Digital Image Analysis Applied to Tumor Cell Proliferation, Aggressiveness, and Migration-Related Protein Synthesis in Neuroblastoma 3D Models
Abstract
:1. Introduction
2. Results
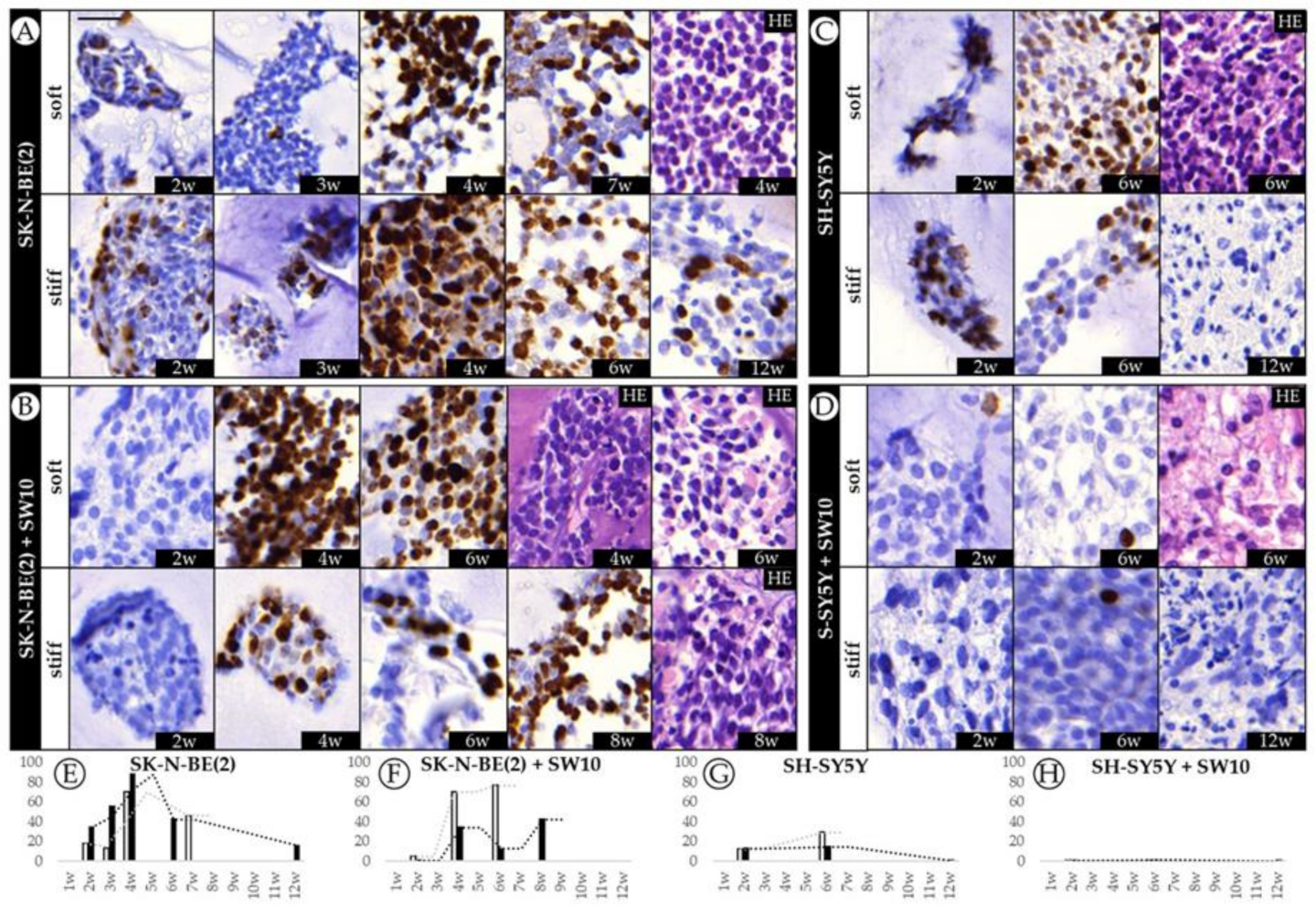
2.1. Differential Effect of 3D Hydrogel Stiffness on Cell Proliferation in SK-N-BE(2) and SH-SY5Y NB Cell Lines
2.2. The Contribution of Co-Cultured Stromal Schwann Cells to SK-N-BE(2) Proliferation Is Dependent on Substrate Stiffness
2.3. Correlation between Mitosis–Karyorrhexis Index and Cell Number Differs with Growth Conditions
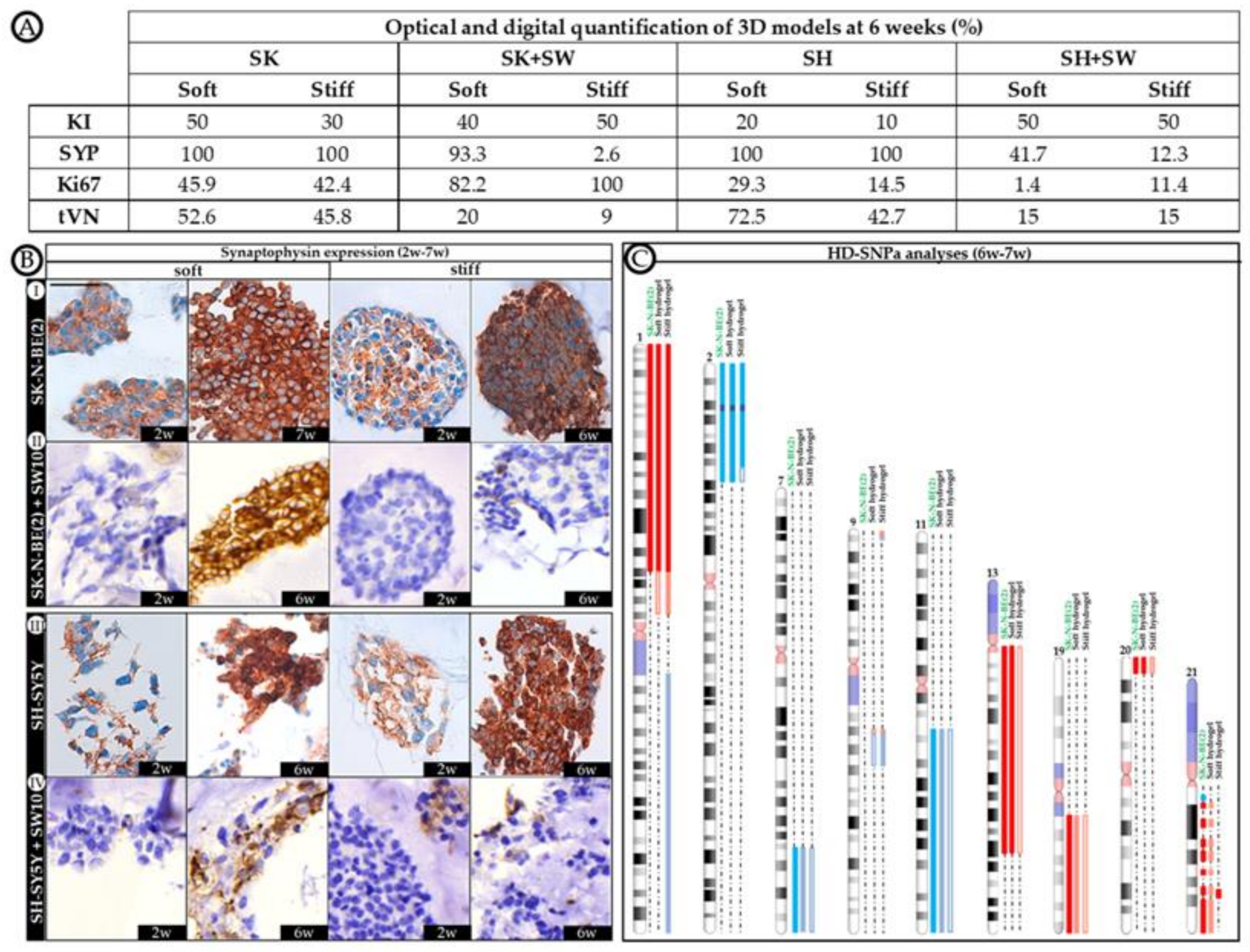
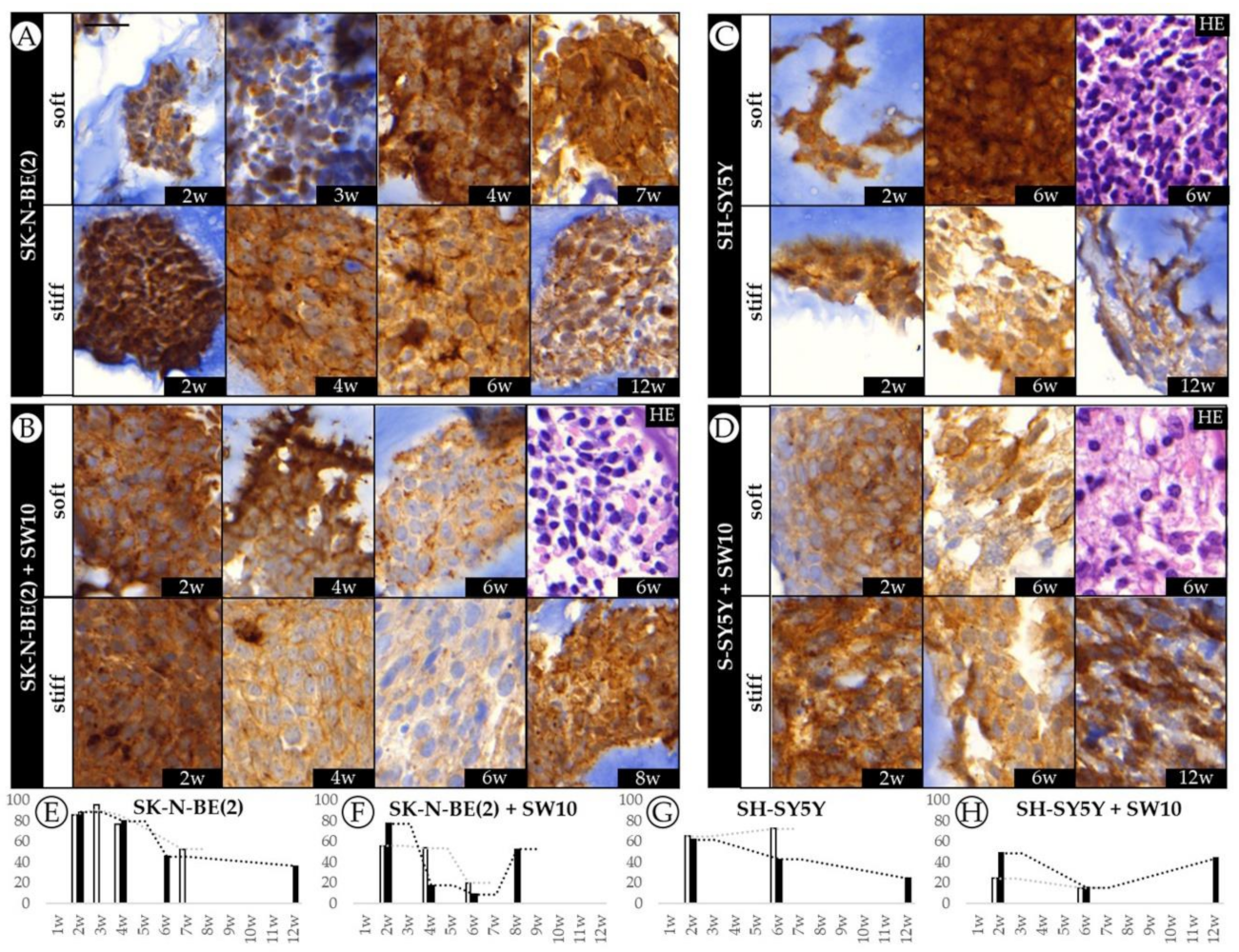
2.4. Differential 3D Culture Stimulation of tVN Expression Is Dependent on Cell Growth Conditions
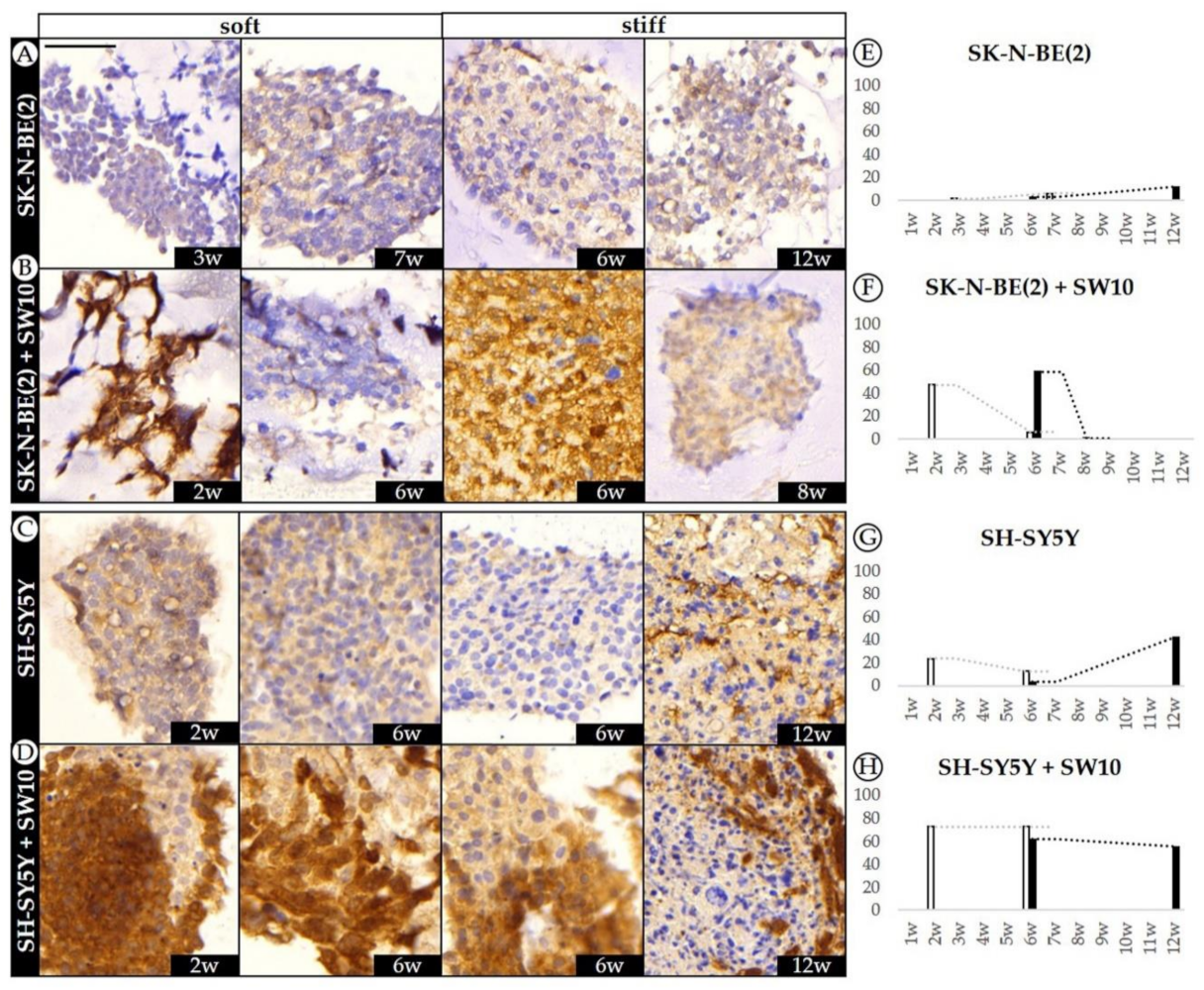
2.5. Co-Culture Condition with SW10 Modifies DOCK8 Expression Dynamics
2.6. Robust Expression of KANK1 in Models Including SH-SY5Y Cells Correlates Negatively with Proliferation
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Synthesis of Hydrogels
4.3. Immunocytochemistry and Immunohistochemistry
4.4. Image Analysis
4.5. Data Treatment
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ECM | extracellular matrix |
| IHC | immunohistochemistry |
| PV | Pannoramic Viewer |
| SW10 | mouse Schwann cell line |
| SYP | synaptophysin |
| NB | neuroblastoma |
| VN | vitronectin |
| 2D | 2 dimensional |
| 3D | 3 dimensional |
References
- Doyle, A.D.; Yamada, K.M. Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp. Cell Res. 2016, 343, 60–66. [Google Scholar] [CrossRef] [Green Version]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [Green Version]
- Wells, R.G. The role of matrix stiffness in regulating cell behavior. Hepatology 2008, 47, 1394–1400. [Google Scholar] [CrossRef]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef] [Green Version]
