L1CAM as an E-selectin Ligand in Colon Cancer
Abstract
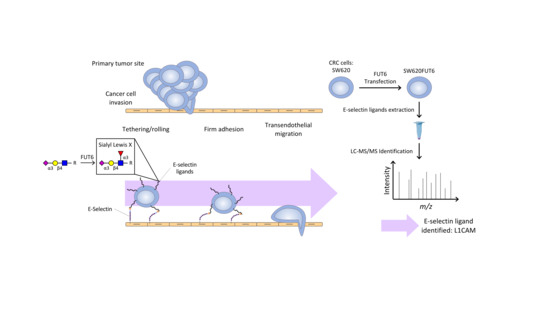
:1. Introduction
2. Results
2.1. Characterization of SW620 Colon Cancer Cell Line Overexpressing FUT6
2.2. N-glycan Profiles of FUT6 vs. Mock Transfected SW620 Cells
2.3. FUT6 Overexpression Increases Migration Ability in SW620 Cells
2.4. SW620FUT6 Cell Line Present High Expression of E-selectin Ligands
2.5. Identification of E-selectin Ligands from SW620FUT6
2.6. L1CAM, Integrin α6 and Integrin β1 Are Expressed on the Surface of SW620Mock and SW620FUT6 Cells
2.7. L1CAM Is an E-selectin Ligand in SW620FUT6 Cells
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Cell Culture
4.3. Flow Cytometry
4.4. Membrane Proteins Extraction
4.5. Immunoprecipitation
4.6. SDS-PAGE and Western Blot
4.7. Mass Spectrometry
4.7.1. E-selectin Ligand Identification
Mass Spectrometry
Data Analysis
4.7.2. Glycoanalysis
N-glycan Release
Glycan Labeling
N-glycan Clean-Up
HILIC-UHPLC Analysis on a Dionex Ultimate 3000 with Inline MS
4.8. RT-qPCR
4.9. Wound Healing Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| EMT | Epithelial–mesenchymal transition |
| FITC | Fluorescein Isothiocyanate |
| FUT | fucosyltransferase |
| ITGA6 | Integrin α6 |
| ITGB1 | Integrin β1 |
| mAb | monoclonal antibody |
| MDPI | Multidisciplinary Digital Publishing Institute |
| PTPRJ | Receptor-type tyrosine-protein phosphatase eta |
| sLeA | sialyl Lewis A |
| sLeX | sialyl Lewis X |
| VLA-6 | Very late antigen 6 |
References
- World Health Organization Cancer Key Facts. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 27 February 2019).
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Meester, R.G.S.; Barzi, A.; Jemal, A. Colorectal cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 177–193. [Google Scholar] [CrossRef] [PubMed]
- van der Geest, L.G.M.; Lam-Boer, J.; Koopman, M.; Verhoef, C.; Elferink, M.A.G.; de Wilt, J.H.W. Nationwide trends in incidence, treatment and survival of colorectal cancer patients with synchronous metastases. Clin. Exp. Metastasis 2015, 32, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Hu, J.; Yang, D.; Cosgrove, D.P.; Xu, R. Pattern of distant metastases in colorectal cancer: A SEER based study. Oncotarget 2015, 6, 38658–38666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeung, K.T.; Yang, J. Epithelial-mesenchymal transition in tumor metastasis. Mol. Oncol. 2017, 11, 28–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buhrmann, C.; Shayan, P.; Goel, A.; Shakibaei, M. Resveratrol Regulates Colorectal Cancer Cell Invasion by Modulation of Focal Adhesion Molecules. Nutrients 2017, 9, 1073. [Google Scholar] [CrossRef] [PubMed]
- Buhrmann, C.; Shayan, P.; Banik, K.; Kunnumakkara, A.B.; Kubatka, P.; Koklesova, L.; Shakibaei, M. Targeting NF-κB Signaling by Calebin A, a Compound of Turmeric, in Multicellular Tumor Microenvironment: Potential Role of Apoptosis Induction in CRC Cells. Biomedicines 2020, 8, 236. [Google Scholar] [CrossRef] [PubMed]
- Baldawa, P.; Shirol, P.; Alur, J.; Kulkarni, V.V. Metastasis: To and fro. J. Oral Maxillofac. Pathol. 2017, 21, 463–464. [Google Scholar] [CrossRef]
- Valastyan, S.; Weinberg, R.A. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; Liang, Y.; Gao, W. Clinicopathological and prognostic significance of sialyl Lewis X overexpression in patients with cancer: A meta-analysis. OncoTargets Ther. 2016, 9, 3113–3125. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, I.G.; Carrascal, M.; Mineiro, A.G.; Bugalho, A.; Borralho, P.; Silva, Z.; Dall’olio, F.; Videira, P.A. Carcinoembryonic antigen is a sialyl Lewis x/a carrier and an E-selectin ligand in non-small cell lung cancer. Int. J. Oncol. 2019, 55, 1033–1048. [Google Scholar] [CrossRef] [PubMed]
