How Elongator Acetylates tRNA Bases
Abstract
:1. Introduction
2. Elp3 Is the Catalytic Core of Elongator
2.1. Elp3 Exists in Three Domains of Life
2.2. Elp3 Protein from All Domains of Life Have the Same Structure
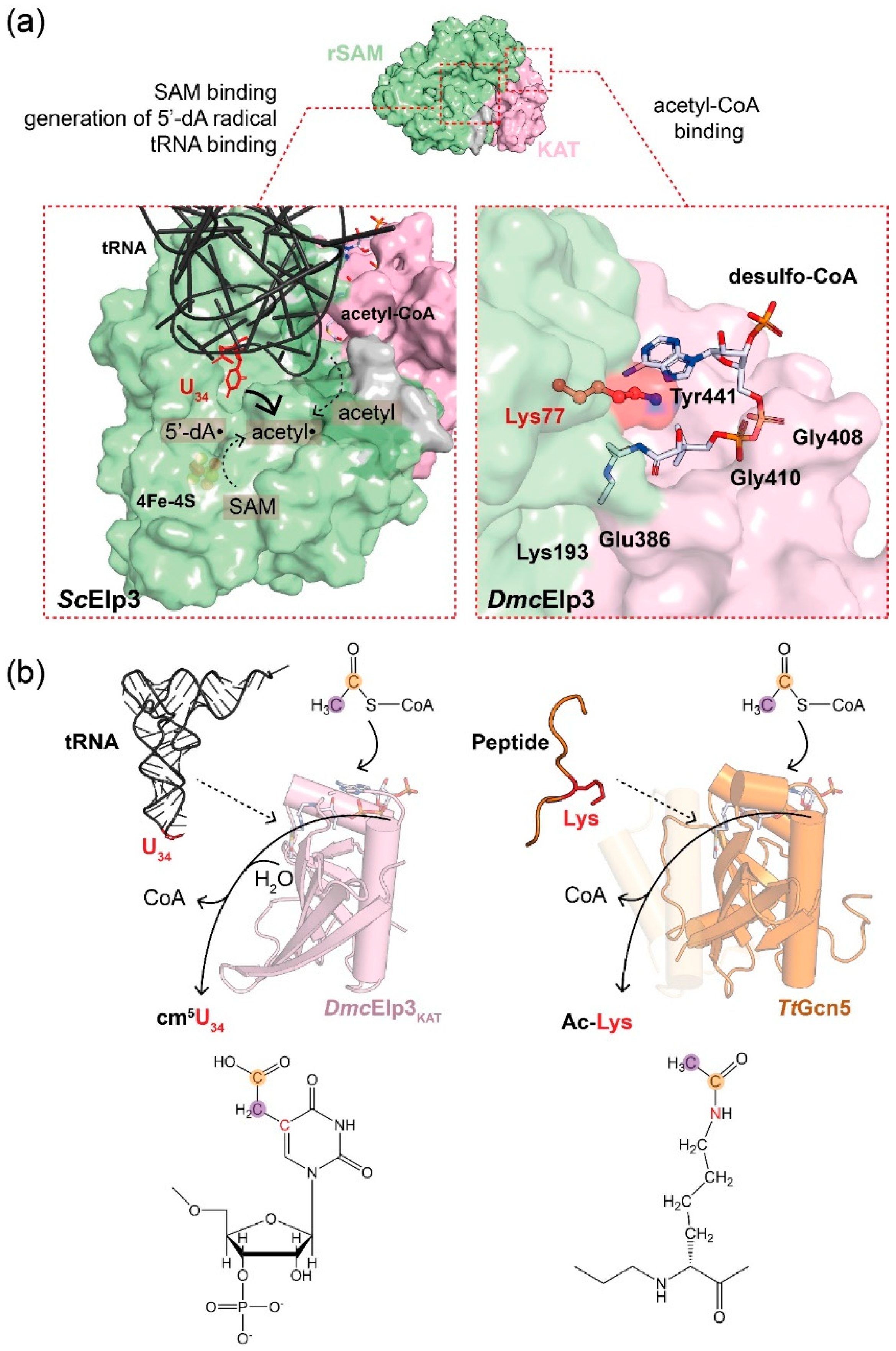
2.3. tRNA Triggers Elp3-Mediated Acetyl-CoA Hydrolysis
3. Elongator Dysfunction Is Linked to Disease
3.1. Defects in Elongator Disturbs Proteome Balance and Is Associated with Neurodegenerative Diseases
3.2. Elongator in Cancers
3.3. Other Mechanisms Regulating Elongator Activity
4. Conclusions and Future Aspects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Acetyl-CoA | Acetyl coenzyme A |
| ALS | Amyotrophic lateral sclerosis |
| ASL | Anti-codon stem loop |
| COSMIC | Catalog of Somatic Mutations in Cancer |
| CTD | C-terminus domain |
| Dmc | Dehalococcoides mccartyi |
| Elp3 | Elongator protein 3 |
| FD | Familial dysautonomia |
| GNAT | Gcn5 related N-acetyltransferases |
| HAT | Histone acetyltransferase |
| ICGC | International Cancer Genome Consortium |
| KAT | Lysine acetyltransferase |
| KDAC | Lysine deacetylase |
| Min | Methanocaldococcus infernus |
| mRNA | Messenger RNA |
| PTM | post-translation modifications |
| rRNA | ribosomal RNA |
| Sc | Sacchomyces cerevisiae |
| SAM | S-adenosyl-methionine |
| rSAM | radical S-adenosyl-methionine |
| TCGA | The Cancer Genome Atlas program |
| tRNA | transfer Ribonucleic Acid |
References
- Shi, L.; Tu, B.P. Acetyl-CoA and the regulation of metabolism: Mechanisms and consequences. Curr. Opin. Cell Biol. 2015, 33, 125–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, S.; McTiernan, N.; Wei, X.; Arnesen, T.; Marmorstein, R. Molecular basis for N-terminal acetylation by human NatE and its modulation by HYPK. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta Proteins Proteom. 2016, 1864, 1372–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janke, C.; Montagnac, G. Causes and Consequences of Microtubule Acetylation. Curr. Biol. 2017, 27, R1287–R1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montgomery, D.C.; Sorum, A.W.; Meier, J.L. Defining the orphan functions of lysine acetyltransferases. ACS Chem. Biol. 2015, 10, 85–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berndsen, C.E.; Albaugh, B.N.; Tan, S.; Denu, J.M. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry 2007, 46, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, L.; Zhao, K.; Thompson, P.R.; Hwang, Y.; Marmorstein, R.; Cole, P.A. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 2008, 451, 846–850. [Google Scholar] [CrossRef]
- Jin, G.; Xu, M.; Zou, M.; Duan, S. The Processing, Gene Regulation, Biological Functions, and Clinical Relevance of N4-Acetylcytidine on RNA: A Systematic Review. Mol. Ther. Nucleic Acids 2020, 20, 13–24. [Google Scholar] [CrossRef]
- Huang, B.; Johansson, M.J.O.; Byström, A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 2005, 11, 424–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dauden, M.I.; Jaciuk, M.; Weis, F.; Lin, T.-Y.; Kleindienst, C.; Abbassi, N.E.H.; Khatter, H.; Krutyholowa, R.; Breunig, K.D.; Kosinski, J.; et al. Molecular basis of tRNA recognition by the Elongator complex. Sci. Adv. 2019, 5, eaaw2326. [Google Scholar] [CrossRef] [Green Version]