- Xiong, G.-F.; Xu, R. Function of cancer cell-derived extracellular matrix in tumor progression. J. Cancer Metastasis Treat. 2016, 2, 357. [Google Scholar] [CrossRef]
- Estofolete, C.F.; Botelho-Machado, C.; Taboga, S.R.; Zucoloto, S.; Polli-Lopes, A.C.; Gil, C.D. Effects of myenteric denervation on extracellular matrix fibers and mast cell distribution in normal stomach and gastric lesions. Cancer Cell Int. 2010, 10, 18. [Google Scholar] [CrossRef] [Green Version]
- Giraldo, N.A.; Sanchez-Salas, R.; Peske, J.D.; Vano, Y.; Becht, E.; Petitprez, F.; Validire, P.; Ingels, A.; Cathelineau, X.; Fridman, W.H.; et al. The clinical role of the TME in solid cancer. Br. J. Cancer 2019, 120, 45–53. [Google Scholar] [CrossRef]
- Kenny, P.A.; Bissell, M.J. Tumor reversion: Correction of malignant behavior by microenvironmental cues. Int. J. Cancer 2003, 107, 688–695. [Google Scholar] [CrossRef] [Green Version]
- Lejeune, M.; Álvaro, T. Clinicobiological, prognostic and therapeutic implications of the tumor microenvironment in follicular lymphoma. Haematologica 2009, 94, 16–21. [Google Scholar] [CrossRef]
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three-dimensional cell culture: A powerful tool in tumor research and drug discovery (Review). Oncol. Lett. 2017, 14, 6999–7010. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.; Wang, D.-A. Macroporous Hydrogel Scaffolds for Three-Dimensional Cell Culture and Tissue Engineering. Tissue Eng. Part B Rev. 2017, 23, 451–461. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, X.-F.; Gao, G.; Yonezawa, T.; Cui, X. 3D bioprinting and the current applications in tissue engineering. Biotechnol. J. 2017, 12, 1600734. [Google Scholar] [CrossRef]