- Holst, S.; Wuhrer, M.; Rombouts, Y. Glycosylation characteristics of colorectal cancer. Adv. Cancer Res. 2015, 126, 203–256. [Google Scholar] [CrossRef] [PubMed]
- Majuri, M.L.; Niemelä, R.; Tiisala, S.; Renkonen, O.; Renkonen, R. Expression and function of alpha 2,3-sialyl- and alpha 1,3/1,4-fucosyltransferases in colon adenocarcinoma cell lines: Role in synthesis of E-selectin counter-receptors. Int. J. Cancer 1995, 63, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Hiraiwa, N.; Sawada-Kasugai, M.; Akamatsu, S.; Tachikawa, T.; Kasai, Y.; Akiyama, S.; Ito, K.; Takagi, H.; Kannagi, R. Altered mRNA expression of specific molecular species of fucosyl- and sialyl-transferases in human colorectal cancer tissues. Int. J. Cancer 1997, 71, 556–564. [Google Scholar] [CrossRef]
- Carvalho, A.S.; Harduin-Lepers, A.; Magalhães, A.; Machado, E.; Mendes, N.; Costa, L.T.; Matthiesen, R.; Almeida, R.; Costa, J.; Reis, C.A. Differential expression of alpha-2,3-sialyltransferases and alpha-1,3/4-fucosyltransferases regulates the levels of sialyl Lewis a and sialyl Lewis x in gastrointestinal carcinoma cells. Int. J. Biochem. Cell Biol. 2010, 42, 80–89. [Google Scholar] [CrossRef]
- Kudo, T.; Ikehara, Y.; Togayachi, A.; Morozumi, K.; Watanabe, M.; Nakamura, M.; Nishihara, S.; Narimatsu, H. Up-regulation of a set of glycosyltransferase genes in human colorectal cancer. Lab. Investig. J. Tech. Methods Pathol. 1998, 78, 797–811. [Google Scholar]
- Köhler, S.; Ullrich, S.; Richter, U.; Schumacher, U. E-/P-selectins and colon carcinoma metastasis: First in vivo evidence for their crucial role in a clinically relevant model of spontaneous metastasis formation in the lung. Br. J. Cancer 2010, 102, 602–609. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, K.; Matsumoto, S.; Morishima, T.; Kawabe, T.; Okamoto, T. Cimetidine inhibits cancer cell adhesion to endothelial cells and prevents metastasis by blocking E-selectin expression. Cancer Res. 2000, 60, 3978–3984. [Google Scholar] [PubMed]
- Sackstein, R. The lymphocyte homing receptors: Gatekeepers of the multistep paradigm. Curr. Opin. Hematol. 2005, 12, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, A.; Peired, A.J.; Wild, M.; Vestweber, D.; Frenette, P.S. Complete identification of E-selectin ligands on neutrophils reveals distinct functions of PSGL-1, ESL-1, and CD44. Immunity 2007, 26, 477–489. [Google Scholar] [CrossRef] [Green Version]
- Sperandio, M.; Gleissner, C.A.; Ley, K. Glycosylation in immune cell trafficking. Immunol. Rev. 2009, 230, 97–113. [Google Scholar] [CrossRef] [Green Version]
- Carrascal, M.A.; Talina, C.; Borralho, P.; Gonçalo Mineiro, A.; Henriques, A.R.; Pen, C.; Martins, M.; Braga, S.; Sackstein, R.; Videira, P.A. Staining of E-selectin ligands on paraffin-embedded sections of tumor tissue. BMC Cancer 2018, 18, 495. [Google Scholar] [CrossRef] [PubMed]
- Dimitroff, C.J.; Descheny, L.; Trujillo, N.; Kim, R.; Nguyen, V.; Huang, W.; Pienta, K.J.; Kutok, J.L.; Rubin, M.A. Identification of leukocyte E-selectin ligands, P-selectin glycoprotein ligand-1 and E-selectin ligand-1, on human metastatic prostate tumor cells. Cancer Res. 2005, 65, 5750–5760. [Google Scholar] [CrossRef] [Green Version]
