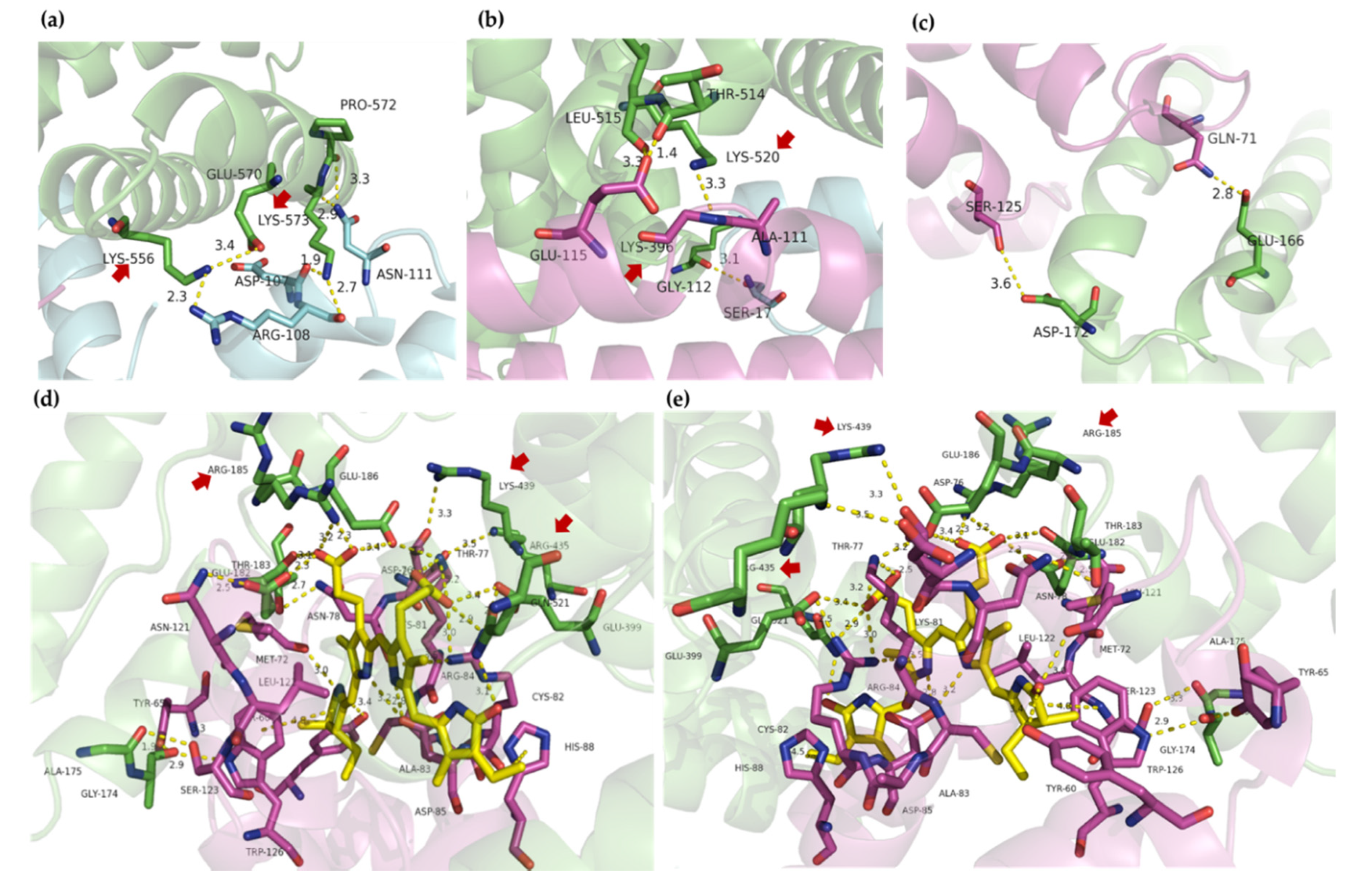
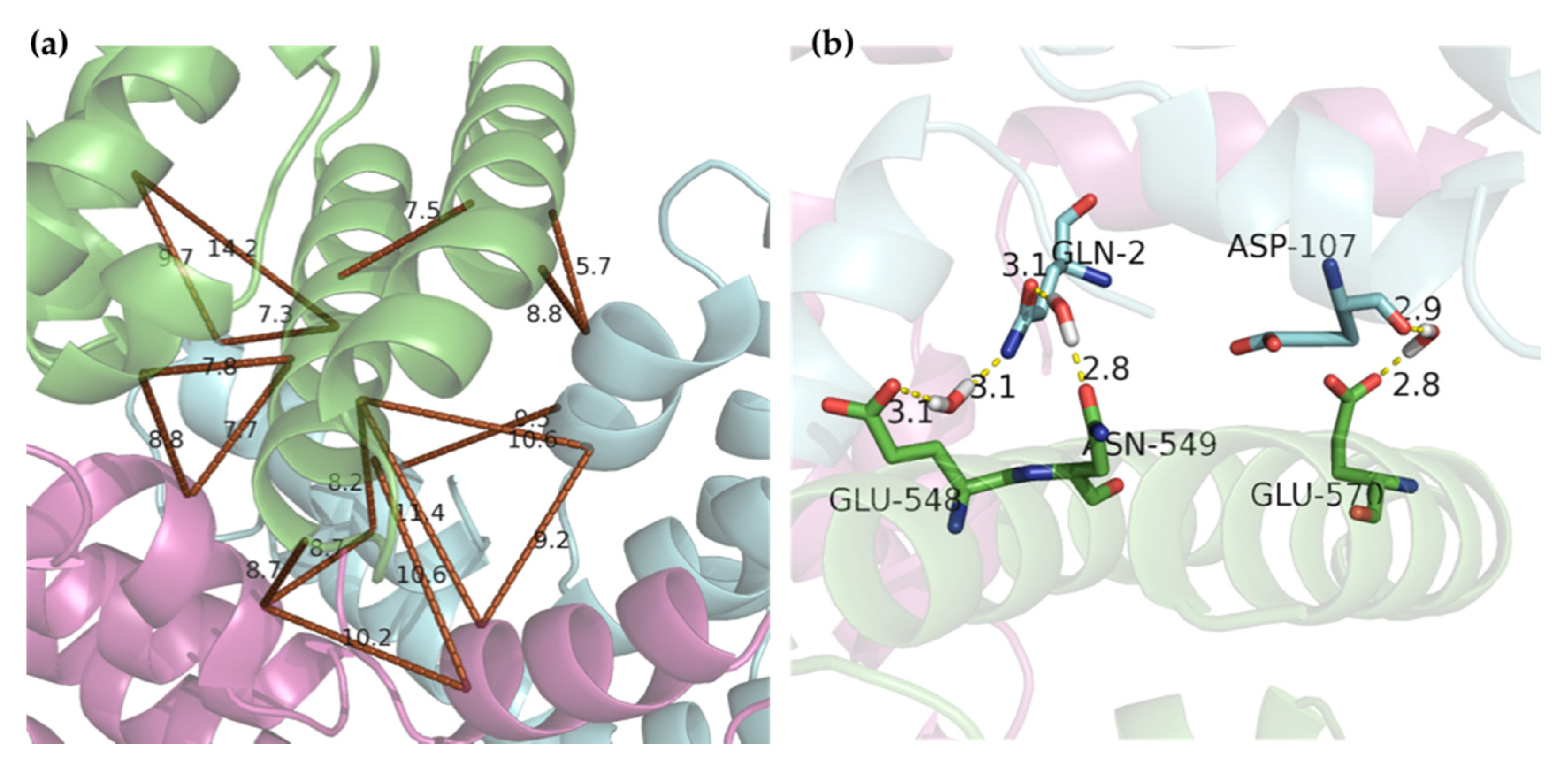
2.4. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Inhibitory Effect of BSA on Amyloid Formation In Vitro
4.3. Kinetic Analysis
4.4. Circular Dichroism Analysis
4.5. Molecular Docking
4.6. Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BSA | Bovine serum albumin |
| CD | Circular dichroism |
| ThT | Thioflavine T |
References
- Dobson, C.M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999, 24, 329–332. [Google Scholar] [CrossRef]
- McLean, C.A.; Cherny, R.A.; Fraser, F.W.; Fuller, S.J.; Smith, M.J.; Vbeyreuther, K.; Bush, A.I.; Masters, C.L. Soluble pool of Aβ amyloid as a determinant of severity of neurodegeneration in Alzheimer’s disease. Ann. Neurol. 1999, 46, 860–866. [Google Scholar] [CrossRef]
- Kirkitadze, M.D.; Condron, M.M.; Teplow, D.B. Identification and characterization of key kinetic intermediates in amyloid β-protein fibrillogenesis. J. Mol. Biol. 2001, 312, 1103–1119. [Google Scholar] [CrossRef] [PubMed]
- Iannuzzi, C.; Borriello, M.; Carafa, V.; Altucci, L.; Vitiello, M.; Balestrieri, M.L.; Ricci, G.; Irace, G.; Sirangelo, I. D-ribose-glycation of insulin prevents amyloid aggregation and produces cytotoxic adducts. Biochim. Biophys. Acta 2016, 1862, 93–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulson, J.B.; Ramsden, M.; Forster, C.; Sherman, M.A.; McGowan, E.; Ashe, K.H. Amyloid plaque and neurofibrillary tangle pathology in a regulatable mouse model of Alzheimer’s disease. Am. J. Pathol. 2008, 173, 762–772. [Google Scholar] [CrossRef] [Green Version]
- Agerschou, E.D.; Flagmeier, P.; Saridaki, T.; Galvagnion, C.; Komnig, D.; Heid, L.; Prasad, V.; Shaykhalishahi, H.; Willbold, D.; Dobson, C.M.; et al. An engineered monomer binding-protein for alpha-synuclein efficiently inhibits the proliferation of amyloid fibrils. Elife 2019, 8, e46112. [Google Scholar] [CrossRef] [PubMed]
- Saelices, L.; Nguyen, B.A.; Chung, K.; Wang, Y.; Ortega, A.; Lee, J.H.; Coelho, T.; Bijzet, J.; Benson, M.D.; Eisenberg, D.S. A pair of peptides inhibits seeding of the hormone transporter transthyretin into amyloid fibrils. J. Biol. Chem. 2019, 294, 6130–6141. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Xing, Y.-F.; Huang, B.; Zhang, Y.-Z.; Zeng, C.-M. Tea Catechins Induce the Conversion of Preformed Lysozyme Amyloid Fibrils to Amorphous Aggregates. J. Agric. Food Chem. 2009, 57, 11391–11396. [Google Scholar] [CrossRef]
