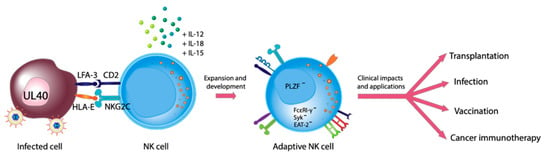
Deciphering the Immunological Phenomenon of Adaptive Natural Killer (NK) Cells and Cytomegalovirus (CMV)
Abstract
:1. Introduction
2. Biology of NK Cells
3. NK Cells and CMV
4. History of Adaptive NK Cells
5. Current Status on the Development of Adaptive NK Cells
6. Current Status on the Role of Adaptive NK Cells in Transplantation
7. Current Status on the Role of Adaptive NK Cells in Viral Infection
8. Current Status on the Role of Adaptive NK Cells in Vaccination
9. Current Status on the Role of Adaptive NK Cells in Cancer Immunotherapy
10. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALL | Acute lymphoblastic leukemia |
| AML | Acute myeloid leukemia |
| ADCC | Antibody-dependent cellular cytotoxicity |
| CMV | Cytomegalovirus |
| EBV | Epstein-Barr virus |
| HIV | Human immunodeficiency virus |
| HLA | Human leukocyte antigen |
| HSCT | Hematopoietic stem cell transplantation |
| HSV | Herpes simplex virus |
| IFNγ | Interferon gamma |
| IL | Interleukin |
| KIR | Killer immunoglobulin-like receptor |
| LIR | Leukocyte-immunoglobulin-like receptor |
| MCMV | Murine cytomegalovirus |
| NK | Natural killer cell |
| PLZF | Promyelocytic leukemia zinc finger |
| SOT | Solid organ transplantation |
| TNFα | Tumor necrosis factor |
| UCB | Umbilical cord blood |
References
- Huygens, A.; Dauby, N.; Vermijlen, D.; Marchant, A. Immunity to cytomegalovirus in early life. Front. Immunol. 2014, 5, 552. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, R.; Klein, E.; Pross, H.; Wigzell, H. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur. J. Immunol. 1975, 5, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Wilk, E.; Kalippke, K.; Buyny, S.; Schmidt, R.E.; Jacobs, R. New aspects of NK cell subset identification and inference of NK cells’ regulatory capacity by assessing functional and genomic profiles. Immunobiology (1979) 2008, 213, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Fauriat, C.; Long, E.O.; Ljunggren, H.-G.; Bryceson, Y.T. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood 2010, 115, 2167–2176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immunol. 2008, 9, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.M.; Yokoyama, W.M. Unifying concepts of MHC-dependent natural killer cell education. Trends Immunol. 2011, 32, 364–372. [Google Scholar] [CrossRef] [Green Version]
- Pegram, H.J.; Andrews, D.M.; Smyth, M.J.; Darcy, P.K.; Kershaw, M.H. Activating and inhibitory receptors of natural killer cells. Immunol. Cell Biol. 2011, 89, 216–224. [Google Scholar] [CrossRef]
- Boudreau, J.E.; Hsu, K.C. Natural Killer Cell Education and the Response to Infection and Cancer Therapy: Stay Tuned. Trends Immunol. 2018, 39, 222–239. [Google Scholar] [CrossRef]
- Zingoni, A.; Molfetta, R.; Fionda, C.; Soriani, A.; Paolini, R.; Cippitelli, M.; Cerboni, C.; Santoni, A. NKG2D and Its Ligands: “One for All, All for One”. Front. Immunol. 2018, 9, 476. [Google Scholar] [CrossRef]
- Barrow, A.D.; Martin, C.J.; Colonna, M. The Natural Cytotoxicity Receptors in Health and Disease. Front. Immunol. 2019, 10, 909. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Wang, G.; Huang, D.; Sui, M.; Xu, Y. Cancer Immunotherapy Based on Natural Killer Cells: Current Progress and New Opportunities. Front. Immunol. 2019, 10, 1205. [Google Scholar] [CrossRef] [PubMed]