- Nonomura, C.; Kikuchi, J.; Kiyokawa, N.; Ozaki, H.; Mitsunaga, K.; Ando, H.; Kanamori, A.; Kannagi, R.; Fujimoto, J.; Muroi, K.; et al. CD43, but not P-selectin glycoprotein ligand-1, functions as an E-selectin counter-receptor in human pre-B-cell leukemia NALL-1. Cancer Res. 2008, 68, 790–799. [Google Scholar] [CrossRef] [Green Version]
- Carrascal, M.A.; Silva, M.; Ferreira, J.A.; Azevedo, R.; Ferreira, D.; Silva, A.M.N.; Ligeiro, D.; Santos, L.L.; Sackstein, R.; Videira, P.A. A functional glycoproteomics approach identifies CD13 as a novel E-selectin ligand in breast cancer. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2069–2080. [Google Scholar] [CrossRef] [PubMed]
- Burdick, M.M.; Chu, J.T.; Godar, S.; Sackstein, R. HCELL is the major E- and L-selectin ligand expressed on LS174T colon carcinoma cells. J. Biol. Chem. 2006, 281, 13899–13905. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.N.; Zhu, F.; Schnaar, R.L.; Alves, C.S.; Konstantopoulos, K. Carcinoembryonic antigen and CD44 variant isoforms cooperate to mediate colon carcinoma cell adhesion to E- and L-selectin in shear flow. J. Biol. Chem. 2008, 283, 15647–15655. [Google Scholar] [CrossRef] [Green Version]
- Tomlinson, J.; Wang, J.L.; Barsky, S.H.; Lee, M.C.; Bischoff, J.; Nguyen, M. Human colon cancer cells express multiple glycoprotein ligands for E-selectin. Int. J. Oncol. 2000, 16, 347–353. [Google Scholar] [CrossRef]
- Trinchera, M.; Malagolini, N.; Chiricolo, M.; Santini, D.; Minni, F.; Caretti, A.; Dall’olio, F. The biosynthesis of the selectin-ligand sialyl Lewis x in colorectal cancer tissues is regulated by fucosyltransferase VI and can be inhibited by an RNA interference-based approach. Int. J. Biochem. Cell Biol. 2011, 43, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Berg, E.L.; Robinson, M.K.; Mansson, O.; Butcher, E.C.; Magnani, J.L. A carbohydrate domain common to both sialyl Le(a) and sialyl Le(X) is recognized by the endothelial cell leukocyte adhesion molecule ELAM-1. J. Biol. Chem. 1991, 266, 14869–14872. [Google Scholar]
- Berg, E.L.; Yoshino, T.; Rott, L.S.; Robinson, M.K.; Warnock, R.A.; Kishimoto, T.K.; Picker, L.J.; Butcher, E.C. The cutaneous lymphocyte antigen is a skin lymphocyte homing receptor for the vascular lectin endothelial cell-leukocyte adhesion molecule 1. J. Exp. Med. 1991, 174, 1461–1466. [Google Scholar] [CrossRef] [Green Version]
- St Hill, C.A. Interactions between endothelial selectins and cancer cells regulate metastasis. Front. Biosci. Landmark Ed. 2011, 16, 3233–3251. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, T.; Asao, T.; Yamauchi, H.; Ide, M.; Tabe, Y.; Fujii, T.; Yamaguchi, S.; Tsutsumi, S.; Yazawa, S.; Kuwano, H. Associated expression of α2,3sialylated type 2 chain structures with lymph node metastasis in distal colorectal cancer. Surg. Today 2013, 43, 155–162. [Google Scholar] [CrossRef]
- Yamadera, M.; Shinto, E.; Tsuda, H.; Kajiwara, Y.; Naito, Y.; Hase, K.; Yamamoto, J.; Ueno, H. Sialyl Lewisx expression at the invasive front as a predictive marker of liver recurrence in stage II colorectal cancer. Oncol. Lett. 2018, 15, 221–228. [Google Scholar] [CrossRef]
- Chik, J.H.L.; Zhou, J.; Moh, E.S.X.; Christopherson, R.; Clarke, S.J.; Molloy, M.P.; Packer, N.H. Comprehensive glycomics comparison between colon cancer cell cultures and tumours: Implications for biomarker studies. J. Proteomics 2014, 108, 146–162. [Google Scholar] [CrossRef]
- Liang, L.; Gao, C.; Li, Y.; Sun, M.; Xu, J.; Li, H.; Jia, L.; Zhao, Y. miR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death Dis. 2017, 8, e2968. [Google Scholar] [CrossRef] [Green Version]
