LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: ceRNA and Alternative Mechanisms
Abstract
:1. Introduction
2. ceRNA Hypothesis and New Methods for Its Unresolved Questions
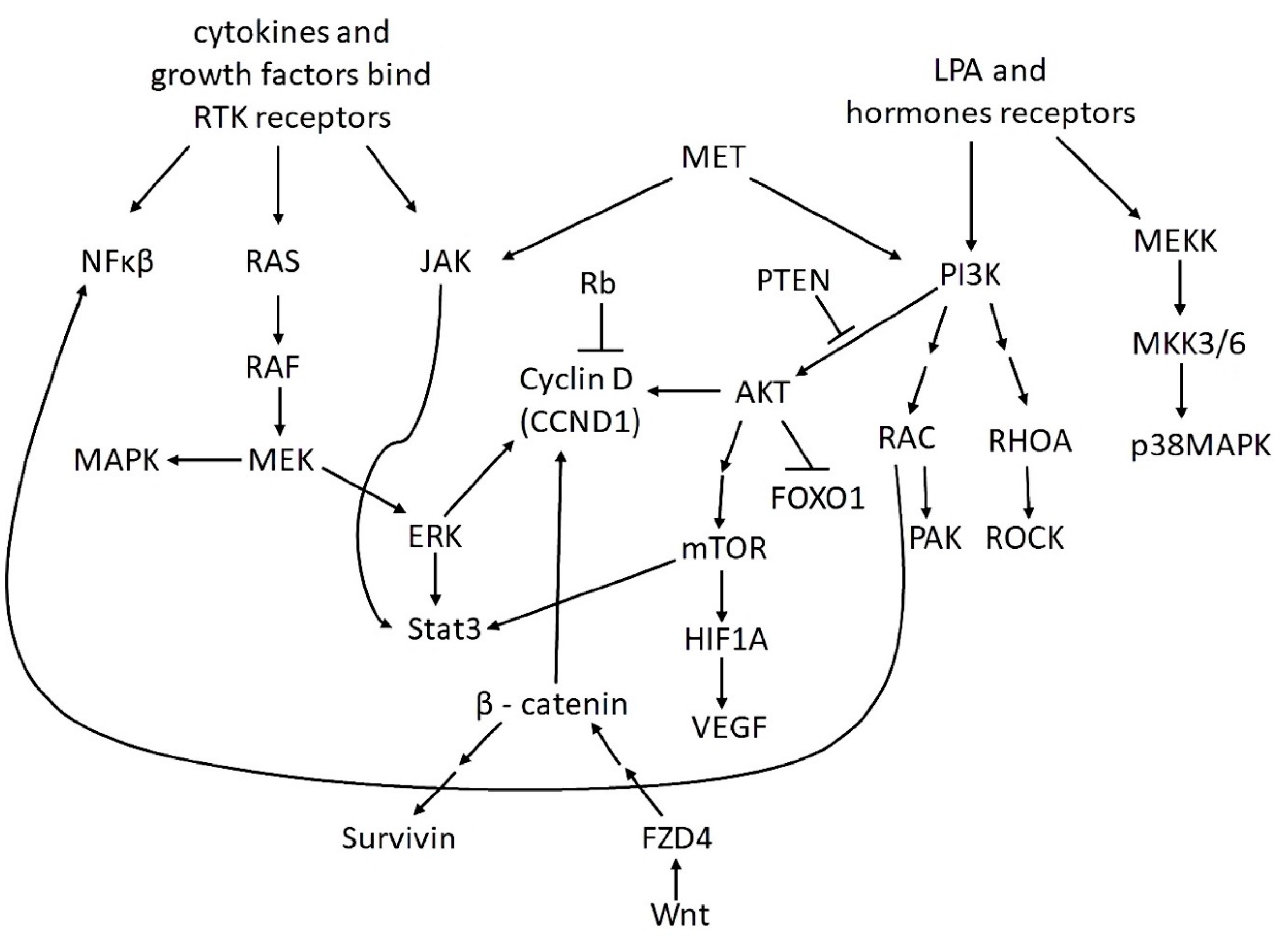
3. The Most Important Signaling Pathways in OvCa
4. Mechanisms and Pathways Associated with OvCa Metastasis
5. Mechanisms and Pathways Associated with Drug Resistance in OvCa
6. LncRNAs in the Development and Progression of OvCa via ceRNA Model
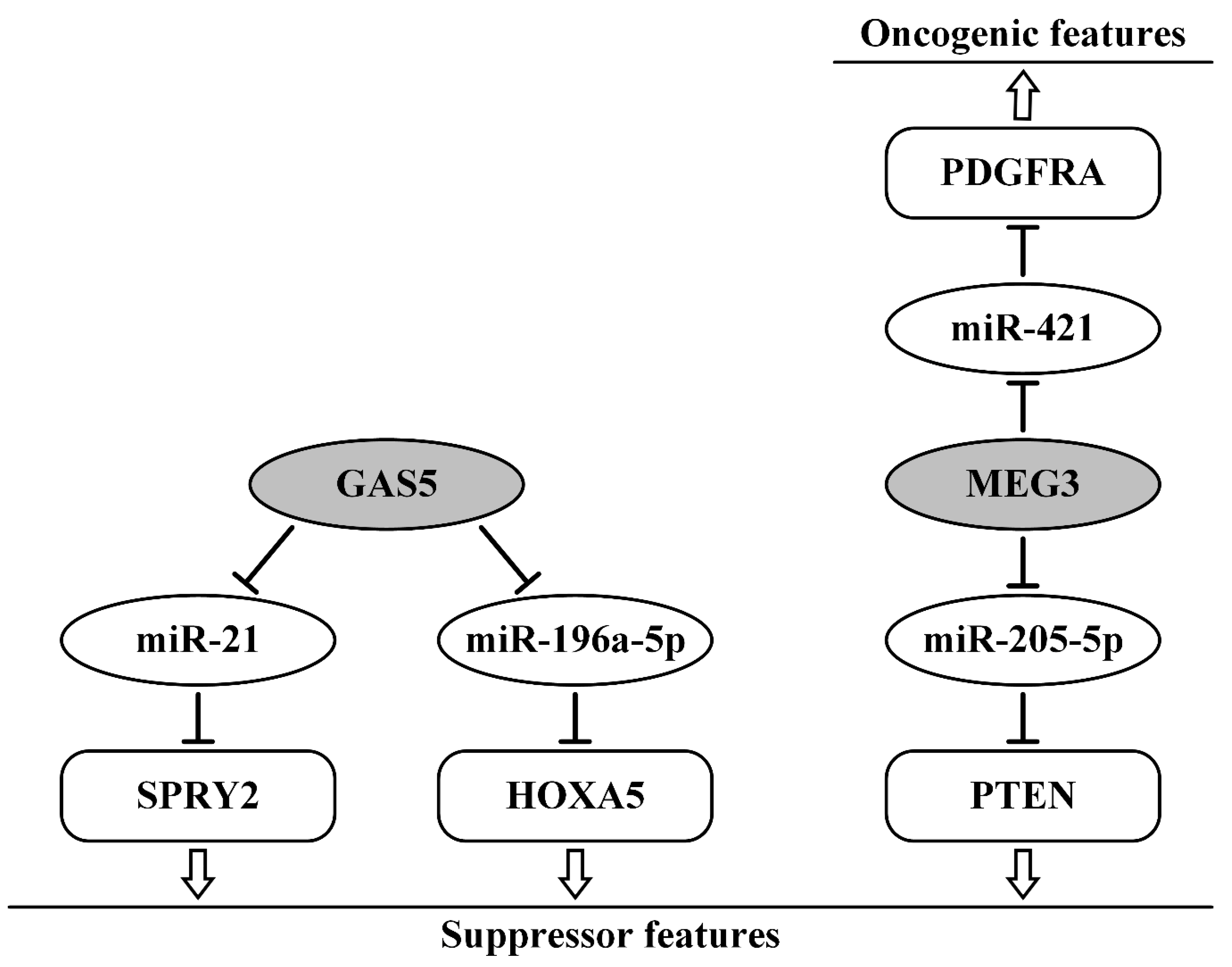
7. Suppressor lncRNAs as ceRNAs in Ovarian Cancer
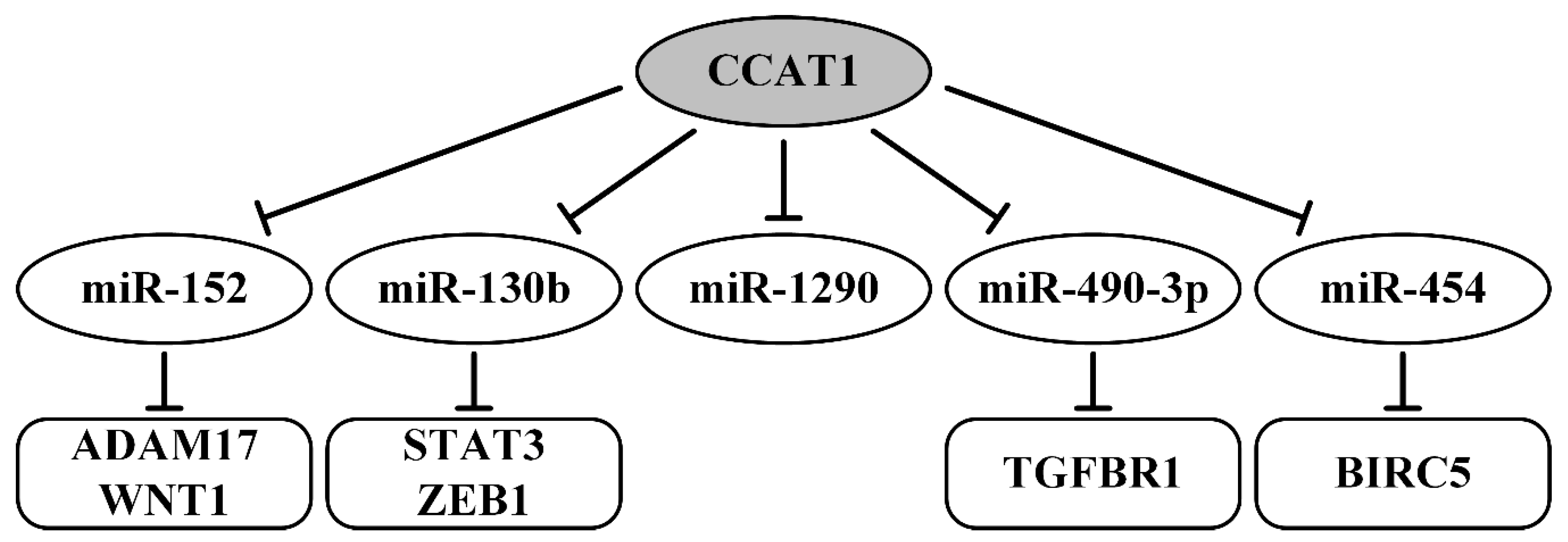
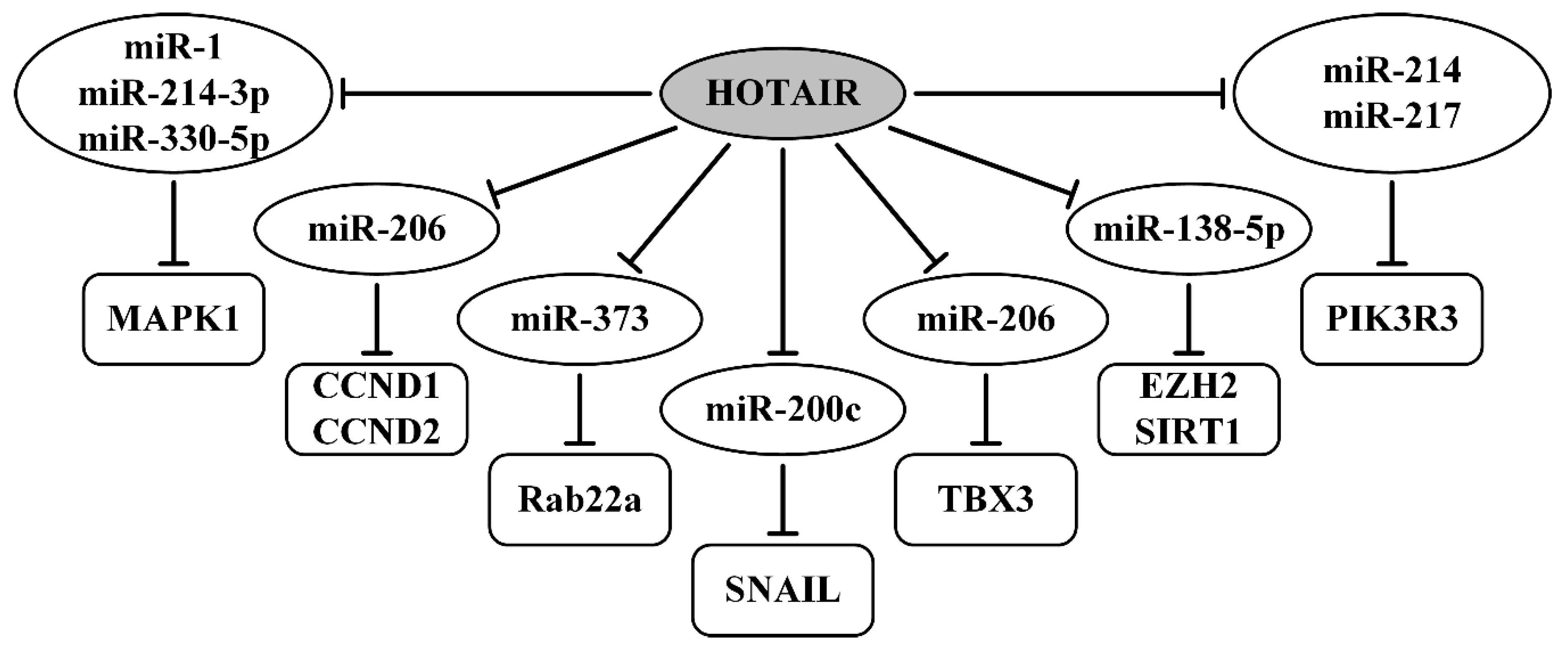
8. Oncogenic lncRNAs as ceRNAs in Ovarian Cancer
9. Examples of Alternative Mechanisms of Action of lncRNAs in OvCa
10. Effect of lncRNA on Signaling Pathways in OvCa Development, Metastasis, and Resistance to Therapy
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| OvCa | ovarian cancer |
| ceRNA | competitive endogenous RNA |
| lncRNA | long non-coding RNA |
| EMT | epithelial-mesenchymal transition |
| CCAT1 | colon cancer-associated transcript 1 |
| HOTAIR | HOX transcript antisense intergenic RNA |
| DANCR | differentiation antagonizing non-protein coding RNA |
| GAS5 | growth arrest-specific transcript 5 |
| MALAT1 | metastasis-associated lung adenocarcinoma transcript 1 |
| MEG3 | maternally expressed 3 |
| NEAT1 | nuclear-enriched abundant transcript 1 |
| TUG1 | taurine upregulated 1 |
| UCA1 | urothelial carcinoma-associated 1 |
| 3′-UTR | 3′-untranslated region |
References
- Yang, D.; Sun, L.; Li, Z.; Gao, P. Noncoding RNAs in Regulation of Cancer Metabolic Reprogramming. Adv. Exp. Med. Biol. 2016, 927, 191–215. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Villen, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerstein, M.B.; Kundaje, A.; Hariharan, M.; Landt, S.G.; Yan, K.K.; Cheng, C.; Mu, X.J.; Khurana, E.; Rozowsky, J.; Alexander, R.; et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012, 489, 91–100. [Google Scholar] [CrossRef]
- Chan, S.H.; Wang, L.H. Regulation of cancer metastasis by microRNAs. J. Biomed. Sci. 2015, 22, 9. [Google Scholar] [CrossRef] [Green Version]
- Deb, B.; Uddin, A.; Chakraborty, S. miRNAs and ovarian cancer: An overview. J. Cell. Physiol. 2018, 233, 3846–3854. [Google Scholar] [CrossRef]
- Loginov, V.I.; Rykov, S.V.; Fridman, M.V.; Braga, E.A. Methylation of miRNA genes and oncogenesis. Biochem. Biokhimiia 2015, 80, 145–162. [Google Scholar] [CrossRef]
- Sanchez Calle, A.; Kawamura, Y.; Yamamoto, Y.; Takeshita, F.; Ochiya, T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018, 109, 2093–2100. [Google Scholar] [CrossRef]
- Zhao, X.; Cai, Y.; Xu, J. Circular RNAs: Biogenesis, Mechanism, and Function in Human Cancers. Int. J. Mol. Sci. 2019, 20, 3926. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leygue, E. Steroid receptor RNA activator (SRA1): Unusual bifaceted gene products with suspected relevance to breast cancer. Nucl. Recept. Signal. 2007, 5, e006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Bure, I.V.; Kuznetsova, E.B.; Zaletaev, D.V. Long Noncoding RNAs and Their Role in Oncogenesis. Mol. Biol. 2018, 52, 907–920. [Google Scholar] [CrossRef]
- Jin, S.J.; Jin, M.Z.; Xia, B.R.; Jin, W.L. Long Non-coding RNA DANCR as an Emerging Therapeutic Target in Human Cancers. Front. Oncol. 2019, 9, 1225. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Wang, Y.; Zhang, X.; Ren, Q.; Li, R.; Huang, Y.; Lu, H.; Chen, J. Calycosin inhibits the in vitro and in vivo growth of breast cancer cells through WDR7-7-GPR30 Signaling. J. Exp. Clin. Cancer Res. 2017, 36, 153. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zheng, X.; Xu, B.; Hu, W.; Huang, T.; Jiang, J. The Identification and Analysis of mRNA-lncRNA-miRNA Cliques From the Integrative Network of Ovarian Cancer. Front. Genet. 2019, 10, 751. [Google Scholar] [CrossRef] [Green Version]
- Abildgaard, C.; Do Canto, L.M.; Steffensen, K.D.; Rogatto, S.R. Long Non-coding RNAs Involved in Resistance to Chemotherapy in Ovarian Cancer. Front. Oncol. 2019, 9, 1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salamini-Montemurri, M.; Lamas-Maceiras, M.; Barreiro-Alonso, A.; Vizoso-Vazquez, A.; Rodriguez-Belmonte, E.; Quindos-Varela, M.; Cerdan, M.E. The Challenges and Opportunities of LncRNAs in Ovarian Cancer Research and Clinical Use. Cancers 2020, 12, 1020. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Tang, D.; Zhao, M.; Kajiyama, H.; Kikkawa, F.; Kondo, Y. Long non-coding RNA: A recently accentuated molecule in chemoresistance in cancer. Cancer Metastasis Rev. 2020, 39, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Vogell, A.; Evans, M.L. Cancer Screening in Women. Obstet. Gynecol. Clin. N. Am. 2019, 46, 485–499. [Google Scholar] [CrossRef] [PubMed]
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef] [Green Version]
- Bosson, A.D.; Zamudio, J.R.; Sharp, P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell 2014, 56, 347–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366. [Google Scholar] [CrossRef]
- Chandradoss, S.D.; Schirle, N.T.; Szczepaniak, M.; MacRae, I.J.; Joo, C. A Dynamic Search Process Underlies MicroRNA Targeting. Cell 2015, 162, 96–107. [Google Scholar] [CrossRef] [Green Version]
- Salomon, W.E.; Jolly, S.M.; Moore, M.J.; Zamore, P.D.; Serebrov, V. Single-Molecule Imaging Reveals that Argonaute Reshapes the Binding Properties of Its Nucleic Acid Guides. Cell 2016, 166, 517–520. [Google Scholar] [CrossRef] [PubMed]
- Braga, E.A.; Fridman, M.V.; Kushlinskii, N.E. Molecular Mechanisms of Ovarian Carcinoma Metastasis: Key Genes and Regulatory MicroRNAs. Biochem. Biokhimiia 2017, 82, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Toss, A.; De Matteis, E.; Rossi, E.; Casa, L.D.; Iannone, A.; Federico, M.; Cortesi, L. Ovarian cancer: Can proteomics give new insights for therapy and diagnosis? Int. J. Mol. Sci. 2013, 14, 8271–8290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, P.; Zheng, C.H.; Xia, J. Identification of ovarian cancer subtype-specific network modules and candidate drivers through an integrative genomics approach. Oncotarget 2016, 7, 4298–4309. [Google Scholar] [CrossRef] [Green Version]
- Ghoneum, A.; Said, N. PI3K-AKT-mTOR and NFkappaB Pathways in Ovarian Cancer: Implications for Targeted Therapeutics. Cancers 2019, 11, 949. [Google Scholar] [CrossRef] [Green Version]
- Malik, M.Z.; Chirom, K.; Ali, S.; Ishrat, R.; Somvanshi, P.; Singh, R.K.B. Methodology of predicting novel key regulators in ovarian cancer network: A network theoretical approach. BMC Cancer 2019, 19, 1129. [Google Scholar] [CrossRef] [Green Version]
- Titone, R.; Morani, F.; Follo, C.; Vidoni, C.; Mezzanzanica, D.; Isidoro, C. Epigenetic control of autophagy by microRNAs in ovarian cancer. BioMed Res. Int. 2014, 2014, 343542. [Google Scholar] [CrossRef]
- Bunkholt Elstrand, M.; Dong, H.P.; Odegaard, E.; Holth, A.; Elloul, S.; Reich, R.; Trope, C.G.; Davidson, B. Mammalian target of rapamycin is a biomarker of poor survival in metastatic serous ovarian carcinoma. Hum. Pathol. 2010, 41, 794–804. [Google Scholar] [CrossRef]
- Binju, M.; Amaya-Padilla, M.A.; Wan, G.; Gunosewoyo, H.; Suryo Rahmanto, Y.; Yu, Y. Therapeutic Inducers of Apoptosis in Ovarian Cancer. Cancers 2019, 11, 1786. [Google Scholar] [CrossRef] [Green Version]
- Chou, J.L.; Chen, L.Y.; Lai, H.C.; Chan, M.W. TGF-beta: Friend or foe? The role of TGF-beta/SMAD signaling in epigenetic silencing of ovarian cancer and its implication in epigenetic therapy. Expert Opin. Ther. Targets 2010, 14, 1213–1223. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Guan, W.; Zhang, L.; Sun, W.; Zhou, D.; Lin, Q.; Ren, W.; Nadeem, L.; Xu, G. Physical interaction of STAT1 isoforms with TGF-beta receptors leads to functional crosstalk between two signaling pathways in epithelial ovarian cancer. J. Exp. Clin. Cancer Res. 2018, 37, 103. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Baker, D.; Ten Dijke, P. TGF-beta-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int. J. Mol. Sci. 2019, 20, 2767. [Google Scholar] [CrossRef] [Green Version]