- Tadeo, I.; Berbegall, A.P.; Castel, V.; García-Miguel, P.; Callaghan, R.; Påhlman, S.; Navarro, S.; Noguera, R. Extracellular matrix composition defines an ultra-high-risk group of neuroblastoma within the high-risk patient cohort. Br. J. Cancer 2016, 115, 480–489. [Google Scholar] [CrossRef] [Green Version]
- Tadeo, I.; Berbegall, A.P.; Navarro, S.; Castel, V.; Noguera, R. A stiff extracellular matrix is associated with malignancy in peripheral neuroblastic tumors. Pediatr. Blood Cancer 2017, 64, e26449. [Google Scholar] [CrossRef]
- Tadeo, I.; Gamero-Sandemetrio, E.; Berbegall, A.P.; Gironella, M.; Ritort, F.; Cañete, A.; Bueno, G.; Navarro, S.; Noguera, R. Lymph microvascularization as a prognostic indicator in neuroblastoma. Oncotarget 2018, 9, 26157–26170. [Google Scholar] [CrossRef] [Green Version]
- Tadeo, I.; Gamero-Sandemetrio, E.; Berbegall, A.P.; Navarro, S.; Cañete, A.; Noguera, R. 1p36 Deletion Results in a Decrease in Glycosaminoglycans Which Is Associated with Aggressiveness in Neuroblastic Tumors. Histol. Histopathol. 2018, 33, 487–495. [Google Scholar]
- Park, J.R.; Eggert, A.; Caron, H. Neuroblastoma: Biology, Prognosis, and Treatment. Hematol. Clin. N. Am. 2010, 24, 65–86. [Google Scholar] [CrossRef]
- Kruger, T.M.; Bell, K.J.; Lansakara, T.I.; Tivanski, A.V.; Doorn, J.A.; Stevens, L.L. A Soft Mechanical Phenotype of SH-SY5Y Neuroblastoma and Primary Human Neurons Is Resilient to Oligomeric Aβ(1–42) Injury. ACS Chem. Neurosci. 2020, 11, 840–850. [Google Scholar] [CrossRef]
- Vicente-Munuera, P.; Burgos--Panadero, R.; Noguera, I.; Navarro, S.; Noguera, R.; Escudero, L.M. The topology of vitronectin: A complementary feature for neuroblastoma risk classification based on computer-aided detection. Int. J. Cancer 2019, 146, 553–565. [Google Scholar] [CrossRef]
- Burgos-Panadero, R.; Noguera, I.; Cañete, A.; Navarro, S.; Noguera, R. Vitronectin as a molecular player of the tumor microenvironment in neuroblastoma. BMC Cancer 2019, 19, 479. [Google Scholar] [CrossRef]