- Prasanna, G.; Jing, P. Cyanidin-3-O-glucoside functions like chemical chaperone and attenuates the glycation mediated amyloid formation in albumin. Arch. Biochem. Biophys. 2018, 643, 50–56. [Google Scholar] [CrossRef]
- Hwang, J.-H.; Lee, I.T.; Jeng, K.-C.; Wang, M.-F.; Hou, R.C.-W.; Wu, S.-M.; Chan, Y.-C. Spirulina Prevents Memory Dysfunction, Reduces Oxidative Stress Damage and Augments Antioxidant Activity in Senescence-Accelerated Mice. J. Nutr. Sci. Vitaminol. 2011, 57, 186–191. [Google Scholar] [CrossRef] [Green Version]
- Min, S.K.; Park, J.S.; Luo, L.; Kwon, Y.S.; Lee, H.C.; Shim, H.J.; Kim, I.D.; Lee, J.K.; Shin, H.S. Assessment of C-phycocyanin effect on astrocytes-mediated neuroprotection against oxidative brain injury using 2D and 3D astrocyte tissue model. Sci. Rep. 2015, 5, 14418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Fan, X.; Qi, P.; Zhang, X. Identification of anti-diabetes peptides from Spirulina platensis. J. Funct. Foods 2019, 56, 333–341. [Google Scholar] [CrossRef]
- Liu, Y.; Jovcevski, B.; Pukala, T.L. C-Phycocyanin from Spirulina Inhibits alpha-Synuclein and Amyloid-beta Fibril Formation but Not Amorphous Aggregation. J. Nat. Prod. 2019, 82, 66–73. [Google Scholar] [CrossRef]
- Singh, N.K.; Hasan, S.S.; Kumar, J.; Raj, I.; Pathan, A.A.; Parmar, A.; Shakil, S.; Gourinath, S.; Madamwar, D. Crystal structure and interaction of phycocyanin with beta-secretase: A putative therapy for Alzheimer’s disease. CNS Neurol. Disord. Drug Targets 2014, 13, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, I.; Harris, D.; Levi-Kalisman, Y.; Yochelis, S.; Shemesh, A.; Ben-Nissan, G.; Sharon, M.; Raviv, U.; Adir, N.; Keren, N.; et al. Concentration-based self-assembly of phycocyanin. Photosynth. Res. 2017, 134, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Schluchter, W.M.; Bryant, D.A. Biogenesis of phycobiliproteins: I. cpcS-I and cpcU mutants of the cyanobacterium Synechococcus sp. PCC 7002 define a heterodimeric phyococyanobilin lyase specific for beta-phycocyanin and allophycocyanin subunits. J. Biol. Chem. 2008, 283, 7503–7512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rondeau, P.; Bourdon, E. The glycation of albumin: Structural and functional impacts. Biochimie 2011, 93, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Stege, G.J.J.; Renkawek, K.; Overkamp, P.S.G.; Verschuure, P.; van Rijk, A.F.; Reijnen-Aalbers, A.; Boelens, W.C.; Bosman, G.J.C.G.M.; de Jong, W.W. The molecular chaperone αB-crystallin enhances amyloid β neurotoxicity. Biochem. Biophys. Res. Commun. 1999, 262, 152–156. [Google Scholar] [CrossRef]
- Benson, M.D.; Scheinberg, M.A.; Shirahama, T.; Cathcart, E.S.; Skinner, M. Kinetics of serum amyloid protein A in casein-induced murine amyloidosis. J. Clin. Investig. 1977, 59, 412–417. [Google Scholar] [CrossRef] [Green Version]
- Feng, B.Y.; Toyama, B.H.; Wille, H.; Colby, D.W.; Collins, S.R.; May, B.C.H.; Prusiner, S.B.; Weissman, J.; Shoichet, B.K. Small-molecule aggregates inhibit amyloid polymerization. Nat. Chem. Biol. 2008, 4, 197–199. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Tiwari, S.; Karmakar, S.; Anki Reddy, K.; Pandey, L.M. The effect of the stoichiometric ratio of zinc towards the fibrillation of Bovine Serum Albumin (BSA): A mechanistic insight. Int. J. Biol. Macromol. 2019, 123, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Dubey, K.; Anand, B.G.; Shekhawat, D.S.; Kar, K. Eugenol prevents amyloid formation of proteins and inhibits amyloid-induced hemolysis. Sci. Rep. 2017, 7, 40744. [Google Scholar] [CrossRef] [PubMed]
- Freire, S.; de Araujo, M.H.; Al-Soufi, W.; Novo, M. Photophysical study of Thioflavin T as fluorescence marker of amyloid fibrils. Dyes Pigment. 2014, 110, 97–105. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Holm, N.K.; Jespersen, S.K.; Thomassen, L.V.; Wolff, T.Y.; Sehgal, P.; Thomsen, L.A.; Christiansen, G.; Andersen, C.B.; Knudsen, A.D.; Otzen, D.E. Aggregation and fibrillation of bovine serum albumin. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2007, 1774, 1128–1138. [Google Scholar] [CrossRef]
- Sahin, Z.; Demir, Y.K.; Kayser, V. Global kinetic analysis of seeded BSA aggregation. Eur. J. Pharm. Sci. 2016, 86, 115–124. [Google Scholar] [CrossRef]
- Merlini, G.; Bellotti, V. Molecular mechanisms of amyloidosis. N. Engl. J. Med. 2003, 349, 583–596. [Google Scholar] [CrossRef] [Green Version]
- Brachmann, A.; Baxa, U.; Wickner, R.B. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J. 2005, 24, 3082–3092. [Google Scholar] [CrossRef] [Green Version]