- Parham, P. Taking license with natural killer cell maturation and repertoire development. Immunol. Rev. 2006, 214, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Brodin, P.; Jojic, V.; Gao, T.; Bhattacharya, S.; Angel, C.J.; Furman, D.; Shen-Orr, S.; Dekker, C.L.; Swan, G.E.; Butte, A.J.; et al. Variation in the human immune system is largely driven by non-heritable influences. Cell 2015, 160, 37–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knipe, D.M.; Howley, P. Fields Virology, 6th ed.; Wolters Kluwer: Philadelphia, PA, USA, 2013. [Google Scholar]
- Orange, J.S. Unraveling human natural killer cell deficiency. J. Clin. Investig. 2012, 122, 798–801. [Google Scholar] [CrossRef]
- Terrazzini, N.; Kern, F. Cell-mediated immunity to human CMV infection: A brief overview. F1000Prime Rep. 2014, 6, 28. [Google Scholar] [CrossRef]
- Manandhar, T.; Ho, G.T.; Pump, W.C.; Blasczyk, R.; Bade-Doeding, C. Battle between Host Immune Cellular Responses and HCMV Immune Evasion. Int. J. Mol. Sci. 2019, 20, 3626. [Google Scholar] [CrossRef] [Green Version]
- Kwangseog, A.; Ana, A.; Peter, G.; Per, A.P.; Young, Y.; Klaus, F. Human Cytomegalovirus Inhibits Antigen Presentation by a Sequential Multistep Process. Proc. Natl. Acad. Sci. USA 1996, 93, 10990–10995. [Google Scholar] [CrossRef] [Green Version]
- Lehner, P.J.; Hewitt, E.W.; Gupta, S.S. The human cytomegalovirus gene product US6 inhibits ATP binding by TAP. EMBO J. 2001, 20, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Kärre, K.; Welsh, R.M. Viral decoy vetoes killer cell. Nature 1997, 386, 446–447. [Google Scholar] [CrossRef]
- Chapman, T.L.; Heikema, A.P.; Bjorkman, P.J. The Inhibitory Receptor LIR-1 Uses a Common Binding Interaction to Recognize Class I MHC Molecules and the Viral Homolog UL18. Immunity 1999, 11, 603–613. [Google Scholar] [CrossRef] [Green Version]
- Dunn, C.; Chalupny, N.J.; Sutherland, C.L.; Dosch, S.; Sivakumar, P.V.; Johnson, D.C.; Cosman, D. Human Cytomegalovirus Glycoprotein UL16 Causes Intracellular Sequestration of NKG2D Ligands, Protecting Against Natural Killer Cell Cytotoxicity. J. Exp. Med. 2003, 197, 1427–1439. [Google Scholar] [CrossRef] [PubMed]
- Tomasec, P.; Braud, V.M.; Rickards, C.; Powell, M.B.; McSharry, B.P.; Gadola, S.; Cerundolo, V.; Borysiewicz, L.K.; McMichael, A.J.; Wilkinson, G.W. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2000, 287, 1031. [Google Scholar] [CrossRef] [PubMed]
- Heatley, S.L.; Pietra, G.; Lin, J.; Widjaja, J.M.L.; Harpur, C.M.; Lester, S.; Rossjohn, J.; Szer, J.; Schwarer, A.; Bradstock, K.; et al. Polymorphism in human cytomegalovirus UL40 impacts on recognition of human leukocyte antigen-E (HLA-E) by natural killer cells. J. Biol. Chem. 2013, 288, 8679–8690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulbrecht, M.; Martinozzi, S.; Grzeschik, M.; Hengel, H.; Ellwart, J.W.; Pla, M.; Weiss, E.H. Cutting Edge: The Human Cytomegalovirus UL40 Gene Product Contains a Ligand for HLA-E and Prevents NK Cell-Mediated Lysis. J. Immunol. 2000, 164, 5019–5022. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.C.; Beilke, J.N.; Lanier, L.L. Adaptive immune features of natural killer cells. Nature 2009, 457, 557–561. [Google Scholar] [CrossRef]
- Guma, M.; Angulo, A.; Vilches, C.; Gomez-Lozano, N.; Malats, N.; Lopez-Botet, M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 2004, 104, 3664–3671. [Google Scholar] [CrossRef] [Green Version]