- Monferrer, E.; Martín-Vañó, S.; Carretero, A.; García-Lizarribar, A.; Burgos-Panadero, R.; Navarro, S.; Samitier, J.; Noguera, R. A three-dimensional bioprinted model to evaluate the effect of stiffness on neuroblastoma cell cluster dynamics and behavior. Sci. Rep. 2020, 10, 6370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Carrasco, A.; Martín-Vañó, S.; Burgos-Panadero, R.; Monferrer, E.; Berbegall, A.P.; Fernández-Blanco, B.; Navarro, S.; Noguera, R. Impact of Extracellular Matrix Stiffness on Genomic Heterogeneity in MYCN-Amplied Neuroblastoma Cell Line. J. Exp. Clin. Cancer Res. 2020, 39, 226. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M.; Seeger, R.C.; Schwab, M.; Varmus, H.E.; Bishop, J.M. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 1984, 224, 1121–1124. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Takita, J.; Nakagawara, A.; Hayashi, Y.; Mano, H.; Ogawa, S.; Choi, Y.L.; Kato, M.; Ohira, M.; Sanada, M.; et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature 2008, 455, 971–974. [Google Scholar] [CrossRef]
- Bunimovich, Y.L.; Keskinov, A.A.; Shurin, G.V.; Shurin, M.R. Schwann cells: A new player in the tumor microenvironment. Cancer Immunol. Immunother. 2017, 66, 959–968. [Google Scholar] [CrossRef]
- McCormick, D.; Chong, H.; Hobbs, C.; Datta, C.; Hall, P.A. Detection of the Ki-67 antigen in fixed and wax-embedded sections with the monoclonal antibody MIB1. Histopathology 1993, 22, 355–360. [Google Scholar] [CrossRef]
- Scott, R.J.; Hall, P.A.; Haldane, J.S.; Van Noorden, S.; Price, Y.; Lane, D.P.; Wright, N.A. A comparison of immunohistochemical markers of cell proliferation with experimentally determined growth fraction. J. Pathol. 1991, 165, 173–178. [Google Scholar] [CrossRef]