- Davidson, B.; Trope, C.G.; Reich, R. Epithelial-mesenchymal transition in ovarian carcinoma. Front. Oncol. 2012, 2, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaravinos, A. The Regulatory Role of MicroRNAs in EMT and Cancer. J. Oncol. 2015, 2015, 865816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teeuwssen, M.; Fodde, R. Wnt Signaling in Ovarian Cancer Stemness, EMT, and Therapy Resistance. J. Clin. Med. 2019, 8, 1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.H.; Song, J.Y.; Park, H.; Jeong, J.Y.; Kwon, A.Y.; Heo, J.H.; Kang, H.; Kim, G.; An, H.J. miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett. 2015, 356, 937–945. [Google Scholar] [CrossRef]
- Deng, J.; Bai, X.; Feng, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Inhibition of PI3K/Akt/mTOR signaling pathway alleviates ovarian cancer chemoresistance through reversing epithelial-mesenchymal transition and decreasing cancer stem cell marker expression. BMC Cancer 2019, 19, 618. [Google Scholar] [CrossRef]
- Wu, C.J.; Sundararajan, V.; Sheu, B.C.; Huang, R.Y.; Wei, L.H. Activation of STAT3 and STAT5 Signaling in Epithelial Ovarian Cancer Progression: Mechanism and Therapeutic Opportunity. Cancers 2019, 12, 24. [Google Scholar] [CrossRef] [Green Version]
- Imam, J.S.; Plyler, J.R.; Bansal, H.; Prajapati, S.; Bansal, S.; Rebeles, J.; Chen, H.I.; Chang, Y.F.; Panneerdoss, S.; Zoghi, B.; et al. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS ONE 2012, 7, e52397. [Google Scholar] [CrossRef] [Green Version]
- Fang, D.; Chen, H.; Zhu, J.Y.; Wang, W.; Teng, Y.; Ding, H.F.; Jing, Q.; Su, S.B.; Huang, S. Epithelial-mesenchymal transition of ovarian cancer cells is sustained by Rac1 through simultaneous activation of MEK1/2 and Src signaling pathways. Oncogene 2017, 36, 1546–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.; Robertson, G.; Pedersen, L.; Lim, E.; Hernandez-Herrera, A.; Rowat, A.C.; Patil, S.L.; Chan, C.K.; Wen, Y.; Zhang, X.; et al. miR-509-3p is clinically significant and strongly attenuates cellular migration and multi-cellular spheroids in ovarian cancer. Oncotarget 2016, 7, 25930–25948. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Wang, Q.; Lau, W.B.; Lau, B.; Xu, L.; Zhao, L.; Yang, H.; Feng, M.; Xuan, Y.; Yang, Y.; et al. Tumor microenvironment: The culprit for ovarian cancer metastasis? Cancer Lett. 2016, 377, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Lei, Y.; Hu, Y.; Ding, W.; Zhang, C.; Fang, C. miR-448 negatively regulates ovarian cancer cell growth and metastasis by targeting CXCL12. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2015, 17, 903–909. [Google Scholar] [CrossRef]
- Hu, Y.; Zhu, Q.N.; Deng, J.L.; Li, Z.X.; Wang, G.; Zhu, Y.S. Emerging role of long non-coding RNAs in cisplatin resistance. OncoTargets Ther. 2018, 11, 3185–3194. [Google Scholar] [CrossRef] [Green Version]
- Loret, N.; Denys, H.; Tummers, P.; Berx, G. The Role of Epithelial-to-Mesenchymal Plasticity in Ovarian Cancer Progression and Therapy Resistance. Cancers 2019, 11, 838. [Google Scholar] [CrossRef] [Green Version]
- Palma Flores, C.; Garcia-Vazquez, R.; Gallardo Rincon, D.; Ruiz-Garcia, E.; Astudillo de la Vega, H.; Marchat, L.A.; Salinas Vera, Y.M.; Lopez-Camarillo, C. MicroRNAs driving invasion and metastasis in ovarian cancer: Opportunities for translational medicine (Review). Int. J. Oncol. 2017, 50, 1461–1476. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.N.; Chang, R.; Lin, L.T.; Chern, C.U.; Tsai, H.W.; Wen, Z.H.; Li, Y.H.; Li, C.J.; Tsui, K.H. MicroRNA in Ovarian Cancer: Biology, Pathogenesis, and Therapeutic Opportunities. Int. J. Environ. Res. Public Health 2019, 16, 1510. [Google Scholar] [CrossRef] [Green Version]
- Ghafouri-Fard, S.; Shoorei, H.; Taheri, M. miRNA profile in ovarian cancer. Exp. Mol. Pathol. 2020, 113, 104381. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, B. microRNAs as biomarkers of ovarian cancer. Expert Rev. Anticancer Ther. 2020, 20, 373–385. [Google Scholar] [CrossRef]
- Loginov, V.I.; Pronina, I.V.; Burdennyy, A.M.; Filippova, E.A.; Kazubskaya, T.P.; Kushlinsky, D.N.; Utkin, D.O.; Khodyrev, D.S.; Kushlinskii, N.E.; Dmitriev, A.A.; et al. Novel miRNA genes deregulated by aberrant methylation in ovarian carcinoma are involved in metastasis. Gene 2018, 662, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Panoutsopoulou, K.; Avgeris, M.; Scorilas, A. miRNA and long non-coding RNA: Molecular function and clinical value in breast and ovarian cancers. Expert Rev. Mol. Diagn. 2018, 18, 963–979. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Lu, A.Q.; Chen, L.J. LncRNAs in ovarian cancer. Clin. Chim. Acta Int. J. Clin. Chem. 2019, 490, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Li, J.; Wei, B. Long non-coding RNAs in ovarian cancer. J. Exp. Clin. Cancer Res. 2018, 37, 120. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yu, S.; Yang, R.; Wang, Q.; Liu, X.; Ma, M.; Li, Y.; Wu, S. Identification of lncRNA-associated ceRNA network in high-grade serous ovarian cancer metastasis. Epigenomics 2020, 12, 1175–1191. [Google Scholar] [CrossRef]
- Zhao, X.; Tang, D.Y.; Zuo, X.; Zhang, T.D.; Wang, C. Identification of lncRNA-miRNA-mRNA regulatory network associated with epithelial ovarian cancer cisplatin-resistant. J. Cell. Physiol. 2019, 234, 19886–19894. [Google Scholar] [CrossRef]
- Li, G.; Han, L.; Ren, F.; Zhang, R.; Qin, G. Prognostic value of the tumor-specific ceRNA network in epithelial ovarian cancer. J. Cell. Physiol. 2019, 234, 22071–22081. [Google Scholar] [CrossRef]
- Chen, J.; Lin, Y.; Jia, Y.; Xu, T.; Wu, F.; Jin, Y. LncRNA HAND2-AS1 exerts anti-oncogenic effects on ovarian cancer via restoration of BCL2L11 as a sponge of microRNA-340-5p. J. Cell. Physiol. 2019, 234, 23421–23436. [Google Scholar] [CrossRef]
- Chang, H.; Zhang, X.; Li, B.; Meng, X. MAGI2-AS3 suppresses MYC signaling to inhibit cell proliferation and migration in ovarian cancer through targeting miR-525-5p/MXD1 axis. Cancer Med. 2020. [Google Scholar] [CrossRef]
- Wang, J.; Ding, W.; Xu, Y.; Tao, E.; Mo, M.; Xu, W.; Cai, X.; Chen, X.; Yuan, J.; Wu, X. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging 2020, 12, 4558–4572. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, T.; Zhang, X.; Zuo, D.; Liu, C. lncRNA RHPN1-AS1 Promotes Ovarian Cancer Growth and Invasiveness Through Inhibiting miR-1299. OncoTargets Ther. 2020, 13, 5337–5344. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Jin, C.; Li, H.; Qin, Q.; Li, L. LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by targeting miR-182-5p/FOXF2 signaling pathway. Int. J. Biol. Macromol. 2018, 120, 1705–1713. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Yang, P.; Gao, Y. Long non-coding RNA EPB41L4A-AS2 suppresses progression of ovarian cancer by sequestering microRNA-103a to upregulate transcription factor RUNX1T1. Exp. Physiol. 2020, 105, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Li, S.; Zhang, Q.; Wang, H.; Qin, H.; Wang, S. Long non-coding RNA GAS5 inhibits ovarian cancer cell proliferation via the control of microRNA-21 and SPRY2 expression. Exp. Ther. Med. 2018, 16, 73–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Yu, H.; Zheng, J.; Ning, N.; Tang, F.; Yang, Y.; Wang, Y. Lowly-expressed lncRNA GAS5 facilitates progression of ovarian cancer through targeting miR-196-5p and thereby regulating HOXA5. Gynecol. Oncol. 2018, 151, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Gokulnath, P.; de Cristofaro, T.; Manipur, I.; Di Palma, T.; Soriano, A.A.; Guarracino, M.R.; Zannini, M. Long Non-Coding RNA HAND2-AS1 Acts as a Tumor Suppressor in High-Grade Serous Ovarian Carcinoma. Int. J. Mol. Sci. 2020, 21, 4059. [Google Scholar] [CrossRef]
- Chao, H.; Zhang, M.; Hou, H.; Zhang, Z.; Li, N. HOTAIRM1 suppresses cell proliferation and invasion in ovarian cancer through facilitating ARHGAP24 expression by sponging miR-106a-5p. Life Sci. 2020, 243, 117296. [Google Scholar] [CrossRef]
- Zhang, W.; Fei, J.; Yu, S.; Shen, J.; Zhu, X.; Sadhukhan, A.; Lu, W.; Zhou, J. LINC01088 inhibits tumorigenesis of ovarian epithelial cells by targeting miR-24-1-5p. Sci. Rep. 2018, 8, 2876. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Pan, H. Long Noncoding RNA LINC01125 Enhances Cisplatin Sensitivity of Ovarian Cancer via miR-1972. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 9844–9854. [Google Scholar] [CrossRef]
- Liu, M.; Shen, C.; Wang, C. Long Noncoding RNA LINC01133 Confers Tumor-Suppressive Functions in Ovarian Cancer by Regulating Leucine-Rich Repeat Kinase 2 as an miR-205 Sponge. Am. J. Pathol. 2019, 189, 2323–2339. [Google Scholar] [CrossRef]