- Guma, M.; Budt, M.; Saez, A.; Brckalo, T.; Hengel, H.; Angulo, A.; Lopez-Botet, M. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood 2006, 107, 3624–3631. [Google Scholar] [CrossRef] [Green Version]
- Beziat, V.; Liu, L.L.; Malmberg, J.A.; Ivarsson, M.A.; Sohlberg, E.; Bjorklund, A.T.; Retiere, C.; Sverremark-Ekstrom, E.; Traherne, J.; Ljungman, P.; et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 2013, 121, 2678–2688. [Google Scholar] [CrossRef]
- Della Chiesa, M.; Falco, M.; Podesta, M.; Locatelli, F.; Moretta, L.; Frassoni, F.; Moretta, A. Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation: A role for human cytomegalovirus? Blood 2012, 119, 399–410. [Google Scholar] [CrossRef]
- Foley, B.; Cooley, S.; Verneris, M.R.; Curtsinger, J.; Luo, X.; Waller, E.K.; Anasetti, C.; Weisdorf, D.; Miller, J.S. Human cytomegalovirus (CMV)-induced memory-like NKG2C(+) NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J. Immunol. 2012, 189, 5082–5088. [Google Scholar] [CrossRef] [Green Version]
- Foley, B.; Cooley, S.; Verneris, M.R.; Pitt, M.; Curtsinger, J.; Luo, X.; Lopez-Verges, S.; Lanier, L.L.; Weisdorf, D.; Miller, J.S. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 2012, 119, 2665–2674. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Verges, S.; Milush, J.M.; Schwartz, B.S.; Pando, M.J.; Jarjoura, J.; York, V.A.; Houchins, J.P.; Miller, S.; Kang, S.M.; Norris, P.J.; et al. Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 14725–14732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luetke-Eversloh, M.; Hammer, Q.; Durek, P.; Nordstrom, K.; Gasparoni, G.; Pink, M.; Hamann, A.; Walter, J.; Chang, H.D.; Dong, J.; et al. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog. 2014, 10, e1004441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlums, H.; Cichocki, F.; Tesi, B.; Theorell, J.; Beziat, V.; Holmes, T.D.; Han, H.; Chiang, S.C.; Foley, B.; Mattsson, K.; et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 2015, 42, 443–456. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Verges, S.; Milush, J.M.; Pandey, S.; York, V.A.; Arakawa-Hoyt, J.; Pircher, H.; Norris, P.J.; Nixon, D.F.; Lanier, L.L. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood 2010, 116, 3865–3874. [Google Scholar] [CrossRef] [Green Version]
- Foley, B.; Cooley, S.; Verneris, M.R.; Curtsinger, J.; Luo, X.; Waller, E.K.; Weisdorf, D.J.; Miller, J.S. NK cell education after allogeneic transplantation: Dissociation between recovery of cytokine-producing and cytotoxic functions. Blood 2011, 118, 2784–2792. [Google Scholar] [CrossRef] [Green Version]
- Comeau, E.M.; Holder, K.A.; Fudge, N.J.; Grant, M.D. Cytomegalovirus-Driven Adaption of Natural Killer Cells in NKG2C(null) Human Immunodeficiency Virus-Infected Individuals. Viruses 2019, 11, 239. [Google Scholar] [CrossRef] [Green Version]
- Della Chiesa, M.; Falco, M.; Bertaina, A.; Muccio, L.; Alicata, C.; Frassoni, F.; Locatelli, F.; Moretta, L.; Moretta, A. Human cytomegalovirus infection promotes rapid maturation of NK cells expressing activating killer Ig-like receptor in patients transplanted with NKG2C-/- umbilical cord blood. J. Immunol. 2014, 192, 1471–1479. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.L.; Landskron, J.; Ask, E.H.; Enqvist, M.; Sohlberg, E.; Traherne, J.A.; Hammer, Q.; Goodridge, J.P.; Larsson, S.; Jayaraman, J.; et al. Critical Role of CD2 Co-stimulation in Adaptive Natural Killer Cell Responses Revealed in NKG2C-Deficient Humans. Cell Rep. 2016, 15, 1088–1099. [Google Scholar] [CrossRef] [Green Version]