- Glatt, S.; Zabel, R.; Kolaj-Robin, O.; Onuma, O.F.; Baudin, F.; Graziadei, A.; Taverniti, V.; Lin, T.Y.; Baymann, F.; Séraphin, B.; et al. Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat. Struct. Mol. Biol. 2016, 23, 794–802. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.-Y.; Abbassi, N.E.H.; Zakrzewski, K.; Chramiec-Głąbik, A.; Jemioła-Rzemińska, M.; Różycki, J.; Glatt, S. The Elongator subunit Elp3 is a non-canonical tRNA acetyltransferase. Nat. Commun. 2019, 10, 625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvadurai, K.; Wang, P.; Seimetz, J.; Huang, R.H. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat. Chem. Biol. 2014, 10, 810–812. [Google Scholar] [CrossRef] [Green Version]
- Nedialkova, D.D.; Leidel, S.A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef] [Green Version]
- Hawer, H.; Hammermeister, A.; Ravichandran, K.E. Roles of Elongator Dependent tRNA Modification Pathways in Neurodegeneration and Cancer. Genes 2018, 10, 19. [Google Scholar] [CrossRef] [Green Version]
- Kojic, M.; Wainwright, B. The Many Faces of Elongator in Neurodevelopment and Disease. Front. Mol. Neurosci. 2016, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Dauden, M.I.; Jaciuk, M.; Müller, C.W.; Glatt, S. Structural asymmetry in the eukaryotic Elongator complex. FEBS Lett. 2018, 592, 502–515. [Google Scholar] [CrossRef] [Green Version]
- Jarosz, M.; Van Lijsebettens, M.; Woloszynska, M. Plant Elongator—Protein Complex of Diverse Activities Regulates Growth, Development, and Immune Responses. Int. J. Mol. Sci. 2020, 21, 6912. [Google Scholar] [CrossRef]
- Wittschieben, B.O.; Otero, G.; De Bizemont, T.; Fellows, J.; Erdjument-Bromage, H.; Ohba, R.; Li, Y.; Allis, C.D.; Tempst, P.; Svejstrup, J.Q. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 1999, 4, 123–128. [Google Scholar] [CrossRef]
- Winkler, G.S.; Kristjuhan, A.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 3517–3522. [Google Scholar] [CrossRef] [Green Version]
- Creppe, C.; Malinouskaya, L.; Volvert, M.L.; Gillard, M.; Close, P.; Malaise, O.; Laguesse, S.; Cornez, I.; Rahmouni, S.; Ormenese, S.; et al. Elongator Controls the Migration and Differentiation of Cortical Neurons through Acetylation of α-Tubulin. Cell 2009, 136, 551–564. [Google Scholar] [CrossRef] [Green Version]
- Laguesse, S.; Close, P.; Van Hees, L.; Chariot, A.; Malgrange, B.; Nguyen, L. Loss of Elp3 Impairs the Acetylation and Distribution of Connexin-43 in the Developing Cerebral Cortex. Front. Cell. Neurosci. 2017, 11, 122. [Google Scholar] [CrossRef] [PubMed]
- Miśkiewicz, K.; Jose, L.E.; Bento-Abreu, A.; Fislage, M.; Taes, I.; Kasprowicz, J.; Swerts, J.; Sigrist, S.; Versées, W.; Robberecht, W.; et al. ELP3 controls active zone morphology by acetylating the ELKS family member bruchpilot. Neuron 2011, 72, 776–788. [Google Scholar] [CrossRef] [Green Version]