- Schneider, G.; Bryndza, E.; Poniewierska-Baran, A.; Serwin, K.; Suszynska, M.; Sellers, Z.P.; Merchant, M.L.; Kaliappan, A.; Ratajczak, J.; Kucia, M.; et al. Evidence that vitronectin is a potent migration-enhancing factor for cancer cells chaperoned by fibrinogen: A novel view of the metastasis of cancer cells to low-fibrinogen lymphatics and body cavities. Oncotarget 2016, 7, 69829–69843. [Google Scholar] [CrossRef] [Green Version]
- Shi, K.; Lan, R.-L.; Tao, X.; Wu, C.-Y.; Hong, H.-F.; Lin, J.-H. Vitronectin significantly influences prognosis in osteosarcoma. Int. J. Clin. Exp. Pathol. 2015, 8, 11364–11371. [Google Scholar]
- Felding-Habermann, B.; O’Toole, T.E.; Mueller, B.M.; Smith, J.W.; Fransvea, E.; Ruggeri, Z.M.; Ginsberg, M.H.; Hughes, P.E.; Pampori, N.; Shattil, S.J.; et al. Integrin activation controls metastasis in human breast cancer. Proc. Natl. Acad. Sci. USA 2001, 98, 1853–1858. [Google Scholar] [CrossRef] [Green Version]
- Shiraishi, A.; Uruno, T.; Sanematsu, F.; Ushijima, M.; Sakata, D.; Hara, T.; Fukui, Y. DOCK8 Protein Regulates Macrophage Migration through Cdc42 Protein Activation and LRAP35a Protein Interaction. J. Biol. Chem. 2017, 292, 2191–2202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, Y.; Torii, T.; Kawahara, K.; Tanoue, A.; Yamauchi, J. Dock8 interacts with Nck1 in mediating Schwann cell precursor migration. Biochem. Biophys. Rep. 2016, 6, 113–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, Y.; Tanaka, Y.; Terasawa, M.; Pieczyk, M.; Habiro, K.; Katakai, T.; Hanawa-Suetsugu, K.; Kukimoto-Niino, M.; Nishizaki, T.; Shirouzu, M.; et al. DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses. Blood 2012, 119, 4451–4461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Han, L.; Zhao, G.; Xue, S.; Gao, Y.; Xiao, J.; Zhang, S.; Chen, P.; Wu, Z.-Y.; Ding, J.; et al. LRCH1 interferes with DOCK8-Cdc42–induced T cell migration and ameliorates experimental autoimmune encephalomyelitis. J. Exp. Med. 2016, 214, 209–226. [Google Scholar] [CrossRef] [Green Version]