- Muntasell, A.; Pupuleku, A.; Cisneros, E.; Vera, A.; Moraru, M.; Vilches, C.; Lopez-Botet, M. Relationship of NKG2C Copy Number with the Distribution of Distinct Cytomegalovirus-Induced Adaptive NK Cell Subsets. J. Immunol. 2016, 196, 3818–3827. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Zhang, T.; Hwang, I.; Kim, A.; Nitschke, L.; Kim, M.; Scott, J.M.; Kamimura, Y.; Lanier, L.L.; Kim, S. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 2015, 42, 431–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Soderstrom, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Q.; Ruckert, T.; Borst, E.M.; Dunst, J.; Haubner, A.; Durek, P.; Heinrich, F.; Gasparoni, G.; Babic, M.; Tomic, A.; et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 2018, 19, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Rolle, A.; Meyer, M.; Calderazzo, S.; Jager, D.; Momburg, F. Distinct HLA-E Peptide Complexes Modify Antibody-Driven Effector Functions of Adaptive NK Cells. Cell Rep. 2018, 24, 1967–1976.e1964. [Google Scholar] [CrossRef] [Green Version]
- Romee, R.; Rosario, M.; Berrien-Elliott, M.M.; Wagner, J.A.; Jewell, B.A.; Schappe, T.; Leong, J.W.; Abdel-Latif, S.; Schneider, S.E.; Willey, S.; et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci. Transl. Med. 2016, 8, 357ra123. [Google Scholar] [CrossRef] [Green Version]
- La Rosa, C.; Diamond, D.J. The immune response to human CMV. Future Virol. 2012, 7, 279–293. [Google Scholar] [CrossRef] [Green Version]
- Elmaagacli, A.H.; Steckel, N.K.; Koldehoff, M.; Hegerfeldt, Y.; Trenschel, R.; Ditschkowski, M.; Christoph, S.; Gromke, T.; Kordelas, L.; Ottinger, H.D.; et al. Early human cytomegalovirus replication after transplantation is associated with a decreased relapse risk: Evidence for a putative virus-versus-leukemia effect in acute myeloid leukemia patients. Blood 2011, 118, 1402–1412. [Google Scholar] [CrossRef] [Green Version]
- Elmaagacli, A.H.; Koldehoff, M. Cytomegalovirus replication reduces the relapse incidence in patients with acute myeloid leukemia. Blood 2016, 128, 456–459. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, J.; Noguchi, M.; Kurauchi, K.; Tanioka, S.; Fukano, R.; Okamura, J. Effect of Cytomegalovirus Reactivation on Relapse after Allogeneic Hematopoietic Stem Cell Transplantation in Pediatric Acute Leukemia. Biol. Blood Marrow Transpl. 2016, 22, 300–306. [Google Scholar] [CrossRef] [Green Version]
- Manjappa, S.; Bhamidipati, P.K.; Stokerl-Goldstein, K.E.; DiPersio, J.F.; Uy, G.L.; Westervelt, P.; Liu, J.; Schroeder, M.A.; Vij, R.; Abboud, C.N.; et al. Protective effect of cytomegalovirus reactivation on relapse after allogeneic hematopoietic cell transplantation in acute myeloid leukemia patients is influenced by conditioning regimen. Biol. Blood Marrow Transpl. 2014, 20, 46–52. [Google Scholar] [CrossRef] [Green Version]
- Peric, Z.; Wilson, J.; Durakovic, N.; Ostojic, A.; Desnica, L.; Vranjes, V.R.; Marekovic, I.; Serventi-Seiwerth, R.; Vrhovac, R. Early human cytomegalovirus reactivation is associated with lower incidence of relapse of myeloproliferative disorders after allogeneic hematopoietic stem cell transplantation. Bone Marrow Transpl. 2018, 53, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Jeljeli, M.; Guerin-El Khourouj, V.; Porcher, R.; Fahd, M.; Leveille, S.; Yakouben, K.; Ouachee-Chardin, M.; LeGoff, J.; Cordeiro, D.J.; Pedron, B.; et al. Relationship between cytomegalovirus (CMV) reactivation, CMV-driven immunity, overall immune recovery and graft-versus-leukemia effect in children. Br. J. Haematol. 