- Hermand, D. Anticodon Wobble Uridine Modification by Elongator at the Crossroad of Cell Signaling, Differentiation, and Diseases. Epigenomes 2020, 4, 7. [Google Scholar] [CrossRef]
- Dauden, M.I.; Kosinski, J.; Kolaj-Robin, O.; Desfosses, A.; Ori, A.; Faux, C.; Hoffmann, N.A.; Onuma, O.F.; Breunig, K.D.; Beck, M.; et al. Architecture of the yeast Elongator complex. EMBO Rep. 2017, 18, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Di Santo, R.; Bandau, S.; Stark, M.J.R. A conserved and essential basic region mediates tRNA binding to the Elp1 subunit of the Saccharomyces cerevisiae Elongator complex. Mol. Microbiol. 2014, 92, 1227–1242. [Google Scholar] [CrossRef] [Green Version]
- Glatt, S.; Létoquart, J.; Faux, C.; Taylor, N.M.I.; Séraphin, B.; Müller, C.W.; Letoquart, J.; Faux, C.; Seraphin, B. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 2012, 19, 314–320. [Google Scholar] [CrossRef]
- Setiaputra, D.T.; Cheng, D.T.; Lu, S.; Hansen, J.M.; Dalwadi, U.; Lam, C.H.; To, J.L.; Dong, M.; Yip, C.K. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep. 2017, 18, 280–291. [Google Scholar] [CrossRef] [Green Version]
- Van Tran, N.; Muller, L.; Ross, R.L.; Lestini, R.; Létoquart, J.; Ulryck, N.; Limbach, P.A.; de Crécy-Lagard, V.; Cianférani, S.; Graille, M. Evolutionary insights into Trm112-methyltransferase holoenzymes involved in translation between archaea and eukaryotes. Nucleic acids Res. 2018, 46, 8483–8499. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Glatt, S. tRNA Modification by Elongator Protein 3 (Elp3). In Encyclopedia of Inorganic and Bioinorganic Chemistry; Scott, R.A., Ed.; Wiley Online Library: Hoboken, NJ, USA, 2018; Volume eibc2623. [Google Scholar]
- Paraskevopoulou, C.; Fairhurst, S.A.; Lowe, D.J.; Brick, P.; Onesti, S. The elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol. Microbiol. 2006, 59, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Conrad, R.J.; Verdin, E.; Ott, M. Lysine Acetylation Goes Global: From Epigenetics to Metabolism and Therapeutics. Chem. Rev. 2018, 118, 1216–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojas, J.R.; Trievel, R.C.; Zhou, J.; Mo, Y.; Li, X.; Berger, S.L.; Allis, C.D.; Marmorstein, R. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 1999, 401, 93–98. [Google Scholar] [CrossRef]
- Rojas-Benítez, D.; Allende, M.L. Elongator subunit 3 (Elp3) is required for zebrafish trunk development. Int. J. Mol. Sci. 2020, 21, 925. [Google Scholar] [CrossRef] [Green Version]
- Simpson, C.L.; Lemmens, R.; Miskiewicz, K.; Broom, W.J.; Hansen, V.K.; van Vught, P.W.J.; Landers, J.E.; Sapp, P.; van Den Bosch, L.; Knight, J.; et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 2009, 18, 472–481. [Google Scholar] [CrossRef] [Green Version]
- Yoo, H.; Son, D.; Jang, Y.J.; Hong, K. Indispensable role for mouse ELP3 in embryonic stem cell maintenance and early development. Biochem. Biophys. Res. Commun. 2016, 478, 631–636. [Google Scholar] [CrossRef]