- Videira, P.A.; Silva, M.; Martin, K.C.; Sackstein, R. Ligation of the CD44 Glycoform HCELL on Culture-Expanded Human Monocyte-Derived Dendritic Cells Programs Transendothelial Migration. J. Immunol. Baltim. Md 1950 2018, 201, 1030–1043. [Google Scholar] [CrossRef]
- Sackstein, R. Engineering cellular trafficking via glycosyltransferase-programmed stereosubstitution. Ann. N. Y. Acad. Sci. 2012, 1253, 193–200. [Google Scholar] [CrossRef]
- Mondal, N.; Dykstra, B.; Lee, J.; Ashline, D.J.; Reinhold, V.N.; Rossi, D.J.; Sackstein, R. Distinct human α(1,3)-fucosyltransferases drive Lewis-X/sialyl Lewis-X assembly in human cells. J. Biol. Chem. 2018, 293, 7300–7314. [Google Scholar] [CrossRef] [Green Version]
- Pastor, D.M.; Poritz, L.S.; Olson, T.L.; Kline, C.L.; Harris, L.R.; Koltun, W.A.; Chinchilli, V.M.; Irby, R.B. Primary cell lines: False representation or model system? A comparison of four human colorectal tumors and their coordinately established cell lines. Int. J. Clin. Exp. Med. 2010, 3, 69–83. [Google Scholar]
- Holst, S.; Deuss, A.J.M.; van Pelt, G.W.; van Vliet, S.J.; Garcia-Vallejo, J.J.; Koeleman, C.A.M.; Deelder, A.M.; Mesker, W.E.; Tollenaar, R.A.; Rombouts, Y.; et al. N-glycosylation Profiling of Colorectal Cancer Cell Lines Reveals Association of Fucosylation with Differentiation and Caudal Type Homebox 1 (CDX1)/Villin mRNA Expression. Mol. Cell. Proteom. MCP 2016, 15, 124–140. [Google Scholar] [CrossRef] [Green Version]
- Sethi, M.K.; Kim, H.; Park, C.K.; Baker, M.S.; Paik, Y.-K.; Packer, N.H.; Hancock, W.S.; Fanayan, S.; Thaysen-Andersen, M. In-depth N-glycome profiling of paired colorectal cancer and non-tumorigenic tissues reveals cancer-, stage- and EGFR-specific protein N-glycosylation. Glycobiology 2015, 25, 1064–1078. [Google Scholar] [CrossRef] [Green Version]
- Kaprio, T.; Satomaa, T.; Heiskanen, A.; Hokke, C.H.; Deelder, A.M.; Mustonen, H.; Hagström, J.; Carpen, O.; Saarinen, J.; Haglund, C. N-glycomic profiling as a tool to separate rectal adenomas from carcinomas. Mol. Cell. Proteom. MCP 2015, 14, 277–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balog, C.I.A.; Stavenhagen, K.; Fung, W.L.J.; Koeleman, C.A.; McDonnell, L.A.; Verhoeven, A.; Mesker, W.E.; Tollenaar, R.A.E.M.; Deelder, A.M.; Wuhrer, M. N-glycosylation of colorectal cancer tissues: A liquid chromatography and mass spectrometry-based investigation. Mol. Cell. Proteom. MCP 2012, 11, 571–585. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Xie, Q.; Wang, Y.; Miao, J.; Li, L.; Zhang, T.; Cao, X.; Li, Y. Mass spectrometry analysis reveals aberrant N-glycans in colorectal cancer tissues. Glycobiology 2019. [Google Scholar] [CrossRef]
- Świderska, M.; Choromańska, B.; Dąbrowska, E.; Konarzewska-Duchnowska, E.; Choromańska, K.; Szczurko, G.; Myśliwiec, P.; Dadan, J.; Ładny, J.R.; Zwierz, K. The diagnostics of colorectal cancer. Contemp. Oncol. 2014, 18, 1–6. [Google Scholar] [CrossRef]
- Baldus, S.E.; Mönig, S.P.; Zirbes, T.K.; Thakran, J.; Köthe, D.; Köppel, M.; Hanisch, F.-G.; Thiele, J.; Schneider, P.M.; Hölscher, A.H.; et al. Lewis(y) antigen (CD174) and apoptosis in gastric and colorectal carcinomas: Correlations with clinical and prognostic parameters. Histol. Histopathol. 2006, 21, 503–510. [Google Scholar] [CrossRef]
- Nonaka, M.; Imaeda, H.; Matsumoto, S.; Yong Ma, B.; Kawasaki, N.; Mekata, E.; Andoh, A.; Saito, Y.; Tani, T.; Fujiyama, Y.; et al. Mannan-binding protein, a C-type serum lectin, recognizes primary colorectal carcinomas through tumor-associated Lewis glycans. J. Immunol. Baltim. Md 2014, 192, 1294–1301. [Google Scholar] [CrossRef] [Green Version]