2014, 166, 229–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariotti, J.; Maura, F.; Spina, F.; Roncari, L.; Dodero, A.; Farina, L.; Montefusco, V.; Carniti, C.; Sarina, B.; Patriarca, F.; et al. Impact of cytomegalovirus replication and cytomegalovirus serostatus on the outcome of patients with B cell lymphoma after allogeneic stem cell transplantation. Biol. Blood Marrow Transpl. 2014, 20, 885–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanada, M.; Yamamoto, K.; Emi, N.; Naoe, T.; Suzuki, R.; Taji, H.; Iida, H.; Shimokawa, T.; Kohno, A.; Mizuta, S.; et al. Cytomegalovirus antigenemia and outcome of patients treated with pre-emptive ganciclovir: Retrospective analysis of 241 consecutive patients undergoing allogeneic hematopoietic stem cell transplantation. Bone Marrow Transpl. 2003, 32, 801–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teira, P.; Battiwalla, M.; Ramanathan, M.; Barrett, A.J.; Ahn, K.W.; Chen, M.; Green, J.S.; Saad, A.; Antin, J.H.; Savani, B.N.; et al. Early cytomegalovirus reactivation remains associated with increased transplant-related mortality in the current era: A CIBMTR analysis. Blood 2016, 127, 2427–2438. [Google Scholar] [CrossRef]
- Cichocki, F.; Cooley, S.; Davis, Z.; DeFor, T.E.; Schlums, H.; Zhang, B.; Brunstein, C.G.; Blazar, B.R.; Wagner, J.; Diamond, D.J.; et al. CD56dimCD57+NKG2C+ NK cell expansion is associated with reduced leukemia relapse after reduced intensity HCT. Leukemia 2016, 30, 456–463. [Google Scholar] [CrossRef]
- Cichocki, F.; Taras, E.; Chiuppesi, F.; Wagner, J.E.; Blazar, B.R.; Brunstein, C.; Luo, X.; Diamond, D.J.; Cooley, S.; Weisdorf, D.J.; et al. Adaptive NK cell reconstitution is associated with better clinical outcomes. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Ruggeri, L.; Capanni, M.; Mancusi, A.; Urbani, E.; Perruccio, K.; Burchielli, E.; Tosti, A.; Topini, F.; Aversa, F.; Martelli, M.F.; et al. Alloreactive natural killer cells in mismatched hematopoietic stem cell transplantation. Blood Cells Mol. Dis. 2004, 33, 216–221. [Google Scholar] [CrossRef]
- Park, K.H.; Ryu, J.H.; Bae, H.; Yun, S.; Jang, J.H.; Han, K.; Cho, B.S.; Kim, H.J.; Lee, H.; Oh, E.J. Delayed NK Cell Reconstitution and Reduced NK Activity Increased the Risks of CMV Disease in Allogeneic-Hematopoietic Stem Cell Transplantation. Int. J. Mol. Sci. 2020, 21, 3663. [Google Scholar] [CrossRef]
- Ataya, M.; Redondo-Pachon, D.; Llinas-Mallol, L.; Yelamos, J.; Heredia, G.; Perez-Saez, M.J.; Vila, J.; Costa-Garcia, M.; Raich-Regue, D.; Vilches, C.; et al. Pretransplant adaptive NKG2C+ NK cells protect against cytomegalovirus infection in kidney transplant recipients. Am. J. Transpl. 2020, 20, 663–676. [Google Scholar] [CrossRef]
- Kuijpers, T.W.; Baars, P.A.; Dantin, C.; van den Burg, M.; van Lier, R.A.; Roosnek, E. Human NK cells can control CMV infection in the absence of T cells. Blood 2008, 112, 914–915. [Google Scholar] [CrossRef] [Green Version]