- Bento-Abreu, A.; Jager, G.; Swinnen, B.; Rué, L.; Hendrickx, S.; Jones, A.; Staats, K.A.; Taes, I.; Eykens, C.; Nonneman, A.; et al. Elongator subunit 3 (ELP3) modifies ALS through tRNA modification. Hum. Mol. Genet. 2018, 27, 1276–1289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laguesse, S.; Creppe, C.; Nedialkova, D.D.; Prévot, P.P.; Borgs, L.; Huysseune, S.; Franco, B.; Duysens, G.; Krusy, N.; Lee, G.; et al. A Dynamic Unfolded Protein Response Contributes to the Control of Cortical Neurogenesis. Dev. Cell 2015, 35, 553–567. [Google Scholar] [CrossRef] [Green Version]
- Ladang, A.; Rapino, F.; Heukamp, L.C.; Tharun, L.; Shostak, K.; Hermand, D.; Delaunay, S.; Klevernic, I.; Jiang, Z.; Jacques, N.; et al. Elp3 drives Wnt-dependent tumor initiation and regeneration in the intestine. J. Exp. Med. 2015, 212, 2057–2075. [Google Scholar] [CrossRef] [Green Version]
- Freeman, S.; Mateo Sánchez, S.; Pouyo, R.; Van Lerberghe, P.; Hanon, K.; Thelen, N.; Thiry, M.; Morelli, G.; Van Hees, L.; Laguesse, S.; et al. Proteostasis is essential during cochlear development for neuron survival and hair cell polarity. EMBO Rep. 2019, 20. [Google Scholar] [CrossRef]
- Rapino, F.; Delaunay, S.; Rambow, F.; Zhou, Z.; Tharun, L.; De Tullio, P.; Sin, O.; Shostak, K.; Schmitz, S.; Piepers, J.; et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 2018, 558, 605–609. [Google Scholar] [CrossRef]
- Tielens, S.; Huysseune, S.; Godin, J.D.; Chariot, A.; Malgrange, B.; Nguyen, L. Elongator controls cortical interneuron migration by regulating actomyosin dynamics. Cell Res. 2016, 26, 1131–1148. [Google Scholar] [CrossRef]
- Goffena, J.; Lefcort, F.; Zhang, Y.; Lehrmann, E.; Chaverra, M.; Felig, J.; Walters, J.; Buksch, R.; Becker, K.G.; George, L. Elongator and codon bias regulate protein levels in mammalian peripheral neurons. Nat. Commun. 2018, 9, 889. [Google Scholar] [CrossRef] [Green Version]
- Morini, E.; Gao, D.; Montgomery, C.M.; Salani, M.; Mazzasette, C.; Krussig, T.A.; Swain, B.; Dietrich, P.; Narasimhan, J.; Gabbeta, V.; et al. ELP1 Splicing Correction Reverses Proprioceptive Sensory Loss in Familial Dysautonomia. Am. J. Hum. Genet. 2019, 104, 638–650. [Google Scholar] [CrossRef] [Green Version]
- Addis, L.; Ahn, J.W.; Dobson, R.; Dixit, A.; Ogilvie, C.M.; Pinto, D.; Vaags, A.K.; Coon, H.; Chaste, P.; Wilson, S.; et al. Microdeletions of ELP4 Are Associated with Language Impairment, Autism Spectrum Disorder, and Mental Retardation. Hum. Mutat. 2015, 36, 842–850. [Google Scholar] [CrossRef]
- Cohen, J.S.; Srivastava, S.; Farwell, K.D.; Lu, H.M.; Zeng, W.; Lu, H.M.; Chao, E.C.; Fatemi, A. ELP2 is a novel gene implicated in neurodevelopmental disabilities. Am. J. Med. Genet. A 2015, 167, 1391–1395. [Google Scholar] [CrossRef]
- Kojic, M.; Gaik, M.; Kiska, B.; Salerno-Kochan, A.; Hunt, S.; Tedoldi, A.; Mureev, S.; Jones, A.; Whittle, B.; Genovesi, L.A.; et al. Elongator mutation in mice induces neurodegeneration and ataxia-like behavior. Nat. Commun. 2018, 9, 3195. [Google Scholar] [CrossRef]
- Reinthaler, E.M.; Lal, D.; Jurkowski, W.; Feucht, M.; Steinböck, H.; Gruber-Sedlmayr, U.; Ronen, G.M.; Geldner, J.; Haberlandt, E.; Neophytou, B.; et al. Analysis of ELP4, SRPX2, and interacting genes in typical and atypical rolandic epilepsy. Epilepsia 2014, 55, e89–e93. [Google Scholar] [CrossRef] [PubMed]