- Sasai, K.; Ikeda, Y.; Eguchi, H.; Tsuda, T.; Honke, K.; Taniguchi, N. The action of N-acetylglucosaminyltransferase-V is prevented by the bisecting GlcNAc residue at the catalytic step. FEBS Lett. 2002, 522, 151–155. [Google Scholar] [CrossRef] [Green Version]
- Sethi, M.K.; Thaysen-Andersen, M.; Smith, J.T.; Baker, M.S.; Packer, N.H.; Hancock, W.S.; Fanayan, S. Comparative N-glycan profiling of colorectal cancer cell lines reveals unique bisecting GlcNAc and α-2,3-linked sialic acid determinants are associated with membrane proteins of the more metastatic/aggressive cell lines. J. Proteome Res. 2014, 13, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Bresalier, R.S.; Ho, S.B.; Schoeppner, H.L.; Kim, Y.S.; Sleisenger, M.H.; Brodt, P.; Byrd, J.C. Enhanced sialylation of mucin-associated carbohydrate structures in human colon cancer metastasis. Gastroenterology 1996, 110, 1354–1367. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Guillebon, A.D.; Hsu, J.-W.; Barthel, S.R.; Dimitroff, C.J.; Lee, Y.-F.; King, M.R. Human fucosyltransferase 6 enables prostate cancer metastasis to bone. Br. J. Cancer 2013, 109, 3014–3022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Garay, M.; Arteta, B.; Llop, E.; Cobler, L.; Pagès, L.; Ortiz, R.; Ferri, M.J.; de Bolós, C.; Figueras, J.; de Llorens, R.; et al. α2,3-Sialyltransferase ST3Gal IV promotes migration and metastasis in pancreatic adenocarcinoma cells and tends to be highly expressed in pancreatic adenocarcinoma tissues. Int. J. Biochem. Cell Biol. 2013, 45, 1748–1757. [Google Scholar] [CrossRef]
- Carrascal, M.A.; Silva, M.; Ramalho, J.S.; Pen, C.; Martins, M.; Pascoal, C.; Amaral, C.; Serrano, I.; Oliveira, M.J.; Sackstein, R.; et al. Inhibition of fucosylation in human invasive ductal carcinoma reduces E-selectin ligand expression, cell proliferation, and ERK1/2 and p38 MAPK activation. Mol. Oncol. 2018, 12, 579–593. [Google Scholar] [CrossRef] [Green Version]
- Hirakawa, M.; Takimoto, R.; Tamura, F.; Yoshida, M.; Ono, M.; Murase, K.; Sato, Y.; Osuga, T.; Sato, T.; Iyama, S.; et al. Fucosylated TGF-β receptors transduces a signal for epithelial-mesenchymal transition in colorectal cancer cells. Br. J. Cancer 2014, 110, 156–163. [Google Scholar] [CrossRef] [Green Version]
- Severino, P.F.; Silva, M.; Carrascal, M.; Malagolini, N.; Chiricolo, M.; Venturi, G.; Astolfi, A.; Catera, M.; Videira, P.A.; Dall’Olio, F. Expression of sialyl-Tn sugar antigen in bladder cancer cells affects response to Bacillus Calmette Guérin (BCG) and to oxidative damage. Oncotarget 2017, 8, 54506–54517. [Google Scholar] [CrossRef] [Green Version]
- Carrascal, M.A.; Severino, P.F.; Guadalupe Cabral, M.; Silva, M.; Ferreira, J.A.; Calais, F.; Quinto, H.; Pen, C.; Ligeiro, D.; Santos, L.L.; et al. Sialyl Tn-expressing bladder cancer cells induce a tolerogenic phenotype in innate and adaptive immune cells. Mol. Oncol. 2014, 8, 753–765. [Google Scholar] [CrossRef]
- Buhrmann, C.; Kraehe, P.; Lueders, C.; Shayan, P.; Goel, A.; Shakibaei, M. Curcumin Suppresses Crosstalk between Colon Cancer Stem Cells and Stromal Fibroblasts in the Tumor Microenvironment: Potential Role of EMT. PLoS ONE 2014, 9, e107514. [Google Scholar] [CrossRef] [Green Version]
- Sawada, R.; Lowe, J.B.; Fukuda, M. E-selectin-dependent adhesion efficiency of colonic carcinoma cells is increased by genetic manipulation of their cell surface lysosomal membrane glycoprotein-1 expression levels. J. Biol. Chem. 1993, 268, 12675–12681. [Google Scholar]