- Gokulnath, P.; de Cristofaro, T.; Manipur, I.; Di Palma, T.; Soriano, A.A.; Guarracino, M.R.; Zannini, M. Long Non-Coding RNA MAGI2-AS3 is a New Player with a Tumor Suppressive Role in High Grade Serous Ovarian Carcinoma. Cancers 2019, 11, 2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Huang, X.; Liang, F.; Zhao, L. LncRNA miR503HG interacts with miR-31-5p through multiple ways to regulate cancer cell invasion and migration in ovarian cancer. J. Ovarian Res. 2020, 13, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Wu, W.; Cao, X.; Zhao, X.; Li, W.; Deng, C.; Huang, Z. lncRNA mortal obligate RNA transcript was downregulated in ovarian carcinoma and inhibits cancer cell proliferation by downregulating miRNA-21. J. Cell. Biochem. 2019. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ma, S.; Bai, X.; Pan, W.; Ai, L.; Tan, W. Long noncoding RNA WDFY3-AS2 suppresses tumor progression by acting as a competing endogenous RNA of microRNA-18a in ovarian cancer. J. Cell. Physiol. 2020, 235, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Qi, S.; Xie, C.; Li, C.; Wang, P.; Liu, D. Upregulation of long non-coding RNA XIST has anticancer effects on epithelial ovarian cancer cells through inverse downregulation of hsa-miR-214-3p. J. Gynecol. Oncol. 2018, 29, e99. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, W.; Zhang, H.; Liu, T.; Zhao, L. miR-214 targets the PTEN-mediated PI3K/Akt signaling pathway and regulates cell proliferation and apoptosis in ovarian cancer. Oncol. Lett. 2017, 14, 5711–5718. [Google Scholar] [CrossRef] [Green Version]
- Ye, W.; Ni, Z.; Yicheng, S.; Pan, H.; Huang, Y.; Xiong, Y.; Liu, T. Anisomycin inhibits angiogenesis in ovarian cancer by attenuating the molecular sponge effect of the lncRNAMeg3/miR421/PDGFRA axis. Int. J. Oncol. 2019, 55, 1296–1312. [Google Scholar] [CrossRef]
- Tao, P.; Yang, B.; Zhang, H.; Sun, L.; Wang, Y.; Zheng, W. The overexpression of lncRNA MEG3 inhibits cell viability and invasion and promotes apoptosis in ovarian cancer by sponging miR-205-5p. Int. J. Clin. Exp. Pathol. 2020, 13, 869–879. [Google Scholar]
- Shi, X.; Xiao, L.; Mao, X.; He, J.; Ding, Y.; Huang, J.; Peng, C.; Xu, Z. miR-205-5p Mediated Downregulation of PTEN Contributes to Cisplatin Resistance in C13K Human Ovarian Cancer Cells. Front. Genet. 2018, 9, 555. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Sheng, X.; Li, J.; Yang, L.; Chen, Z.; Zhao, Q.; Tan, L.; Zhou, Y.; Li, J. Promoter hypermethylation influences the suppressive role of maternally expressed 3, a long non-coding RNA, in the development of epithelial ovarian cancer. Oncol. Rep. 2014, 32, 277–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Z.H.; Wu, D.M.; Fan, S.H.; Wen, X.; Han, X.R.; Wang, S.; Wang, Y.J.; Zhang, Z.F.; Shan, Q.; Li, M.Q.; et al. LncRNA AB209371 up-regulated Survivin gene by down-regulating miR-203 in ovarian carcinoma. J. Ovarian Res. 2019, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Qian, H.; Guo, T.; Ye, J.; Jin, C.; Liu, X.; Jiang, L.; Wang, X.; Lin, M.; Yu, H. LncRNA-ATB Promotes the Tumorigenesis of Ovarian Cancer via Targeting miR-204-3p. OncoTargets Ther. 2020, 13, 573–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Li, Y.; Kong, D.; Hu, L.; Liu, D.; Wu, J. Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. J. Cell. Physiol. 2019, 234, 10800–10808. [Google Scholar] [CrossRef]
- Cao, Y.; Shi, H.; Ren, F.; Jia, Y.; Zhang, R. Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in epithelial ovarian cancer. Exp. Cell Res. 2017, 359, 185–194. [Google Scholar] [CrossRef]
- Lai, X.J.; Cheng, H.F. LncRNA colon cancer-associated transcript 1 (CCAT1) promotes proliferation and metastasis of ovarian cancer via miR-1290. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 322–328. [Google Scholar] [CrossRef]
- Mu, Y.; Li, N.; Cui, Y.L. The lncRNA CCAT1 upregulates TGFbetaR1 via sponging miR-490-3p to promote TGFbeta1-induced EMT of ovarian cancer cells. Cancer Cell Int. 2018, 18, 145. [Google Scholar] [CrossRef]
- Wang, D.Y.; Li, N.; Cui, Y.L. Long Non-coding RNA CCAT1 Sponges miR-454 to Promote Chemoresistance of Ovarian Cancer Cells to Cisplatin by Regulation of Surviving. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2020, 52, 798–814. [Google Scholar] [CrossRef] [Green Version]
- Hua, F.; Li, C.H.; Chen, X.G.; Liu, X.P. Long Noncoding RNA CCAT2 Knockdown Suppresses Tumorous Progression by Sponging miR-424 in Epithelial Ovarian Cancer. Oncol. Res. 2018, 26, 241–247. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, Y.; Liu, H.; Su, D.; Luo, F.; Zhou, F. Long noncoding RNA CDKN2B-AS1 interacts with miR-411-3p to regulate ovarian cancer in vitro and in vivo through HIF-1a/VEGF/P38 pathway. Biochem. Biophys. Res. Commun. 2019, 514, 44–50. [Google Scholar] [CrossRef]
- Xu, C.; Zhai, J.; Fu, Y. LncRNA CDKN2B-AS1 promotes the progression of ovarian cancer by miR-143-3p/SMAD3 axis and predicts a poor prognosis. Neoplasma 2020, 67, 782–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Yang, F.; Qi, X.; Li, Q.; Wang, D.; Yi, T.; Yin, R.; Zhao, X.; Zhong, X.; Bian, C. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol. Carcinog. 2019, 58, 2286–2296. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Sun, K.X.; Wu, D.D.; Xiu, Y.L.; Chen, X.; Chen, S.; Zong, Z.H.; Sang, X.B.; Liu, Y.; Zhao, Y. DLEU1 contributes to ovarian carcinoma tumourigenesis and development by interacting with miR-490-3p and altering CDK1 expression. J. Cell. Mol. Med. 2017, 21, 3055–3065. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, C. LncRNA DLX6-AS1 aggravates the development of ovarian cancer via modulating FHL2 by sponging miR-195-5p. Cancer Cell Int. 2020, 20, 370. [Google Scholar] [CrossRef]
- Yan, H.; Silva, M.A.; Li, H.; Zhu, L.; Li, P.; Li, X.; Wang, X.; Gao, J.; Wang, P.; Zhang, Z. Long noncoding RNA DQ786243 interacts with miR-506 and promotes progression of ovarian cancer through targeting cAMP responsive element binding protein 1. J. Cell. Biochem. 2018, 119, 9764–9780. [Google Scholar] [CrossRef]
- Duan, M.; Fang, M.; Wang, C.; Wang, H.; Li, M. LncRNA EMX2OS Induces Proliferation, Invasion and Sphere Formation of Ovarian Cancer Cells via Regulating the miR-654-3p/AKT3/PD-L1 Axis. Cancer Manag. Res. 2020, 12, 2141–2154. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Li, H.; Silva, M.A.; Guan, Y.; Yang, L.; Zhu, L.; Zhang, Z.; Li, G.; Ren, C. LncRNA FLVCR1-AS1 mediates miR-513/YAP1 signaling to promote cell progression, migration, invasion and EMT process in ovarian cancer. J. Exp. Clin. Cancer Res. 2019, 38, 356. [Google Scholar] [CrossRef] [Green Version]
- Yao, N.; Yu, L.; Zhu, B.; Gan, H.Y.; Guo, B.Q. LncRNA GIHCG promotes development of ovarian cancer by regulating microRNA-429. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8127–8134. [Google Scholar] [CrossRef]
- Li, J.; Huang, Y.; Deng, X.; Luo, M.; Wang, X.; Hu, H.; Liu, C.; Zhong, M. Long noncoding RNA H19 promotes transforming growth factor-beta-induced epithelial-mesenchymal transition by acting as a competing endogenous RNA of miR-370-3p in ovarian cancer cells. OncoTargets Ther. 2018, 11, 427–440. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Zhou, Y.; Chen, W.; Chen, L.; Lu, J.; He, F.; Li, X.; Zhao, L. Ginsenoside 20(S)-Rg3 Prevents PKM2-Targeting miR-324-5p from H19 Sponging to Antagonize the Warburg Effect in Ovarian Cancer Cells. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 51, 1340–1353. [Google Scholar] [CrossRef]
- Yang, M.; Zhai, Z.; Zhang, Y.; Wang, Y. Clinical significance and oncogene function of long noncoding RNA HAGLROS overexpression in ovarian cancer. Arch. Gynecol. Obstet. 2019, 300, 703–710. [Google Scholar] [CrossRef]
- Tong, L.; Wang, Y.; Ao, Y.; Sun, X. CREB1 induced lncRNA HAS2-AS1 promotes epithelial ovarian cancer proliferation and invasion via the miR-466/RUNX2 axis. Biomed. Pharmacother. 2019, 115, 108891. [Google Scholar] [CrossRef]
- Gao, Y.; Meng, H.; Liu, S.; Hu, J.; Zhang, Y.; Jiao, T.; Liu, Y.; Ou, J.; Wang, D.; Yao, L.; et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum. Mol. Genet. 2015, 24, 841–852. [Google Scholar] [CrossRef] [Green Version]