- Miyashita, R.; Tsuchiya, N.; Hikami, K.; Kuroki, K.; Fukazawa, T.; Bijl, M.; Kallenberg, C.G.; Hashimoto, H.; Yabe, T.; Tokunaga, K. Molecular genetic analyses of human NKG2C (KLRC2) gene deletion. Int. Immunol. 2004, 16, 163–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moraru, M.; Cisneros, E.; Gomez-Lozano, N.; de Pablo, R.; Portero, F.; Canizares, M.; Vaquero, M.; Roustan, G.; Millan, I.; Lopez-Botet, M.; et al. Host genetic factors in susceptibility to herpes simplex type 1 virus infection: Contribution of polymorphic genes at the interface of innate and adaptive immunity. J. Immunol. 2012, 188, 4412–4420. [Google Scholar] [CrossRef] [Green Version]
- Muntasell, A.; Lopez-Montanes, M.; Vera, A.; Heredia, G.; Romo, N.; Penafiel, J.; Moraru, M.; Vila, J.; Vilches, C.; Lopez-Botet, M. NKG2C zygosity influences CD94/NKG2C receptor function and the NK-cell compartment redistribution in response to human cytomegalovirus. Eur. J. Immunol. 2013, 43, 3268–3278. [Google Scholar] [CrossRef] [PubMed]
- Noyola, D.E.; Fortuny, C.; Muntasell, A.; Noguera-Julian, A.; Munoz-Almagro, C.; Alarcon, A.; Juncosa, T.; Moraru, M.; Vilches, C.; Lopez-Botet, M. Influence of congenital human cytomegalovirus infection and the NKG2C genotype on NK-cell subset distribution in children. Eur. J. Immunol. 2012, 42, 3256–3266. [Google Scholar] [CrossRef] [PubMed]
- Kared, H.; Martelli, S.; Tan, S.W.; Simoni, Y.; Chong, M.L.; Yap, S.H.; Newell, E.W.; Pender, S.L.F.; Kamarulzaman, A.; Rajasuriar, R.; et al. Adaptive NKG2C(+)CD57(+) Natural Killer Cell and Tim-3 Expression During Viral Infections. Front. Immunol. 2018, 9, 686. [Google Scholar] [CrossRef]
- Mahapatra, S.; Shearer, W.T.; Minard, C.G.; Mace, E.; Paul, M.; Orange, J.S. NK cells in treated HIV-infected children display altered phenotype and function. J. Allergy Clin. Immunol. 2019, 144, 294–303.e213. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Amran, F.S.; Kramski, M.; Angelovich, T.A.; Elliott, J.; Hearps, A.C.; Price, P.; Jaworowski, A. An NK Cell Population Lacking FcRgamma Is Expanded in Chronically Infected HIV Patients. J. Immunol. 2015, 194, 4688–4697. [Google Scholar] [CrossRef] [Green Version]
- Guma, M.; Cabrera, C.; Erkizia, I.; Bofill, M.; Clotet, B.; Ruiz, L.; Lopez-Botet, M. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J. Infect. Dis. 2006, 194, 38–41. [Google Scholar] [CrossRef]
- Mela, C.M.; Burton, C.T.; Imami, N.; Nelson, M.; Steel, A.; Gazzard, B.G.; Gotch, F.M.; Goodier, M.R. Switch from inhibitory to activating NKG2 receptor expression in HIV-1 infection: Lack of reversion with highly active antiretroviral therapy. AIDS 2005, 19, 1761–1769. [Google Scholar] [CrossRef]
- Ma, M.; Wang, Z.; Chen, X.; Tao, A.; He, L.; Fu, S.; Zhang, Z.; Fu, Y.; Guo, C.; Liu, J.; et al. NKG2C(+)NKG2A(-) Natural Killer Cells are Associated with a Lower Viral Set Point and may Predict Disease Progression in Individuals with Primary HIV Infection. Front. Immunol. 2017, 8, 1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gondois-Rey, F.; Cheret, A.; Granjeaud, S.; Mallet, F.; Bidaut, G.; Lecuroux, C.; Ploquin, M.; Muller-Trutwin, M.; Rouzioux, C.; Avettand-Fenoel, V.; et al. NKG2C(+) memory-like NK cells contribute to the control of HIV viremia during primary infection: Optiprim-ANRS 147. Clin. Transl. Immunol. 2017, 6, e150. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Low, H.Z.; Kniesch, K.; Jacobs, R.; Schmidt, R.E.; Witte, T. NKG2C deletion is a risk factor of HIV infection. AIDS Res. Hum. Retrovir. 2012, 28, 844–851. [Google Scholar] [CrossRef] [Green Version]
- Bjorkstrom, N.K.; Lindgren, T.; Stoltz, M.; Fauriat, C.; Braun, M.; Evander, M.; Michaelsson, J.; Malmberg, K.J.; Klingstrom, J.; Ahlm, C.; et al. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J. Exp. Med. 2011, 208, 13–21. [Google Scholar] [CrossRef]