- Kitayama, J.; Ikeda, S.; Kumagai, K.; Saito, H.; Nagawa, H. Alpha 6 beta 1 integrin (VLA-6) mediates leukocyte tether and arrest on laminin under physiological shear flow. Cell. Immunol. 2000, 199, 97–103. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Tu, R.; Li, K.; Ye, P.; Cui, X. Tumor Suppressor PTPRJ Is a Target of miR-155 in Colorectal Cancer. J. Cell. Biochem. 2017, 118, 3391–3400. [Google Scholar] [CrossRef]
- Luo, L.; Shen, G.-Q.; Stiffler, K.A.; Wang, Q.K.; Pretlow, T.G.; Pretlow, T.P. Loss of heterozygosity in human aberrant crypt foci (ACF), a putative precursor of colon cancer. Carcinogenesis 2006, 27, 1153–1159. [Google Scholar] [CrossRef] [Green Version]
- Ruivenkamp, C.; Hermsen, M.; Postma, C.; Klous, A.; Baak, J.; Meijer, G.; Demant, P. LOH of PTPRJ occurs early in colorectal cancer and is associated with chromosomal loss of 18q12-21. Oncogene 2003, 22, 3472–3474. [Google Scholar] [CrossRef] [Green Version]
- McGuire, J.C.; Greene, L.A.; Furano, A.V. NGF stimulates incorporation of fucose or glucosamine into an external glycoprotein in cultured rat PC12 pheochromocytoma cells. Cell 1978, 15, 357–365. [Google Scholar] [CrossRef]
- Wolff, J.M.; Frank, R.; Mujoo, K.; Spiro, R.C.; Reisfeld, R.A.; Rathjen, F.G. A human brain glycoprotein related to the mouse cell adhesion molecule L1. J. Biol. Chem. 1988, 263, 11943–11947. [Google Scholar]
- Schultheis, M.; Diestel, S.; Schmitz, B. The role of cytoplasmic serine residues of the cell adhesion molecule L1 in neurite outgrowth, endocytosis, and cell migration. Cell. Mol. Neurobiol. 2007, 27, 11–31. [Google Scholar] [CrossRef]
- Yamasaki, M.; Thompson, P.; Lemmon, V. CRASH syndrome: Mutations in L1CAM correlate with severity of the disease. Neuropediatrics 1997, 28, 175–178. [Google Scholar] [CrossRef]
- Bateman, A.; Jouet, M.; MacFarlane, J.; Du, J.S.; Kenwrick, S.; Chothia, C. Outline structure of the human L1 cell adhesion molecule and the sites where mutations cause neurological disorders. EMBO J. 1996, 15, 6050–6059. [Google Scholar] [CrossRef]
- Klat, J.; Mladenka, A.; Dvorackova, J.; Bajsova, S.; Simetka, O. L1CAM as a Negative Prognostic Factor in Endometrioid Endometrial Adenocarcinoma FIGO Stage IA-IB. Anticancer Res. 2019, 39, 421–424. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, F.; Ding, Y.; Zhen, L.; Han, X.; Jiao, F.; Tang, J. Overexpression of L1 cell adhesion molecule correlates with aggressive tumor progression of patients with breast cancer and promotes motility of breast cancer cells. Int. J. Clin. Exp. Pathol. 2015, 8, 9240–9247. [Google Scholar]
- Liu, X.; Min, S.; Wu, N.; Liu, H.; Wang, T.; Li, W.; Shen, Y.; Zhao, C.; Wang, H.; Qian, Z.; et al. miR-193a-3p inhibition of the Slug activator PAK4 suppresses non-small cell lung cancer aggressiveness via the p53/Slug/L1CAM pathway. Cancer Lett. 2019, 447, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Ernst, A.-K.; Putscher, A.; Samatov, T.R.; Suling, A.; Galatenko, V.V.; Shkurnikov, M.Y.; Knyazev, E.N.; Tonevitsky, A.G.; Haalck, T.; Lange, T.; et al. Knockdown of L1CAM significantly reduces metastasis in a xenograft model of human melanoma: L1CAM is a potential target for anti-melanoma therapy. PLoS ONE 2018, 13, e0192525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.-C.; Xie, Y.-M.; Ran, L.-Q.; Cao, H.-H.; Sun, C.; Wu, J.-Y.; Wu, Z.-Y.; Liao, L.-D.; Zhao, W.-J.; Fang, W.-K.; et al. L1CAM drives oncogenicity in esophageal squamous cell carcinoma by stimulation of ezrin transcription. J. Mol. Med. Berl. Ger. 2017, 95, 1355–1368. [Google Scholar] [CrossRef]