- Li, C.-C.; Kuo, J.-C.; Waterman, C.M.; Kiyama, R.; Moss, J.; Vaughan, M. Effects of brefeldin A-inhibited guanine nucleotide-exchange (BIG) 1 and KANK1 proteins on cell polarity and directed migration during wound healing. Proc. Natl. Acad. Sci. USA 2011, 108, 19228–19233. [Google Scholar] [CrossRef] [Green Version]
- Pu, J.; Shen, J.; Zhong, Z.; Yanling, M.; Gao, J. KANK1 regulates paclitaxel resistance in lung adenocarcinoma A549 cells. Artif. Cells Nanomed. Biotechnol. 2020, 48, 639–647. [Google Scholar] [CrossRef] [Green Version]
- Bouchet, B.P.; Gough, R.E.; Ammon, Y.C.; van de Willige, D.; Post, H.; Jacquemet, G.; Altelaar, A.F.M.; Heck, A.J.R.; Goult, B.T.; Akhmanova, A. Talin-KANK1 Interaction Controls the Recruitment of Cortical Microtubule Stabilizing Complexes to Focal Adhesions. eLife 2016, 5, e18124. [Google Scholar] [CrossRef]
- Kakinuma, N.; Roy, B.C.; Zhu, Y.; Wang, Y.; Kiyama, R. Kank regulates RhoA-dependent formation of actin stress fibers and cell migration via 14-3-3 in PI3K–Akt signaling. J. Cell Biol. 2008, 181, 537–549. [Google Scholar] [CrossRef]
- Cui, Z.; Shen, Y.; Chen, K.H.; Mittal, S.K.; Yang, J.-Y.; Zhang, G. KANK1 inhibits cell growth by inducing apoptosis through regulating CXXC5 in human malignant peripheral nerve sheath tumors. Sci. Rep. 2017, 7, 40325. [Google Scholar] [CrossRef]
- Gu, Y.; Zhang, M. Upregulation of the Kank1 gene inhibits human lung cancer progression in vitro and in vivo. Oncol. Rep. 2018, 40, 1243–1250. [Google Scholar] [CrossRef] [Green Version]