- Petitdemange, C.; Becquart, P.; Wauquier, N.; Beziat, V.; Debre, P.; Leroy, E.M.; Vieillard, V. Unconventional repertoire profile is imprinted during acute chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS Pathog. 2011, 7, e1002268. [Google Scholar] [CrossRef] [Green Version]
- Petitdemange, C.; Wauquier, N.; Devilliers, H.; Yssel, H.; Mombo, I.; Caron, M.; Nkoghe, D.; Debre, P.; Leroy, E.; Vieillard, V. Longitudinal Analysis of Natural Killer Cells in Dengue Virus-Infected Patients in Comparison to Chikungunya and Chikungunya/Dengue Virus-Infected Patients. PLoS Negl. Trop. Dis. 2016, 10, e0004499. [Google Scholar] [CrossRef] [Green Version]
- Malone, D.F.G.; Lunemann, S.; Hengst, J.; Ljunggren, H.G.; Manns, M.P.; Sandberg, J.K.; Cornberg, M.; Wedemeyer, H.; Bjorkstrom, N.K. Cytomegalovirus-Driven Adaptive-Like Natural Killer Cell Expansions Are Unaffected by Concurrent Chronic Hepatitis Virus Infections. Front. Immunol. 2017, 8, 525. [Google Scholar] [CrossRef] [Green Version]
- Saghafian-Hedengren, S.; Sohlberg, E.; Theorell, J.; Carvalho-Queiroz, C.; Nagy, N.; Persson, J.O.; Nilsson, C.; Bryceson, Y.T.; Sverremark-Ekstrom, E. Epstein-Barr virus coinfection in children boosts cytomegalovirus-induced differentiation of natural killer cells. J. Virol. 2013, 87, 13446–13455. [Google Scholar] [CrossRef] [Green Version]
- Hendricks, D.W.; Balfour, H.H., Jr.; Dunmire, S.K.; Schmeling, D.O.; Hogquist, K.A.; Lanier, L.L. Cutting edge: NKG2C(hi)CD57+ NK cells respond specifically to acute infection with cytomegalovirus and not Epstein-Barr virus. J. Immunol. 2014, 192, 4492–4496. [Google Scholar] [CrossRef] [Green Version]
- Hart, G.T.; Tran, T.M.; Theorell, J.; Schlums, H.; Arora, G.; Rajagopalan, S.; Sangala, A.D.J.; Welsh, K.J.; Traore, B.; Pierce, S.K.; et al. Adaptive NK cells in people exposed to Plasmodium falciparum correlate with protection from malaria. J. Exp. Med. 2019, 216, 1280–1290. [Google Scholar] [CrossRef] [Green Version]
- Draghi, M.; Pashine, A.; Sanjanwala, B.; Gendzekhadze, K.; Cantoni, C.; Cosman, D.; Moretta, A.; Valiante, N.M.; Parham, P. NKp46 and NKG2D recognition of infected dendritic cells is necessary for NK cell activation in the human response to influenza infection. J. Immunol. 2007, 178, 2688–2698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maucourant, C.; Filipovic, I.; Ponzetta, A.; Aleman, S.; Cornillet, M.; Hertwig, L.; Strunz, B.; Lentini, A.; Reinius, B.; Brownlie, D.; et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, A.; Behrens, R.H.; Okell, L.; Fooks, A.R.; Riley, E.M. NK cells as effectors of acquired immune responses: Effector CD4+ T cell-dependent activation of NK cells following vaccination. J. Immunol. 2010, 185, 2808–2818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, C.M.; White, M.J.; Bottomley, C.; Lusa, C.; Rodriguez-Galan, A.; Turner, S.E.; Goodier, M.R.; Riley, E.M. Impaired NK Cell Responses to Pertussis and H1N1 Influenza Vaccine Antigens in Human Cytomegalovirus-Infected Individuals. J. Immunol. 2015, 194, 4657–4667. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, A.; Hafalla, J.C.; King, E.; Lusingu, J.; Dekker, D.; Leach, A.; Moris, P.; Cohen, J.; Vekemans, J.; Villafana, T.; et al. Antigen-specific IL-2 secretion correlates with NK cell responses after immunization of Tanzanian children with the RTS,S/AS01 malaria vaccine. J. Immunol. 2012, 188, 5054–5062. [Google Scholar] [CrossRef] [PubMed]