- Yiwei, T.; Hua, H.; Hui, G.; Mao, M.; Xiang, L. HOTAIR Interacting with MAPK1 Regulates Ovarian Cancer skov3 Cell Proliferation, Migration, and Invasion. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2015, 21, 1856–1863. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.; Hui, L. HOTAIR Promotes Proliferation, Migration, and Invasion of Ovarian Cancer SKOV3 Cells Through Regulating PIK3R3. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 325–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Cheng, J.; Wu, Y.; Qiu, J.; Sun, Y.; Tong, X. LncRNA HOTAIR controls the expression of Rab22a by sponging miR-373 in ovarian cancer. Mol. Med. Rep. 2016, 14, 2465–2472. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.; Guo, R.; Yuan, Z.; Shi, H.; Zhang, D. LncRNA HOTAIR Regulates CCND1 and CCND2 Expression by Sponging miR-206 in Ovarian Cancer. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 49, 1289–1303. [Google Scholar] [CrossRef]
- Yang, C.; Li, H.; Zhang, T.; Chu, Y.; Chen, D.; Zuo, J. miR-200c overexpression inhibits the invasion and tumorigenicity of epithelial ovarian cancer cells by suppressing lncRNA HOTAIR in mice. J. Cell. Biochem. 2020, 121, 1514–1523. [Google Scholar] [CrossRef]
- Zhang, Y.; Ai, H.; Fan, X.; Chen, S.; Wang, Y.; Liu, L. Knockdown of long non-coding RNA HOTAIR reverses cisplatin resistance of ovarian cancer cells through inhibiting miR-138-5p-regulated EZH2 and SIRT1. Biol. Res. 2020, 53, 18. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, J.; Cai, E.; Cai, J.; Wen, Y.; Lu, S.; Li, X.; Han, Q.; Jiang, J.; Li, T.; et al. HOTAIR maintains the stemness of ovarian cancer stem cells via the miR-206/TBX3 axis. Exp. Cell Res. 2020, 395, 112218. [Google Scholar] [CrossRef]
- Zhang, Y.; Dun, Y.; Zhou, S.; Huang, X.H. LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells proliferation and invasion by targeting miR-133a-3p and activating Wnt/beta-catenin signaling pathway. Biomed. Pharmacother. 2017, 96, 1216–1221. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.; Wang, Y.; Wang, S. HOXD-AS1 promotes cell proliferation, migration and invasion through miR-608/FZD4 axis in ovarian cancer. Am. J. Cancer Res. 2018, 8, 170–182. [Google Scholar] [PubMed]
- Dong, S.; Wang, R.; Wang, H.; Ding, Q.; Zhou, X.; Wang, J.; Zhang, K.; Long, Y.; Lu, S.; Hong, T.; et al. HOXD-AS1 promotes the epithelial to mesenchymal transition of ovarian cancer cells by regulating miR-186-5p and PIK3R3. J. Exp. Clin. Cancer Res. 2019, 38, 110. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.; Xu, L.; Su, H. HULC functions as an oncogene in ovarian carcinoma cells by negatively modulating miR-125a-3p. J. Physiol. Biochem. 2019, 75, 163–171. [Google Scholar] [CrossRef]
- Lu, X.; Wang, F.; Fu, M.; Li, Y.; Wang, L. Long Noncoding RNA KCNQ1OT1 Accelerates the Progression of Ovarian Cancer via MicroRNA-212-3/LCN2 Axis. Oncol. Res. 2020, 28, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, R.; Kang, F.; Lai, H.; Wang, Y. KCNQ1OT1 promotes ovarian cancer progression via modulating MIR-142-5p/CAPN10 axis. Mol. Genet. Genom. Med. 2020, 8, e1077. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Ruan, F. LncRNA LEF1-AS1 Promotes Ovarian Cancer Development Through Interacting with miR-1285-3p. Cancer Manag. Res. 2020, 12, 687–694. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Fang, X.; Xia, B.; Zhao, Y.; Li, Q.; Wu, X. Long noncoding RNA LINC00152 promotes cell proliferation through competitively binding endogenous miR-125b with MCL-1 by regulating mitochondrial apoptosis pathways in ovarian cancer. Cancer Med. 2018, 7, 4530–4541. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Zhou, K.; Wu, Y.; Wang, L.; Lu, S. Linc00161 regulated the drug resistance of ovarian cancer by sponging microRNA-128 and modulating MAPK1. Mol. Carcinog. 2019, 58, 577–587. [Google Scholar] [CrossRef]
- Du, W.; Feng, Z.; Sun, Q. LncRNA LINC00319 accelerates ovarian cancer progression through miR-423-5p/NACC1 pathway. Biochem. Biophys. Res. Commun. 2018, 507, 198–202. [Google Scholar] [CrossRef]
- Pan, L.; Meng, Q.; Li, H.; Liang, K.; Li, B. LINC00339 promotes cell proliferation, migration, and invasion of ovarian cancer cells via miR-148a-3p/ROCK1 axes. Biomed. Pharmacother. 2019, 120, 109423. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wen, J.; Wang, H.; Wang, Y. Long non-coding RNA LINC00460 promotes epithelial ovarian cancer progression by regulating microRNA-338-3p. Biomed. Pharmacother. 2018, 108, 1022–1028. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, X.; Chen, Y.; Cao, D. Long non-coding RNA LINC00504 regulates the Warburg effect in ovarian cancer through inhibition of miR-1244. Mol. Cell. Biochem. 2020, 464, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhu, G.; Bao, S.; Chen, S. Long Non-Coding RNA LINC00511 Mediates the Effects of ESR1 on Proliferation and Invasion of Ovarian Cancer Through miR-424-5p and miR-370-5p. Cancer Manag. Res. 2019, 11, 10807–10819. [Google Scholar] [CrossRef]
- Liu, W.; Yang, Y.J.; An, Q. LINC00963 Promotes Ovarian Cancer Proliferation, Migration and EMT via the miR-378g /CHI3L1 Axis. Cancer Manag. Res. 2020, 12, 463–473. [Google Scholar] [CrossRef] [Green Version]
- Shi, C.; Wang, M. LINC01118 Modulates Paclitaxel Resistance of Epithelial Ovarian Cancer by Regulating miR-134/ABCC1. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 8831–8839. [Google Scholar] [CrossRef]
- Yu, H.; Xu, Y.; Zhang, D.; Liu, G. Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of miR-612/HOXA13 pathway. Biochem. Biophys. Res. Commun. 2018, 503, 2095–2100. [Google Scholar] [CrossRef]
- Liu, H.Z.; Liu, G.Y.; Pang, W.W.; Zhang, H.; Zeng, Z.J.; Wang, H.J. LncRNA LUCAT1 promotes proliferation of ovarian cancer cells by regulating miR-199a-5p expression. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1682–1687. [Google Scholar] [CrossRef]
- Lei, R.; Xue, M.; Zhang, L.; Lin, Z. Long noncoding RNA MALAT1-regulated microRNA 506 modulates ovarian cancer growth by targeting iASPP. OncoTargets Ther. 2017, 10, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Pa, M.; Naizaer, G.; Seyiti, A.; Kuerbang, G. Long Noncoding RNA MALAT1 Functions as a Sponge of MiR-200c in Ovarian Cancer. Oncol. Res. 2017. [Google Scholar] [CrossRef]
- Lin, Q.; Guan, W.; Ren, W.; Zhang, L.; Zhang, J.; Xu, G. MALAT1 affects ovarian cancer cell behavior and patient survival. Oncol. Rep. 2018, 39, 2644–2652. [Google Scholar] [CrossRef] [PubMed]
- Tao, F.; Tian, X.; Ruan, S.; Shen, M.; Zhang, Z. miR-211 sponges lncRNA MALAT1 to suppress tumor growth and progression through inhibiting PHF19 in ovarian carcinoma. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Li, Q.; Xie, F. LncRNA-MALAT1 regulates proliferation and apoptosis of ovarian cancer cells by targeting miR-503-5p. OncoTargets Ther. 2019, 12, 6297–6307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.; Wang, L.; Han, X.C.; Ma, H.Y.; Zhang, N.; Zhe, L. LncRNA MIF-AS1 aggravates the progression of ovarian cancer by sponging miRNA-31-5p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2248–2255. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Li, H.; Li, P.; Li, X.; Lin, J.; Zhu, L.; Silva, M.A.; Wang, X.; Wang, P.; Zhang, Z. Long noncoding RNA MLK7-AS1 promotes ovarian cancer cells progression by modulating miR-375/YAP1 axis. J. Exp. Clin. Cancer Res. 2018, 37, 237. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, A.; Gao, M.; Duan, X.; Li, Z. LncRNA MIR4435-2HG triggers ovarian cancer progression by regulating miR-128-3p/CKD14 axis. Cancer Cell Int. 2020, 20, 145. [Google Scholar] [CrossRef]
- Chang, H.; Li, B.; Zhang, X.; Meng, X. NCK1-AS1 promotes NCK1 expression to facilitate tumorigenesis and chemo-resistance in ovarian cancer. Biochem. Biophys. Res. Commun. 2020, 522, 292–299. [Google Scholar] [CrossRef]
- Chai, Y.; Liu, J.; Zhang, Z.; Liu, L. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med. 2016, 5, 1588–1598. [Google Scholar] [CrossRef]
- Ding, N.; Wu, H.; Tao, T.; Peng, E. NEAT1 regulates cell proliferation and apoptosis of ovarian cancer by miR-34a-5p/BCL2. OncoTargets Ther. 2017, 10, 4905–4915. [Google Scholar] [CrossRef] [Green Version]