- Na’ara, S.; Amit, M.; Gil, Z. L1CAM induces perineural invasion of pancreas cancer cells by upregulation of metalloproteinase expression. Oncogene 2019, 38, 596–608. [Google Scholar] [CrossRef]
- Doberstein, K.; Wieland, A.; Lee, S.B.B.; Blaheta, R.A.A.; Wedel, S.; Moch, H.; Schraml, P.; Pfeilschifter, J.; Kristiansen, G.; Gutwein, P. L1-CAM expression in ccRCC correlates with shorter patients survival times and confers chemoresistance in renal cell carcinoma cells. Carcinogenesis 2011, 32, 262–270. [Google Scholar] [CrossRef] [Green Version]
- Fang, Q.-X.; Lü, L.-Z.; Yang, B.; Zhao, Z.-S.; Wu, Y.; Zheng, X.-C. L1, β-catenin, and E-cadherin expression in patients with colorectal cancer: Correlation with clinicopathologic features and its prognostic significance. J. Surg. Oncol. 2010, 102, 433–442. [Google Scholar] [CrossRef]
- Boo, Y.-J.; Park, J.-M.; Kim, J.; Chae, Y.-S.; Min, B.-W.; Um, J.-W.; Moon, H.-Y. L1 expression as a marker for poor prognosis, tumor progression, and short survival in patients with colorectal cancer. Ann. Surg. Oncol. 2007, 14, 1703–1711. [Google Scholar] [CrossRef]
- Kaifi, J.T.; Reichelt, U.; Quaas, A.; Schurr, P.G.; Wachowiak, R.; Yekebas, E.F.; Strate, T.; Schneider, C.; Pantel, K.; Schachner, M.; et al. L1 is associated with micrometastatic spread and poor outcome in colorectal cancer. Mod. Pathol. Off. J. 2007, 20, 1183–1190. [Google Scholar] [CrossRef] [Green Version]
- Haase, G.; Gavert, N.; Brabletz, T.; Ben-Ze’ev, A. A point mutation in the extracellular domain of L1 blocks its capacity to confer metastasis in colon cancer cells via CD10. Oncogene 2017, 36, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Brodt, P.; Fallavollita, L.; Bresalier, R.S.; Meterissian, S.; Norton, C.R.; Wolitzky, B.A. Liver endothelial E-selectin mediates carcinoma cell adhesion and promotes liver metastasis. Int. J. Cancer 1997, 71, 612–619. [Google Scholar] [CrossRef]
- Duchalais, E.; Guilluy, C.; Nedellec, S.; Touvron, M.; Bessard, A.; Touchefeu, Y.; Bossard, C.; Boudin, H.; Louarn, G.; Neunlist, M.; et al. Colorectal Cancer Cells Adhere to and Migrate Along the Neurons of the Enteric Nervous System. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 31–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebeling, O.; Duczmal, A.; Aigner, S.; Geiger, C.; Schöllhammer, S.; Kemshead, J.T.; Möller, P.; Schwartz-Albiez, R.; Altevogt, P. L1 adhesion molecule on human lymphocytes and monocytes: Expression and involvement in binding to alpha v beta 3 integrin. Eur. J. Immunol. 1996, 26, 2508–2516. [Google Scholar] [CrossRef]
- Magrini, E.; Villa, A.; Angiolini, F.; Doni, A.; Mazzarol, G.; Rudini, N.; Maddaluno, L.; Komuta, M.; Topal, B.; Prenen, H.; et al. Endothelial deficiency of L1 reduces tumor angiogenesis and promotes vessel normalization. J. Clin. Invest. 2014, 124, 4335–4350. [Google Scholar] [CrossRef] [Green Version]
- Hall, H.; Djonov, V.; Ehrbar, M.; Hoechli, M.; Hubbell, J.A. Heterophilic interactions between cell adhesion molecule L1 and alphavbeta3-integrin induce HUVEC process extension in vitro and angiogenesis in vivo. Angiogenesis 2004, 7, 213–223. [Google Scholar] [CrossRef]
- Shi, G.; Du, Y.; Li, Y.; An, Y.; He, Z.; Lin, Y.; Zhang, R.; Yan, X.; Zhao, J.; Yang, S.; et al. Cell Recognition Molecule L1 Regulates Cell Surface Glycosylation to Modulate Cell Survival and Migration. Int. J. Med. Sci. 2017, 14, 1276–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozak, R.P.; Tortosa, C.B.; Fernandes, D.L.; Spencer, D.I.R. Comparison of procainamide and 2-aminobenzamide labeling for profiling and identification of glycans by liquid chromatography with fluorescence detection coupled to electrospray ionization-mass spectrometry. Anal. Biochem. 2015, 486, 38–40. [Google Scholar] [CrossRef]