- Xue, W.-F.; Homans, S.W.; Radford, S.E. Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc. Natl. Acad. Sci. USA 2008, 105, 8926. [Google Scholar] [CrossRef] [Green Version]
- Harper, J.D.; Lansbury, P.T. Models of amyloid seeding in Alzheimer’s disease and scrapie: Mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 1997, 66, 385–407. [Google Scholar] [CrossRef]
- Jahanban-Esfahlan, A.; Panahi-Azar, V. Interaction of glutathione with bovine serum albumin: Spectroscopy and molecular docking. Food Chem. 2016, 202, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, G.; Jing, P. Spectroscopic and molecular modelling studies on glycation modified bovine serum albumin with cyanidin-3-O-glucoside. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 204, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Ahmad, B.; Rabbani, G.; Khan, R.H. 2,2,2-Trifluroethanol induces simultaneous increase in α-helicity and aggregation in alkaline unfolded state of bovine serum albumin. Int. J. Biol. Macromol. 2010, 46, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-G.; Lim, S. Characterization of anti-advanced glycation end product antibodies to nonenzymatically lysine-derived and arginine-derived glycated products. J. Immunoass. Immunochem. 2009, 30, 386–399. [Google Scholar] [CrossRef] [PubMed]
- Vitek, M.P.; Bhattacharya, K.; Glendening, J.M.; Stopa, E.; Vlassara, H.; Bucala, R.; Manogue, K.; Cerami, A. Advanced glycation end products contribute to amyloidosis in Alzheimer disease. Proc. Natl. Acad. Sci. USA 1994, 91, 4766–4770. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zheng, T.; Sang, S.; Lv, L. Quercetin inhibits advanced glycation end product formation by trapping methylglyoxal and glyoxal. J. Agric. Food Chem. 2014, 62, 12152–12158. [Google Scholar] [CrossRef]
- Ma, X.-J.; Zhang, Y.-J.; Zeng, C.-M. Inhibition of amyloid aggregation of bovine serum albumin by sodium dodecyl sulfate at submicellar concentrations. Biochemistry 2018, 83, 60–68. [Google Scholar] [CrossRef]
- Groenning, M.; Norrman, M.; Flink, J.M.; van de Weert, M.; Bukrinsky, J.T.; Schluckebier, G.; Frokjaer, S. Binding mode of Thioflavin T in insulin amyloid fibrils. J. Struct. Biol. 2007, 159, 483–497. [Google Scholar] [CrossRef]