- An, J.; Lv, W.; Zhang, Y. LncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR-194. OncoTargets Ther. 2017, 10, 5377–5390. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wang, Y.; Fu, X.; Lu, Z. Long non-coding RNA NEAT1 promoted ovarian cancer cells’ metastasis through regulation of miR-382-3p/ROCK1 axial. Cancer Sci. 2018, 109, 2188–2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yong, W.; Yu, D.; Jun, Z.; Yachen, D.; Weiwei, W.; Midie, X.; Xingzhu, J.; Xiaohua, W. Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer. Cell Death Dis. 2018, 9, 861. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Yang, L.; Wang, X. NEAT1 Knockdown Suppresses the Cisplatin Resistance in Ovarian Cancer by Regulating miR-770-5p/PARP1 Axis. Cancer Manag. Res. 2020, 12, 7277–7289. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Ao, Y.; Zhang, H.; Wang, K.; Wang, Y.; Ma, Q. Long noncoding RNA NORAD is upregulated in epithelial ovarian cancer and its downregulation suppressed cancer cell functions by competing with miR-155-5p. Cancer Med. 2019, 8, 4782–4791. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhu, L.X.; Sun, D.M.; Yao, H.; Han, D.X. Regulatory mechanism of lncRNA NORAD on proliferation and invasion of ovarian cancer cells through miR-199a-3p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1672–1681. [Google Scholar] [CrossRef]
- Tao, F.; Tian, X.; Lu, M.; Zhang, Z. A novel lncRNA, Lnc-OC1, promotes ovarian cancer cell proliferation and migration by sponging miR-34a and miR-34c. J. Genet. Genom. Yi Chuan Xue Bao 2018, 45, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.Y.; Jiang, X.X.; Tian, H.N.; Guo, H.L.; Guo, H.; Guo, Y. Long non-coding RNA OIP5-AS1 plays an oncogenic role in ovarian cancer through targeting miR-324-3p/NFIB axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7266–7275. [Google Scholar] [CrossRef]
- Guo, L.; Chen, J.; Liu, D.; Liu, L. OIP5-AS1/miR-137/ZNF217 Axis Promotes Malignant Behaviors in Epithelial Ovarian Cancer. Cancer Manag. Res. 2020, 12, 6707–6717. [Google Scholar] [CrossRef]
- Gu, L.P.; Jin, S.; Xu, R.C.; Zhang, J.; Geng, Y.C.; Shao, X.Y.; Qin, L.B. Long non-coding RNA PCAT-1 promotes tumor progression by inhibiting miR-129-5p in human ovarian cancer. Arch. Med. Sci. AMS 2019, 15, 513–521. [Google Scholar] [CrossRef]
- Min, F.; Chu, G. Long noncoding RNA PCAT-1 knockdown prevents the development of ovarian cancer cells via microRNA-124-3p. J. Cell. Biochem. 2020, 121, 1963–1972. [Google Scholar] [CrossRef]
- Liang, H.; Zhao, X.; Wang, C.; Sun, J.; Chen, Y.; Wang, G.; Fang, L.; Yang, R.; Yu, M.; Gu, Y.; et al. Systematic analyses reveal long non-coding RNA (PTAF)-mediated promotion of EMT and invasion-metastasis in serous ovarian cancer. Mol. Cancer 2018, 17, 96. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Yu, T.; Han, Y.; Jiang, H.; Wang, C.; You, T.; Zhao, X.; Shan, H.; Yang, R.; Yang, L.; et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol. Cancer 2018, 17, 119. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Yu, M.; Yang, R.; Zhang, L.; Zhang, L.; Zhu, D.; Luo, H.; Hong, Y.; Yu, T.; Sun, J.; et al. A PTAL-miR-101-FN1 Axis Promotes EMT and Invasion-Metastasis in Serous Ovarian Cancer. Mol. Ther. Oncolytics 2020, 16, 53–62. [Google Scholar] [CrossRef]
- Yang, Q.; Yu, Y.; Sun, Z.; Pan, Y. Long non-coding RNA PVT1 promotes cell proliferation and invasion through regulating miR-133a in ovarian cancer. Biomed. Pharmacother. 2018, 106, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Du, H.; Bao, L.; Liu, W. LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol. Med. 2018, 15, 238–250. [Google Scholar] [CrossRef]
- Ding, Y.; Fang, Q.; Li, Y.; Wang, Y. Amplification of lncRNA PVT1 promotes ovarian cancer proliferation by binding to miR-140. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2019, 30, 217–225. [Google Scholar] [CrossRef]
- Qu, C.; Dai, C.; Guo, Y.; Qin, R.; Liu, J. Long non-coding RNA PVT1-mediated miR-543/SERPINI1 axis plays a key role in the regulatory mechanism of ovarian cancer. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, S.; Wu, L.; Pei, M. Interaction between LncRNA-ROR and miR-145 contributes to epithelial-mesenchymal transition of ovarian cancer cells. Gen. Physiol. Biophys. 2019, 38, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Liu, Z.; Lu, L.; Liu, F.; Zhang, B. Long Noncoding RNA SCAMP1 Targets miR-137/CXCL12 Axis to Boost Cell Invasion and Angiogenesis in Ovarian Cancer. DNA Cell Biol. 2020, 39, 1041–1050. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, A.; Zhang, Z. LncRNA SDHAP1 confers paclitaxel resistance of ovarian cancer by regulating EIF4G2 expression via miR-4465. J. Biochem. 2020, 168, 171–181. [Google Scholar] [CrossRef]
- Sun, D.; Fan, X.H. LncRNA SNHG12 accelerates the progression of ovarian cancer via absorbing miRNA-129 to upregulate SOX4. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2345–2352. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, L.L.; Sun, K.X.; Xiu, Y.L.; Zong, Z.H.; Chen, X.; Zhao, Y. The role of the long non-coding RNA TDRG1 in epithelial ovarian carcinoma tumorigenesis and progression through miR-93/RhoC pathway. Mol. Carcinog. 2018, 57, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, Y.; Xu, Y.; He, X.; Li, Y. Silencing the long noncoding RNA, TINCR, a molecular sponge of miR335, inhibits the malignant phenotype of epithelial ovarian cancer via FGF2 suppression. Int. J. Oncol. 2019, 55, 1110–1124. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, Y.; Cheng, H.; Tian, J.; Yang, S. Roles of a TMPO-AS1/microRNA-200c/TMEFF2 ceRNA network in the malignant behaviors and 5-FU resistance of ovarian cancer cells. Exp. Mol. Pathol. 2020, 115, 104481. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Wen, J.; Qi, T.; Wang, Y. Long non-coding RNA TTN-AS1 promotes tumorigenesis of ovarian cancer through modulating the miR-139-5p/ROCK2 axis. Biomed. Pharmacother. 2020, 125, 109882. [Google Scholar] [CrossRef] [PubMed]
- Miao, S.; Wang, J.; Xuan, L.; Liu, X. LncRNA TTN-AS1 acts as sponge for miR-15b-5p to regulate FBXW7 expression in ovarian cancer. BioFactors 2020. [Google Scholar] [CrossRef]
- Yang, X.; Xin, N.; Qu, H.J.; Wei, L.; Han, Z. Long noncoding RNA TUG1 facilitates cell ovarian cancer progression through targeting MiR-29b-3p/MDM2 axis. Anat. Rec. 2020. [Google Scholar] [CrossRef]
- Gu, L.; Li, Q.; Liu, H.; Lu, X.; Zhu, M. Long Noncoding RNA TUG1 Promotes Autophagy-Associated Paclitaxel Resistance by Sponging miR-29b-3p in Ovarian Cancer Cells. OncoTargets Ther. 2020, 13, 2007–2019. [Google Scholar] [CrossRef] [Green Version]
- Zhan, F.L.; Chen, C.F.; Yao, M.Z. LncRNA TUG1 facilitates proliferation, invasion and stemness of ovarian cancer cell via miR-186-5p/ZEB1 axis. Cell Biochem. Funct. 2020. [Google Scholar] [CrossRef]
- Pei, Y.; Li, K.; Lou, X.; Wu, Y.; Dong, X.; Wang, W.; Li, N.; Zhang, D.; Cui, W. miR1299/NOTCH3/TUG1 feedback loop contributes to the malignant proliferation of ovarian cancer. Oncol. Rep. 2020, 44, 438–448. [Google Scholar] [CrossRef]
- Wang, J.; Ye, C.; Liu, J.; Hu, Y. UCA1 confers paclitaxel resistance to ovarian cancer through miR-129/ABCB1 axis. Biochem. Biophys. Res. Commun. 2018, 501, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Wang, X.L.; Dang, Y.; Zhu, X.Z.; Zhang, Y.H.; Cai, B.X.; Zheng, L. Long non-coding RNA UCA1 promotes the progression of paclitaxel resistance in ovarian cancer by regulating the miR-654-5p/SIK2 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Hou, Y.; Chen, H.; Yang, S.; Liu, T.; Lin, M.; Lou, G. Long non-coding RNA ZFAS1 interacts with miR-150-5p to regulate Sp1 expression and ovarian cancer cell malignancy. Oncotarget 2017, 8, 19534–19546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lupia, M.; Cavallaro, U. Ovarian cancer stem cells: Still an elusive entity? Mol. Cancer 2017, 16, 64. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Feng, S.J.; Qiu, S.; Shao, N.; Zheng, J.H. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3176–3184. [Google Scholar]
- Cao, H.; Huang, S.; Liu, A.; Chen, Z. Up-regulated expression of miR-155 in human colonic cancer. J. Cancer Res. Ther. 2018, 14, 604–607. [Google Scholar] [CrossRef]
- Khalighfard, S.; Alizadeh, A.M.; Irani, S.; Omranipour, R. Plasma miR-21, miR-155, miR-10b, and Let-7a as the potential biomarkers for the monitoring of breast cancer patients. Sci. Rep. 2018, 8, 17981. [Google Scholar] [CrossRef]
- Shao, C.; Yang, F.; Qin, Z.; Jing, X.; Shu, Y.; Shen, H. The value of miR-155 as a biomarker for the diagnosis and prognosis of lung cancer: A systematic review with meta-analysis. BMC Cancer 2019, 19, 1103. [Google Scholar] [CrossRef] [Green Version]