- Strug, L.J.; Clarke, T.; Chiang, T.; Chien, M.C.; Baskurt, Z.; Li, W.L.; Dorfman, R.; Bali, B.; Wirrell, E.; Kugler, S.L.; et al. Centrotemporal sharp wave EEG trait in rolandic epilepsy maps to Elongator Protein Complex 4 (ELP4). Eur. J. Hum. Genet. 2009, 17, 1171–1181. [Google Scholar] [CrossRef] [Green Version]
- Delaunay, S.; Rapino, F.; Tharun, L.; Zhou, Z.; Heukamp, L.; Termathe, M.; Shostak, K.; Klevernic, I.; Florin, A.; Desmecht, H.; et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J. Exp. Med. 2016, 213, 2503–2523. [Google Scholar] [CrossRef] [Green Version]
- Waszak, S.M.; Robinson, G.W.; Gudenas, B.L.; Smith, K.S.; Forget, A.; Kojic, M.; Garcia-Lopez, J.; Hadley, J.; Hamilton, K.V.; Indersie, E.; et al. Germline Elongator mutations in Sonic Hedgehog medulloblastoma. Nature 2020, 1–6. [Google Scholar] [CrossRef]
- Xu, S.; Zhan, M.; Jiang, C.; He, M.; Yang, L.; Shen, H.; Huang, S.; Huang, X.; Lin, R.; Shi, Y.; et al. Genome-wide CRISPR screen identifies ELP5 as a determinant of gemcitabine sensitivity in gallbladder cancer. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Fattah, W.; Jablonowski, D.; Di Santo, R.; Thüring, K.L.; Scheidt, V.; Hammermeister, A.; ten Have, S.; Helm, M.; Schaffrath, R.; Stark, M.J.R.; et al. Phosphorylation of Elp1 by Hrr25 is required for elongator-dependent tRNA modification in yeast. PLoS Genet. 2015, 11, e1004931. [Google Scholar] [CrossRef]
- Krutyhołowa, R.; Hammermeister, A.; Zabel, R.; Abdel-Fattah, W.; Reinhardt-Tews, A.; Helm, M.; Stark, M.J.R.; Breunig, K.D.; Schaffrath, R.; Glatt, S. Kti12, a PSTK-like tRNA dependent ATPase essential for tRNA modification by Elongator. Nucleic Acids Res. 2019, 47, 4814–4830. [Google Scholar] [CrossRef] [PubMed]
- Li, M.T.; Liang, J.Y.; Sun, Y.P.; Jin, J.; Xiong, Y.; Guan, K.L.; Yuan, H.X. ELP3 Acetyltransferase is phosphorylated and regulated by the oncogenic anaplastic lymphoma kinase (ALK). Biochem. J. 2019, 476, 2239–2254. [Google Scholar] [CrossRef]
- Candiracci, J.; Migeot, V.; Chionh, Y.H.; Bauer, F.; Brochier, T.; Russell, B.; Shiozaki, K.; Dedon, P.; Hermand, D. Reciprocal regulation of TORC signaling and tRNA modifications by Elongator enforces nutrient-dependent cell fate. Sci. Adv. 2019, 5. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbassi, N.-e.-H.; Biela, A.; Glatt, S.; Lin, T.-Y. How Elongator Acetylates tRNA Bases. Int. J. Mol. Sci. 2020, 21, 8209. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218209
Abbassi N-e-H, Biela A, Glatt S, Lin T-Y. How Elongator Acetylates tRNA Bases. International Journal of Molecular Sciences. 2020; 21(21):8209. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218209
Chicago/Turabian StyleAbbassi, Nour-el-Hana, Anna Biela, Sebastian Glatt, and Ting-Yu Lin. 2020. "How Elongator Acetylates tRNA Bases" International Journal of Molecular Sciences 21, no. 21: 8209. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218209