- Biancalana, M.; Makabe, K.; Koide, A.; Koide, S. Molecular Mechanism of Thioflavin-T Binding to the Surface of β-Rich Peptide Self-Assemblies. J. Mol. Biol. 2009, 385, 1052–1063. [Google Scholar] [CrossRef] [Green Version]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.-H.; Vreven, T.; Weng, Z. ZDOCK server: Interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef]
- Pierce, B.G.; Hourai, Y.; Weng, Z. Accelerating Protein Docking in ZDOCK Using an Advanced 3D Convolution Library. PLoS ONE 2011, 6, e24657. [Google Scholar] [CrossRef] [PubMed]
- Mintseris, J.; Pierce, B.; Wiehe, K.; Anderson, R.; Chen, R.; Weng, Z. Integrating statistical pair potentials into protein complex prediction. Proteins Struct. Funct. Bioinform. 2007, 69, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Schrodinger, LLC. The PyMOL Molecular Graphics System; Version 1.8; Schrodinger, LLC: New York, NY, USA, 2015. [Google Scholar]



| C-phycocyanin (µg/mL) | k1 | k2 | R2 | Half-Life (t1/2, Days) |
|---|---|---|---|---|
| 0.0 | 1.820 × 10−2 ± 3.14 × 10−3 | 4.198 × 10−5 ± 2.68 × 10−5 | 0.9931 | 12.43 |
| 0.5 | 1.445 × 10−2 ± 2.07 × 10−3 | 4.003 × 10−5 ± 4.28 × 10−6 | 0.9945 | 14.41 |
| 2.5 | 1.256 × 10−2 ± 3.42 × 10−3 | 4.894 × 10−5 ± 8.83 × 10−6 | 0.9804 | 13.89 |
| 5.0 | 1.247 × 10−2 ± 1.77 × 10−4 | 4.444 × 10−5 ± 4.63 × 10−6 | 0.9939 | 14.69 |
| 25.0 | 5.610 × 10−3 ± 9.42 × 10−4 | 5.930 × 10−5 ± 5.04 × 10−6 | 0.9928 | 16.45 |
| 50.0 | 2.620 × 10−3 ± 3.03 × 10−4 | 7.012 × 10−5 ± 3.34 × 10−6 | 0.9969 | 17.73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.-C.; Jing, P. Molecular Interaction of Protein-Pigment C-Phycocyanin with Bovine Serum Albumin in a Gomphosis Structure Inhibiting Amyloid Formation. Int. J. Mol. Sci. 2020, 21, 8207. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218207
Luo Y-C, Jing P. Molecular Interaction of Protein-Pigment C-Phycocyanin with Bovine Serum Albumin in a Gomphosis Structure Inhibiting Amyloid Formation. International Journal of Molecular Sciences. 2020; 21(21):8207. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218207
Chicago/Turabian StyleLuo, Yi-Cong, and Pu Jing. 2020. "Molecular Interaction of Protein-Pigment C-Phycocyanin with Bovine Serum Albumin in a Gomphosis Structure Inhibiting Amyloid Formation" International Journal of Molecular Sciences 21, no. 21: 8207. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218207