- Noguera, R.; Nieto, O.A.; Tadeo, I.; Fariñas, F.; Alvaro, T. Extracellular matrix, biotensegrity and tumor microenvironment. An update and overview. Histol. Histopathol. 2012, 27, 693–705. [Google Scholar] [PubMed]
- Burgos-Panadero, R.; Lucantoni, F.; Gamero-Sandemetrio, E.; De La Cruz-Merino, L.; Álvaro, T.; Noguera, R. The tumour microenvironment as an integrated framework to understand cancer biology. Cancer Lett. 2019, 461, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xue, Q.; Li, J.; Ma, L.; Yao, Y.; Ye, H.; Cui, Z.; Yang, H. 3D bioprinting for artificial cornea: Challenges and perspectives. Med. Eng. Phys. 2019, 71, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Duchamp, M.; Oklu, R.; Ellisen, L.W.; Langer, R.; Khademhosseini, A. Bioprinting the Cancer Microenvironment. ACS Biomater. Sci. Eng. 2016, 2, 1710–1721. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Farach-Carson, M.C.; Jia, X. Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnol. Adv. 2014, 32, 1256–1268. [Google Scholar] [CrossRef] [Green Version]
- Cohn, S.L.; Pearson, A.D.J.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The International Neuroblastoma Risk Group (INRG) Classification System: An INRG Task Force Report. J. Clin. Oncol. 2009, 27, 289–297. [Google Scholar] [CrossRef]
- Brodeur, G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef]
- Innala, M.; Riebe, I.; Kuzmenko, V.; Sundberg, J.; Gatenholm, P.; Hanse, E.; Johannesson, S. 3D Culturing and differentiation of SH-SY5Y neuroblastoma cells on bacterial nanocellulose scaffolds. Artif. Cells Nanomed. Biotechnol. 2013, 42, 302–308. [Google Scholar] [CrossRef]
- Nolan, J.C.; Frawley, T.; Tighe, J.; Soh, H.; Curtin, C.; Piskareva, O. Preclinical models for neuroblastoma: Advances and challenges. Cancer Lett. 2020, 474, 53–62. [Google Scholar] [CrossRef]