- Mitra, A.K.; Zillhardt, M.; Hua, Y.; Tiwari, P.; Murmann, A.E.; Peter, M.E.; Lengyel, E. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012, 2, 1100–1108. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Jia, R.; Zhao, J. Dexmedetomidine Regulates Proliferation, Apoptosis, Migration, and Invasion in Ovarian Cancer Cells via MiR-155-HIF-1alpha Axis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 10164–10172. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Xu, C.; Zhang, X. MicroRNA-139-5p Inhibits Cell Proliferation and Invasion by Targeting RHO-Associated Coiled-Coil-Containing Protein Kinase 2 in Ovarian Cancer. Oncol. Res. 2018, 26, 411–420. [Google Scholar] [CrossRef]
- Wu, B.; Liu, G.; Jin, Y.; Yang, T.; Zhang, D.; Ding, L.; Zhou, F.; Pan, Y.; Wei, Y. miR-15b-5p Promotes Growth and Metastasis in Breast Cancer by Targeting HPSE2. Front. Oncol. 2020, 10, 108. [Google Scholar] [CrossRef] [Green Version]
- Yeh, C.H.; Bellon, M.; Nicot, C. FBXW7: A critical tumor suppressor of human cancers. Mol. Cancer 2018, 17, 115. [Google Scholar] [CrossRef]
- Long, X.; Song, K.; Hu, H.; Tian, Q.; Wang, W.; Dong, Q.; Yin, X.; Di, W. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J. Exp. Clin. Cancer Res. 2019, 38, 345. [Google Scholar] [CrossRef]
- Wang, H.; Fang, L.; Jiang, J.; Kuang, Y.; Wang, B.; Shang, X.; Han, P.; Li, Y.; Liu, M.; Zhang, Z.; et al. The cisplatin-induced lncRNA PANDAR dictates the chemoresistance of ovarian cancer via regulating SFRS2-mediated p53 phosphorylation. Cell Death Dis. 2018, 9, 1103. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, H. Long non-coding RNA GEHT1 promoted the proliferation of ovarian cancer cells via modulating the protein stability of HIF1alpha. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Pei, C.L.; Fei, K.L.; Yuan, X.Y.; Gong, X.J. LncRNA DANCR aggravates the progression of ovarian cancer by downregulating UPF1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10657–10663. [Google Scholar] [CrossRef]
- Huang, K.; Geng, J.; Wang, J. Long non-coding RNA RP11-552M11.4 promotes cells proliferation, migration and invasion by targeting BRCA2 in ovarian cancer. Cancer Sci. 2018, 109, 1428–1446. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, L.; Wang, Z.; Pan, T.; Sahni, N.; Jin, X.; Wang, G.; Li, J.; Zheng, X.; Zhang, Y.; et al. LncMAP: Pan-cancer atlas of long noncoding RNA-mediated transcriptional network perturbations. Nucleic Acids Res. 2018, 46, 1113–1123. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Jiao, Y.; Hao, J.; Xing, H.; Li, C. Long noncoding RNA TP73-AS1 accelerates the epithelial ovarian cancer via epigenetically repressing p21. Am. J. Transl. Res. 2019, 11, 2447–2454. [Google Scholar]
- Guo, L.L.; Wang, S.F. Downregulated Long Noncoding RNA GAS5 Fails to Function as Decoy of CEBPB, Resulting in Increased GDF15 Expression and Rapid Ovarian Cancer Cell Proliferation. Cancer Biother. Radiopharm. 2019, 34, 537–546. [Google Scholar] [CrossRef]
- Lin, X.; Spindler, T.J.; de Souza Fonseca, M.A.; Corona, R.I.; Seo, J.H.; Dezem, F.S.; Li, L.; Lee, J.M.; Long, H.W.; Sellers, T.A.; et al. Super-Enhancer-Associated LncRNA UCA1 Interacts Directly with AMOT to Activate YAP Target Genes in Epithelial Ovarian Cancer. iScience 2019, 17, 242–255. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Wang, Y.; Zhong, W.; Cheng, H.; Tian, Z. The Long Non-Coding RNA MALAT1 Enhances Ovarian Cancer Cell Stemness by Inhibiting YAP Translocation from Nucleus to Cytoplasm. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e922012. [Google Scholar] [CrossRef]
- Bai, L.; Wang, A.; Zhang, Y.; Xu, X.; Zhang, X. Knockdown of MALAT1 enhances chemosensitivity of ovarian cancer cells to cisplatin through inhibiting the Notch1 signaling pathway. Exp. Cell Res. 2018, 366, 161–171. [Google Scholar] [CrossRef]
- Gordon, M.A.; Babbs, B.; Cochrane, D.R.; Bitler, B.G.; Richer, J.K. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol. Carcinog. 2019, 58, 196–205. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, J.; Yu, J.; Ge, W.; Xiao, X.; Dai, S.; Xiang, Q. LncRNA CACS15 accelerates the malignant progression of ovarian cancer through stimulating EZH2-induced inhibition of APC. Am. J. Transl. Res. 2019, 11, 6561–6568. [Google Scholar]
- Wang, L.; Yu, M.; Zhao, S. lncRNA MEG3 modified epithelial-mesenchymal transition of ovarian cancer cells by sponging miR-219a-5p and regulating EGFR. J. Cell. Biochem. 2019, 120, 17709–17722. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, R.; Wu, Y.; Liu, Y.; Su, W.; Xiong, W.; Zeng, Z. PVT1 Promotes Cancer Progression via MicroRNAs. Front. Oncol. 2019, 9, 609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Gu, X.; Yang, X.; Meng, Y. MiR-1204 promotes ovarian squamous cell carcinoma growth by increasing glucose uptake. Biosci. Biotechnol. Biochem. 2019, 83, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Thomas, P. PANTHER pathway: An ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 2009, 563, 123–140. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Huang, H.; Li, Y.; Li, L.; Hou, W.; You, Z. Decreased expression of long non-coding RNA GAS5 promotes cell proliferation, migration and invasion, and indicates a poor prognosis in ovarian cancer. Oncol. Rep. 2016, 36, 3241–3250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Liu, M.; Zou, Y.; Mao, M.; Shen, T.; Zhang, C.; Song, S.; Sun, M.; Zhang, S.; Wang, B.; et al. Long non-coding RNA growth arrest-specific transcript 5 is involved in ovarian cancer cell apoptosis through the mitochondria-mediated apoptosis pathway. Oncol. Rep. 2015, 34, 3212–3221. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.Q.; Cheng, H.Y.; Liu, K.F. Long non-coding RNA DANCR upregulates IGF2 expression and promotes ovarian cancer progression. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3621–3626. [Google Scholar] [CrossRef]
- Chen, S.; Wu, D.D.; Sang, X.B.; Wang, L.L.; Zong, Z.H.; Sun, K.X.; Liu, B.L.; Zhao, Y. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 2017, 8, e3118. [Google Scholar] [CrossRef] [Green Version]
- Zou, H.; Li, H. Knockdown of long non-coding RNA LINC00152 increases cisplatin sensitivity in ovarian cancer cells. Exp. Ther. Med. 2019, 18, 4510–4516. [Google Scholar] [CrossRef]
- Wang, J.; Tian, Y.; Zheng, H.; Ding, Y.; Wang, X. An integrated analysis reveals the oncogenic function of lncRNA LINC00511 in human ovarian cancer. Cancer Med. 2019, 8, 3026–3035. [Google Scholar] [CrossRef] [Green Version]
- Ding, C.; Wei, R.; Rodriguez, R.A.; Del Mar Requena Mullor, M. LncRNA PCAT-1 plays an oncogenic role in epithelial ovarian cancer by modulating cyclinD1/CDK4 expression. Int. J. Clin. Exp. Pathol. 2019, 12, 2148–2156. [Google Scholar]
- Ozes, A.R.; Miller, D.F.; Ozes, O.N.; Fang, F.; Liu, Y.; Matei, D.; Huang, T.; Nephew, K.P. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene 2016, 35, 5350–5361. [Google Scholar] [CrossRef] [Green Version]
- Zou, A.; Liu, R.; Wu, X. Long non-coding RNA MALAT1 is up-regulated in ovarian cancer tissue and promotes SK-OV-3 cell proliferation and invasion. Neoplasma 2016, 63, 865–872. [Google Scholar] [CrossRef]
- Gong, J.; Xu, X.; Zhang, X.; Zhou, Y. LncRNA MIR4435-2HG is a potential early diagnostic marker for ovarian carcinoma. Acta Biochim. Biophys. Sin. 2019, 51, 953–959. [Google Scholar] [CrossRef]
- Yang, X.; Yan, Y.; Chen, Y.; Li, J.; Yang, J. Involvement of NORAD/miR-608/STAT3 axis in carcinostasis effects of physcion 8-O-beta-glucopyranoside on ovarian cancer cells. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2855–2865. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Wang, S.; Wang, Z.; Cai, J.; Han, L.; Xie, L.; Han, Q.; Wang, W.; Zhang, Y.; He, X.; et al. HOTAIR promotes paclitaxel resistance by regulating CHEK1 in ovarian cancer. Cancer Chemother. Pharmacol. 2020, 86, 295–305. [Google Scholar] [CrossRef]
- Luo, Z.P.; Jin, H. Effects of LncRNA KCNQ1OT1 on proliferation and migration of ovarian cancer cells by Wnt/beta-catenin. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8788–8794. [Google Scholar] [CrossRef]
- Guo, C.; Wang, X.; Chen, L.P.; Li, M.; Li, M.; Hu, Y.H.; Ding, W.H.; Wang, X. Long non-coding RNA MALAT1 regulates ovarian cancer cell proliferation, migration and apoptosis through Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3703–3712. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, H.R. Down-regulation of long noncoding RNA DLX6-AS1 defines good prognosis and inhibits proliferation and metastasis in human epithelial ovarian cancer cells via Notch signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3243–3252. [Google Scholar] [CrossRef]