- Guile, G.R.; Rudd, P.M.; Wing, D.R.; Prime, S.B.; Dwek, R.A. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Anal. Biochem. 1996, 240, 210–226. [Google Scholar] [CrossRef]
- Videira, P.A.; Correia, M.; Malagolini, N.; Crespo, H.J.; Ligeiro, D.; Calais, F.M.; Trindade, H.; Dall’Olio, F. ST3Gal.I sialyltransferase relevance in bladder cancer tissues and cell lines. BMC Cancer 2009, 9, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| FUT3 | FUT4 | FUT5 | FUT6 | FUT7 | FUT10 | FUT11 | |
|---|---|---|---|---|---|---|---|
| SW620Mock | 0.001 ± 0.0001 | 12.6 ± 1.1 | 0.01 ± 0.004 | 2.7 ± 0.8 | 0.002 ± 0.0002 | 5.1 ± 0.8 | 1.5 ± 0.4 |
| SW620FUT6 | 0.0002 ± 0.00005 | 14.3 ± 4.0 | 0.002 ± 0.0008 | 107.3 ± 40.0 | 0.002 ± 0.0005 | 4.3 ± 0.8 | 1.4 ± 0.3 |
| p value | ** p = 0.0079 | N.S. | * p = 0.0317 | ** p = 0.0079 | N.S. | N.S. | N.S. |
| Protein Name | Gene Name | MW (kDa) | UniProtKB Entry | Exclusive Unique Peptide Counts § | Total Spectrum Count (sum) § | Subcellular Location ‡ | |||
|---|---|---|---|---|---|---|---|---|---|
| Exp. 1 | Exp. 2 | Exp. 3 | Exp. 4 | ||||||
| Neural cell adhesion molecule L1 | L1CAM | 140 | P32004 | 31 | 20 | 30 | 27 | 118 | Plasma membrane |
| Integrin α6 | ITGA6 | 127 | P23229 | 11 | 31 | 12 | 10 | 91 | Plasma membrane |
| Receptor-type tyrosine-protein phosphatase eta | PTPRJ | 146 | Q12913 | 21 | 9 | 18 | 11 | 58 | Plasma membrane |
| Integrin β1 | ITGB1 | 88 | P05556 | 6 | 11 | 7 | 5 | 39 | Plasma membrane, recycling endosome |
| Cation-independent mannose-6-phosphate receptor (Insulin-like growth factor 2 receptor) | IGF2R | 274 | P11717 | 16 | 3 | 16 | 7 | 39 | Lysosome membrane |
| Receptor-type tyrosine-protein phosphatase alpha | PTPRA | 91 | P18433 | 10 | 3 | 4 | 5 | 22 | Membrane |
| Leucyl-cystinyl aminopeptidase | LNPEP | 117 | Q9UIQ6 | 7 | 9 | 6 | 5 | 22 | Plasma membrane, secreted |
| Carboxypeptidase D | CPD | 153 | O75976 | 2 | 0 | 6 | 6 | 17 | Plasma membrane |
| Lysosome-associated membrane glycoprotein 2 | LAMP2 | 45 | P13473 | 3 | 3 | 3 | 3 | 12 | Lysosome/endosome/plasma membrane |
| CD109 antigen | CD109 | 162 | Q6YHK3 | 3 | 2 | 2 | 4 | 12 | Plasma membrane |
| Golgi membrane protein 1 | GOLM1 | 45 | Q8NBJ4 | 2 | 2 | 2 | 4 | 10 | Golgi apparatus membrane |
| Plexin-D1 | PLXND1 | 212 | Q9Y4D7 | 2 | 0 | 1 | 4 | 7 | Plasma membrane |
| Zymogen granule protein 16 homolog B | ZG16B | 23 | Q96DA0 | 0 | 2 | 0 | 2 | 4 | Secreted |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deschepper, F.M.; Zoppi, R.; Pirro, M.; Hensbergen, P.J.; Dall’Olio, F.; Kotsias, M.; Gardner, R.A.; Spencer, D.I.R.; Videira, P.A. L1CAM as an E-selectin Ligand in Colon Cancer. Int. J. Mol. Sci. 2020, 21, 8286. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218286
Deschepper FM, Zoppi R, Pirro M, Hensbergen PJ, Dall’Olio F, Kotsias M, Gardner RA, Spencer DIR, Videira PA. L1CAM as an E-selectin Ligand in Colon Cancer. International Journal of Molecular Sciences. 2020; 21(21):8286. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218286
Chicago/Turabian StyleDeschepper, Fanny M., Roberta Zoppi, Martina Pirro, Paul J. Hensbergen, Fabio Dall’Olio, Maximillianos Kotsias, Richard A. Gardner, Daniel I.R. Spencer, and Paula A. Videira. 2020. "L1CAM as an E-selectin Ligand in Colon Cancer" International Journal of Molecular Sciences 21, no. 21: 8286. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218286