- Liu, S.; Tian, Y.; Chlenski, A.; Yang, Q.; Salwen, H.R.; Cohn, S.L. ‘Cross-talk’ between Schwannian stroma and neuroblasts promotes neuroblastoma tumor differentiation and inhibits angiogenesis. Cancer Lett. 2005, 228, 125–131. [Google Scholar] [CrossRef]
- Zhu, J.; Thompson, C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef] [PubMed]
- García-Lizarribar, A.; Fernández-Garibay, X.; Velasco-Mallorquí, F.; Castaño, A.G.; Samitier, J.; Ramón-Azcón, J. Composite Biomaterials as Long-Lasting Scaffolds for 3D Bioprinting of Highly Aligned Muscle Tissue. Macromol. Biosci. 2018, 18, e1800167. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monferrer, E.; Sanegre, S.; Martín-Vañó, S.; García-Lizarribar, A.; Burgos-Panadero, R.; López-Carrasco, A.; Navarro, S.; Samitier, J.; Noguera, R. Digital Image Analysis Applied to Tumor Cell Proliferation, Aggressiveness, and Migration-Related Protein Synthesis in Neuroblastoma 3D Models. Int. J. Mol. Sci. 2020, 21, 8676. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228676
Monferrer E, Sanegre S, Martín-Vañó S, García-Lizarribar A, Burgos-Panadero R, López-Carrasco A, Navarro S, Samitier J, Noguera R. Digital Image Analysis Applied to Tumor Cell Proliferation, Aggressiveness, and Migration-Related Protein Synthesis in Neuroblastoma 3D Models. International Journal of Molecular Sciences. 2020; 21(22):8676. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228676
Chicago/Turabian StyleMonferrer, Ezequiel, Sabina Sanegre, Susana Martín-Vañó, Andrea García-Lizarribar, Rebeca Burgos-Panadero, Amparo López-Carrasco, Samuel Navarro, Josep Samitier, and Rosa Noguera. 2020. "Digital Image Analysis Applied to Tumor Cell Proliferation, Aggressiveness, and Migration-Related Protein Synthesis in Neuroblastoma 3D Models" International Journal of Molecular Sciences 21, no. 22: 8676. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228676