- Mitra, R.; Chen, X.; Greenawalt, E.J.; Maulik, U.; Jiang, W.; Zhao, Z.; Eischen, C.M. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat. Commun. 2017, 8, 1604. [Google Scholar] [CrossRef] [Green Version]
- Elsayed, A.M.; Amero, P.; Salama, S.A.; Abdelaziz, A.H.; Lopez-Berestein, G.; Rodriguez-Aguayo, C. Back to the Future: Rethinking the Great Potential of lncRNAS for Optimizing Chemotherapeutic Response in Ovarian Cancer. Cancers 2020, 12, 2406. [Google Scholar] [CrossRef]
- Fu, L.L.; Li, C.J.; Xu, Y.; Li, L.Y.; Zhou, X.; Li, D.D.; Chen, S.X.; Wang, F.G.; Zhang, X.Y.; Zheng, L.W. Role of lncRNAs as Novel Biomarkers and Therapeutic Targets in Ovarian Cancer. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 183–195. [Google Scholar] [CrossRef]



| Axis lncRNA/miRNA/mRNA | LncRNA Expression; Involvement in Suppression of Progression, Influence on Prognosis, Survival, Drug Sensitivity | References |
|---|---|---|
| ADAMTS9-AS2/miR-182-5p/FOXF2 | downregulated; reduced invasion, EMT in vitro, in vivo | [72] |
| EPB41L4A-AS2/miR-103a/RUNX1T1 | inhibited migration, invasion in vitro, in vivo | [73] |
| GAS5/miR-21/SPRY2 | suppressed migration, invasion in vitro, better prognosis | [74] |
| GAS5/miR-196a-5p/HOXA5 | downregulated; suppressed proliferation, EMT, migration | [75] |
| HAND2-AS1/miR-340-5p/BCL2L11 | downregulated, hypermethylated; inhibits migration, invasion, adhesion to extracellular matrix in vitro, in vivo | [68,76] |
| HOTAIRM1/miR-106a-5p/ARHGAP24 | downregulated; inhibited migration, invasion, metastasis | [77] |
| LINC01088/miR-24-1-5p/PAK4 | under-expressed; inhibited proliferation, xenografts | [78] |
| LINC01125/miR-1972 | reduced proliferation, enhanced cisplatin sensitivity | [79] |
| LINC01133/miR-205/LRRK2 | suppresses invasion, migration in vitro, in vivo | [80] |
| MAGI2-AS3/miR-15-5p (miR-374a-5p, miR-374b-5p)/PTEN | downregulated, hypermethylated; decreased migration, viability, adhesion to extra cellular matrix | [81] |
| MAGI2-AS3/miR-525-5p/MXD1 | inhibited cell cycle, migration, invasion; MYC signaling | [69] |
| MIR503HG/miR-31-5p | downregulated; inhibited invasion, migration; methylation of miR-31 gene by MIR503HG | [82] |
| MORT/miR-21 | downregulated; inhibited proliferation, in vitro | [83] |
| WDFY3-AS2/miR-18a/RORA | suppressed migration, invasion, EMT, in vitro, in vivo | [84] |
| XIST/miR-214-3p/PTEN | downregulated; inhibited migration, invasion, in vitro, in vivo, xenografts, increase chemo-sensitivity | [85,86] |
| Suppressor lncRNA with dual functions | ||
| MEG3/miR-421/PDGFRA | upregulated; promoted angiogenesis, invasion | [87] |
| MEG3/miR-205-5p/PTEN | downregulated; inhibited migration, invasion, increase cisplatin sensitivity | [88,89] |
| Axis lncRNA/miRNA/mRNA | LncRNA Expression; Involvement in Progression, Influence on Prognosis, Survival, Drug Resistance, Signaling Pathways | Reference |
|---|---|---|
| AB209371/miR-203/BIRC5 | upregulated; advanced clinical stages | [92] |
| LncRNA-ATB/miR-204-3p/NID1 | tumorigenesis in vitro, in vivo, invasion | [93] |
| CASC9/miR-758-3p/LIN7A | proliferation, migration, invasion, in vitro, in vivo | [94] |
| CCAT1/miR-152/ADAM17 (WNT1) CCAT1/miR-130b/STAT3 (ZEB1) | upregulated; EMT, migration, invasion, metastasis, FIGO stage, poor survival | [95] |
| CCAT1/miR-1290 | tumor size, metastasis, prognosis | [96] |
| CCAT1/miR-490-3p/TGFBR1 | migration, invasion, EMT, metastasis | [97] |
| CCAT1/miR-454/BIRC5 | in vivo tumor formation, cisplatin resistance | [98] |
| CCAT2/miR-424 | upregulated; proliferation, progression | [99] |
| CDKN2B-AS1/miR-411-3p/HIF1A | migration, invasion, metastasis, HIF-1α/VEGF/P38 | [100] |
| CDKN2B-AS1/miR-143-3p/SMAD3 | migration, invasion, in vivo, poor prognosis | [101] |
| DANCR/miR-145/VEGF | invasion, angiogenesis, tube formation | [102] |
| DLEU1/miR-490-3p/CDK1 | upregulated; migration, invasion, in vivo | [103] |
| DLX6-AS1/miR-195-5p/FHL2 | upregulated; migration, invasion, EMT | [104] |
| DQ786243/miR-506/CREB1 | migration, invasion, EMT, in vivo, xenograft | [105] |
| EMX2OS/miR-654/AKT3 | invasion, sphere formation, poorer survival, PD-L1 | [106] |
| FLVCR1-AS1/miR-513/YAP1 | migration, invasion, EMT, in vivo | [107] |
| GIHCG/miR-429 | promoted cell cycle, colony formation, shorter OS | [108] |
| H19/miR-370-3p | upregulated; promotes TGF-β-induced EMT | [109] |
| H19/miR-324-5p/PKM2 | promotes aerobic glycolysis (Warburg effect) | [110] |
| HAGLROS/miR-100/mTOR (ZNRF2) | upregulated; poor prognosis, mTOR pathway | [111] |
| CREB1-HAS2-AS1/miR-466/RUNX2 | invasion, tumor growth in vivo, poor outcome | [112] |
| HOST2/let-7b | upregulated; migration, invasion, metastasis | [113] |
| HOTAIR/miR-1, miR-214-3p, miR-330-5p/MAPK1 | upregulated; migration, invasion | [114] |
| HOTAIR/miR-214, miR-217/PIK3R3 | upregulated; proliferation, migration, invasion | [115] |
| HOTAIR/miR-373/Rab22a | upregulated; migration, invasion, metastasis | [116] |
| HOTAIR/miR-206/CCND1 (CCND2) | upregulated; migration, invasion, metastasis | [117] |
| HOTAIR/miR-200c/SNAIL | EMT, migration, invasion, tumorigenicity in vivo | [118] |
| HOTAIR/miR-138-5p/EZH2, SIRT1 | upregulated; promoted cisplatin resistance | [119] |
| HOTAIR/miR-206/TBX3 | upregulated; cell stemness, cisplatin resistance | [120] |
| HOXD-AS1/miR-133a-3p | EMT, invasion, metastasis, poor OS, Wnt/β-catenin | [121] |
| HOXD-AS1/miR-608/FZD4 | upregulated; migration, invasion, poor prognosis | [122] |
| HOXD-AS1/miR-186-5p/PIK3R3 | migration, invasion, EMT, poor PFS/OS, PIK3R3 | [123] |
| HULC/miR-125a-3p | migration, invasion, PI3K/AKT/mTOR pathway | [124] |
| KCNQ1OT1/miR-212-3p/LCN2 | migration, invasion, in vitro, in vivo, shorter OS | [125] |
| KCNQ1OT1/miR-142-5p/CAPN10 | upregulated; migration in vitro, poor OS | [126] |
| LEF1-AS1/miR-1285-3p | migration, invasion, metastasis, poor prognosis | [127] |
| LINC00152/miR-125b/MCL-1 | upregulated; grade, clinical stage, poor prognosis | [128] |
| LINC00161/miR-128/MAPK1 | xenograft tumor model in vivo, drug resistance | [129] |
| LINC00319/miR-423-5p/NACC1 | upregulated; proliferation, migration, invasion | [130] |
| LINC00339/miR-148a-3p/ROCK1 | upregulated; migration, invasion, poor prognosis | [131] |
| LINC00460/miR-338-3p | migration, invasion, metastasis, shorter OS | [132] |
| LINC00504/miR-1244/PKM2 (HK2, PDK1) | upregulated; aerobic glycolysis/Warburg effect | [133] |
| ESR1-LINC00511/miR-424, miR-370 | proliferation, invasion, poor prognosis, risk model | [134] |
| LINC00963/miR-378g/CHI3L1 | upregulated; migration, EMT | [135] |
| LINC01118/miR-134/ABCC1 | migration, invasion, paclitaxel resistance | [136] |
| LUCAT1/miR-612/HOXA13 | upregulated; metastasis, poorer prognosis | [137] |
| LUCAT1/miR-199a-5p | upregulated; proliferation, colony formation | [138] |
| MALAT1/miR-506/iASPP | upregulated; proliferation, DNA synthesis in vitro | [139] |
| MALAT1/miR-200c | migration, invasion, metastasis, worse prognosis | [140] |
| MALAT1/miR-143-3p/CMPK | cell viability, migration, invasion, OS/PFS | [141] |
| MALAT1/miR-211/PHF19 | proliferation, migration, xenograft growth | [142] |
| MALAT1/miR-503-5p | proliferation, JAK2/STAT3 pathway | [143] |
| MIF-AS1/miR-31-5p/PLCB1 | elevated; migratory, invasive abilities of cells | [144] |
| MLK7-AS1/miR-375/YAP1 | invasion, metastasis, EMT, in vitro, in vivo | [145] |
| MIR4435-2HG/miR-128-3p/CDK14 | upregulated; migration, invasion | [146] |
| NCK1-AS1/miR-137/NCK1 | upregulated; migration, invasion, chemo-resistance | [147] |
| Hur>NEAT1/miR-124-3p | upregulated; migration, invasion, stage, metastasis | [148] |
| NEAT1/miR-34a-5p/BCL2 | increases cells in S phase, suppresses apoptosis | [149] |
| NEAT1/miR-194/ZEB1 | upregulated; paclitaxel resistance in vitro, in vivo | [150] |
| NEAT1/miR-382-3p/ROCK1 | upregulated; migration, invasion, metastasis | [151] |
| LIN28B>NEAT1/miR-506 | migration, invasion in vitro, in vivo, poor prognosis | [152] |
| NEAT1/miR-770-5p/PARP1 | upregulated; cisplatin resistance in vitro, in vivo | [153] |
| NORAD/miR-155-5p | upregulated; chemo-resistance, xenograft growth | [154] |
| NORAD/miR-199a-3p | proliferation, migration, invasion, EMT | [155] |
| Lnc-OC1/miR-34a, miR-34c | migration, invasion, in vitro, in vivo, prognosis | [156] |
| OIP5-AS1/miR-324-3p/NFIB | upregulated; cell viability, migration, invasion | [157] |
| OIP5-AS1/miR-137/ZNF217 | migration, invasion, EMT in vitro, in vivo | [158] |
| PCAT-1/miR-129-5p | upregulated; proliferation, inhibits apoptosis | [159] |
| PCAT-1/miR-124-3p | migration, invasion, Wnt/β-catenin, AKT/mTOR | [160] |
| PTAF(LINC00922)/miR-25/SNAI2 | TGF-β-induced EMT, invasion, metastasis | [161] |
| PTAR (AP000695.4)/miR-101/ZEB1 | migration, EMT, metastasis, in vitro, in vivo | [162] |
| PTAL/miR-101/FN1 | upregulated; EMT, invasion, metastasis | [163] |
| PVT1/miR-133a | proliferation, migration, invasion, worse PFS/OS | [164] |
| PVT1/miR-214 | invasion, EMT, short PFS/OS, PI3K/AKT | [165] |
| FOXO4/PVT1/miR-140 | upregulated; metastasis, poor survival outcomes | [166] |
| PVT1/miR-543/SERPINI1 | migration, invasion, lower 5-year OS | [167] |
| RHPN1-AS1/miR-596/LETM1 | upregulated; metastasis, DFS/OS, FAK/PI3K/AKT | [70] |
| RHPN1-AS1/miR-1299 | upregulated; migration, invasion, poor prognosis | [71] |
| LncRNA-ROR/miR-145/FLNB | upregulated; migration and invasion, EMT | [168] |
| SCAMP1/miR-137/CXCL12 | upregulated; invasion, angiogenesis | [169] |
| SDHAP1/miR-4465/EIF4G2 | upregulation; paclitaxel resistance | [170] |
| SNHG12/miR-129/SOX4 | upregulated; migration, metastasis, stage III-IV | [171] |
| TDRG1/miR-93/RhoC | upregulated; migration, invasion | [172] |
| TINCR/miR-335/FGF2 | tumor size, FIGO stage, lymphatic metastasis | [173] |
| TMPO-AS1/miR-200c/TMEFF2 | EMT, invasion, 5-FU resistance, PI3K/AKT | [174] |
| TTN-AS1/miR-139-5p/ROCK2 | migration, invasion, in vivo, metastasis, poor OS | [175] |
| TTN-AS1/miR-15b-5p/FBXW7 * | inhibits proliferation, promotes apoptosis | [176] |
| TUG1/miR-29b-3p/MDM2 | migration, invasion in vitro, tumor growth in vivo | [177] |
| TUG1/miR-29b-3p | metastasis, autophagy, paclitaxel resistance | [178] |
| TUG1/miR-186-5p/ZEB1 | proliferation, invasion, stemness | [179] |
| TUG1/miR-1299/NOTCH3 | upregulated; proliferation, a feedback loop | [180] |
| UCA1/miR-129/ABCB1 | upregulated; proliferation, paclitaxel resistance | [181] |
| UCA1/miR-654-5p/SIK2 | migration, invasion, paclitaxel resistance | [182] |
| ZFAS1/miR-150-5p/Sp1 | proliferation, migration, chemoresistance | [183] |
| Pathway | LncRNAs and Target Proteins * | References |
|---|---|---|
| Oncosuppressive lncRNAs | ||
| PI3K/AKT/mTOR | GAS5 (CCND1+), GAS5 (CDKN1A+), HAND2-AS1 (BCL2L11+), MAGI2-AS3 (PTEN+), MAGI2-AS3 (MYC+) | [212], [68], [76], [81], [69] |
| NF-κB | GAS5 (PARP1+) | [194] |
| MAPK/ERK | GAS5+, MAGI2-AS3 (MYC+) | [194], [69] |
| HIF/VEGF | GAS5 (CDKN1A+), MAGI2-AS3 (PTEN+) | [212], [81] |
| JAK/STAT | GAS5 (CDKN1A+), MAGI2-AS3 (MYC+) | [212], [69] |
| P53 | GAS5 (APAF1+), GAS5 (CDKN1A+), GAS5 (BAX+), MAGI2-AS3 (PTEN+), MAGI2-AS3 (MYC+) | [212], [212], [213], [81], [69] |
| Wnt/β-catenin | GAS5 (CCND1+), MAGI2-AS3 (MYC+) | [212], [69] |
| TGF-β | GAS5 (GDF15+), MAGI2-AS3 (MYC+) | [201], [69] |
| Hippo | MAGI2-AS3 (MYC+) | [69] |
| RAS | LINC01088 (PAK4+) | [78] |
| Oncogenic lncRNAs | ||
| PI3K/AKT/mTOR | CCAT1 (WNT1+), DANCR (IGF2+), DQ786243 (CREB1+), EMX2OS (AKT3+), HAGLROS (mTOR+), HOTAIR (PIK3R3+), HOTAIR (CCND1+), HOTAIR (CCND2+), HOXD-AS1 (FZD4+), HOXD-AS1 (PIK3R3+), HULC+, HULC (ITGB1+), LEF1-AS1 (MCL-1+), LINC00511 (CDKN1+), MALAT1+, NEAT1 (BCL2+), PCAT-1 (CCND1+), PTAL (FN1+), RHPN1-AS1+, TDRG1 (Bcl-xL+), TINCR (FGF2+), TMPO-AS1+ | [95], [214], [105], [106], [111], [115], [117], [117], [122], [123], [124], [215], [216], [217], [185], [150], [218], [163], [70], [172], [173], [174] |
| NF-κB | HOTAIR (decreasing Iκ-Bα+), NEAT1 (BCL2+), NEAT (PARP1+), SCAMP1 (CXCL12+), TDRG1 (Bcl-xL+) | [219], [150], [153], [169], [172] |
| MAPK/ERK | CCAT1 (TGFBR1+), DANCR (IGF2+), H19 (TGF-β+), HOTAIR (MAPK1+), LINC00161 (MAPK1+), MALAT1+, MIR4435-2HG (TGF-β1+), TINCR (FGF2+) | [97], [214], [109], [114], [129], [220], [221], [173] |
| HIF/VEGF | CCAT1 (STAT3+), CDKN2B-AS1 (HIF1A+), CDKN2B-AS1 (SMAD3+), DANCR (VEGFA+), HOTAIR (MAPK1+), HOTAIR (PIK3R3+), HOXD-AS1 (PIK3R3+), LINC00161 (MAPK1+), LINC00511 (CDKN1+), NEAT1 (BCL2+), NORAD (STAT3+), TDRG1 (P70S6K+) | [95], [100], [101], [102], [114], [115], [123], [129], [217], [150], [222], [172] |
| JAK/STAT | CCAT1 (STAT3+), HOTAIR (CCND1+), HOTAIR (CCND2+), LINC00511 (CDKN1+), MALAT1+, NEAT1 (BCL2+), NORAD (STAT3+), PCAT-1 (CCND1+), TDRG1 (Bcl-xL+) | [95], [117], [117], [217], [143], [150], [222], [218], [172] |
| P53 | DLEU1 (CDK1+), HOTAIR (PIK3R3+), HOTAIR (CCND1+), HOTAIR (CCND2+). HOTAIR (SIRT1+), HOTAIR (CHEK1+), HOXD-AS1 (PIK3R3+), LINC00511 (CDKN1+), NEAT1 (BCL2+), PCAT-1 (CCND1+), TDRG1 (Bcl-xL+), TUG1 (MDM2+) | [103], [115], [117], [117], [119], [223], [123], [217], [150], [218], [172], [177] |
| Wnt/β-catenin | CCAT1 (WNT1+), CCAT1 (TGFBR1+), CDKN2B-AS1 (SMAD3+), HOTAIR (CCND1+), HOTAIR (CCND2+), HOXD-AS1+, HOXD-AS1 (FZD4+), KCNQ1OT1+, MALAT1+, MIF-AS1 (PLCB1+), PCAT-1 (CCND1+), PCAT-1+, TTN-AS1 (ROCK2+) | [95], [97], [101], [117], [117], [121], [122], [224], [225], [144], [218], [160], [169] |
| TGF-β | CCAT1 (TGFBR1+), H19 (TGF-β+), HOTAIR (MAPK1+), LINC00161 (MAPK1+), LINC00339 (ROCK1+), MIR4435-2HG (TGF-β1+), NEAT1 (ROCK1+), TDRG1 (P70S6K+), ZFAS1 (Sp1+) | [97], [109], [114], [129], [131], [221], [151], [172], [183] |
| Notch | CCAT1 (ADAM17+), DLX6-AS1+, MALAT1 (NOTCH1+), TUG1 (NOTCH3+) | [95], [226], [204], [180] |
| Hippo | CCAT1 (WNT1+), CCAT1 (TGFBR1+), CCAT1 (BIRC5+), CDKN2B-AS1 (SMAD3+), FLVCR1-AS1 (YAP1+), H19 (TGF-β+), HOTAIR (CCND1+), HOTAIR (CCND2+), HOXD-AS1 (FZD4+), MALAT1 (YAP1+), MLK7-AS1 (YAP1+), MIR4435-2HG (TGF-β1+), PCAT-1 (CCND1+), PTAF (SNAI2+), UCA1 (YAP+) | [95], [97], [98], [101], [107], [109], [117], [117], [122], [203], [145], [221], [218], [161], [202] |
| RAS | CCAT1 (STAT3+), DANCR (IGF2+), HOTAIR (MAPK1+), LINC00161 (MAPK1+), NEAT1 (BCL2+), NORAD (STAT3+), TDRG1 (RhoC+), TINCR (FGF2+) | [95], [214], [114], [129], [150], [222], [172], [173] |
| LncRNA | Protein | Reference |
|---|---|---|
| Oncosuppressive lncRNAs and protein targets | ||
| ADAMTS9-AS2 | FOXF2 (forkhead-related transcription factor 2) | [72] |
| GAS5 | HOXA5 (homeobox protein Hox-A5) | [75] |
| WDFY3-AS2 | RORA (retinoid-related orphan receptor-alpha) | [84] |
| Oncogenic lncRNAs and protein targets | ||
| CCAT1 | ADAM17 (ADAM metallopeptidase domain 17), WNT1, STAT3, ZEB1 | [95] |
| CCAT1 | TGFBR1 | [97] |
| DLX6-AS1 | FHL2 (downregulated in rhabdomyosarcoma LIM protein) | [104] |
| DQ786243 | CREB1 (active transcription factor CREB) | [105] |
| FLVCR1-AS1 | YAP1 | [107] |
| H19 | TGF-β | [109] |
| HOTAIR | SNAIL | [118] |
| HOXD-AS1 | β-catenin, cyclin D1, c-Myc | [121] |
| HOXD-AS1 | PIK3R3 | [123] |
| LINC00963 | CHI3L1 (chitinase 3-like 1) | [135] |
| MALAT1 | KIF1B (kinesin family member 1B) | [205] |
| MALAT1 | YAP1 | [203] |
| MLK7-AS1 | YAP1 | [145] |
| NEAT1 | ZEB1 | [150] |
| OIP5-AS1 | ZNF217 (zinc finger protein 217) | [158] |
| PTAF | SNAI2 (SNAIL family transcriptional repressor 2) | [161] |
| PTAR | ZEB1 | [162] |
| PTAL | FN1 (fibronectin 1) | [163] |
| LncRNA-ROR | FLNB (filamin B) | [168] |
| TMPO-AS1 | TMEFF2 (transmembrane protein with EGF like and two follistatin-like domains) | [174] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Braga, E.A.; Fridman, M.V.; Moscovtsev, A.A.; Filippova, E.A.; Dmitriev, A.A.; Kushlinskii, N.E. LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: ceRNA and Alternative Mechanisms. Int. J. Mol. Sci. 2020, 21, 8855. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228855
Braga EA, Fridman MV, Moscovtsev AA, Filippova EA, Dmitriev AA, Kushlinskii NE. LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: ceRNA and Alternative Mechanisms. International Journal of Molecular Sciences. 2020; 21(22):8855. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228855
Chicago/Turabian StyleBraga, Eleonora A., Marina V. Fridman, Alexey A. Moscovtsev, Elena A. Filippova, Alexey A. Dmitriev, and Nikolay E. Kushlinskii. 2020. "LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: ceRNA and Alternative Mechanisms" International Journal of Molecular Sciences 21, no. 22: 8855. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228855