- White, M.J.; Nielsen, C.M.; McGregor, R.H.; Riley, E.H.; Goodier, M.R. Differential activation of CD57-defined natural killer cell subsets during recall responses to vaccine antigens. Immunology 2014, 142, 140–150. [Google Scholar] [CrossRef]
- Riese, P.; Trittel, S.; Pathirana, R.D.; Klawonn, F.; Cox, R.J.; Guzman, C.A. Responsiveness to Influenza Vaccination Correlates with NKG2C-Expression on NK Cells. Vaccines (Basel) 2020, 8, 281. [Google Scholar] [CrossRef]
- Wagstaffe, H.R.; Clutterbuck, E.A.; Bockstal, V.; Stoop, J.N.; Luhn, K.; Douoguih, M.; Shukarev, G.; Snape, M.D.; Pollard, A.J.; Riley, E.M.; et al. Antibody-Dependent Natural Killer Cell Activation after Ebola Vaccination. J. Infect. Dis. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Darboe, A.; Danso, E.; Clarke, E.; Umesi, A.; Touray, E.; Wegmuller, R.; Moore, S.E.; Riley, E.M.; Goodier, M.R. Enhancement of cytokine-driven NK cell IFN-gamma production after vaccination of HCMV infected Africans. Eur. J. Immunol. 2017, 47, 1040–1050. [Google Scholar] [CrossRef] [Green Version]
- Goodier, M.R.; Rodriguez-Galan, A.; Lusa, C.; Nielsen, C.M.; Darboe, A.; Moldoveanu, A.L.; White, M.J.; Behrens, R.; Riley, E.M. Influenza Vaccination Generates Cytokine-Induced Memory-like NK Cells: Impact of Human Cytomegalovirus Infection. J. Immunol. 2016, 197, 313–325. [Google Scholar] [CrossRef] [Green Version]
- Goodier, M.R.; Jonjic, S.; Riley, E.M.; Juranic Lisnic, V. CMV and natural killer cells: Shaping the response to vaccination. Eur. J. Immunol. 2018, 48, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, N.; Ivarsson, M.A.; Blom, K.; Gonzalez, V.D.; Braun, M.; Falconer, K.; Gustafsson, R.; Fogdell-Hahn, A.; Sandberg, J.K.; Michaelsson, J. The Human NK Cell Response to Yellow Fever Virus 17D Is Primarily Governed by NK Cell Differentiation Independently of NK Cell Education. J. Immunol. 2015, 195, 3262–3272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suliman, S.; Geldenhuys, H.; Johnson, J.L.; Hughes, J.E.; Smit, E.; Murphy, M.; Toefy, A.; Lerumo, L.; Hopley, C.; Pienaar, B.; et al. Bacillus Calmette-Guerin (BCG) Revaccination of Adults with Latent Mycobacterium tuberculosis Infection Induces Long-Lived BCG-Reactive NK Cell Responses. J. Immunol. 2016, 197, 1100–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Scott, J.M.; Hwang, I.; Kim, S. Cutting edge: Antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. J. Immunol. 2013, 190, 1402–1406. [Google Scholar] [CrossRef] [Green Version]
- Wiernik, A.; Foley, B.; Zhang, B.; Verneris, M.R.; Warlick, E.; Gleason, M.K.; Ross, J.A.; Luo, X.; Weisdorf, D.J.; Walcheck, B.; et al. Targeting natural killer cells to acute myeloid leukemia in vitro with a CD16 x 33 bispecific killer cell engager and ADAM17 inhibition. Clin. Cancer Res. 2013, 19, 3844–3855. [Google Scholar] [CrossRef] [Green Version]
- Cichocki, F.; Wu, C.Y.; Zhang, B.; Felices, M.; Tesi, B.; Tuininga, K.; Dougherty, P.; Taras, E.; Hinderlie, P.; Blazar, B.R.; et al. ARID5B regulates metabolic programming in human adaptive NK cells. J. Exp. Med. 2018, 215, 2379–2395. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.L.; Beziat, V.; Oei, V.Y.S.; Pfefferle, A.; Schaffer, M.; Lehmann, S.; Hellstrom-Lindberg, E.; Soderhall, S.; Heyman, M.; Grander, D.; et al. Ex Vivo Expanded Adaptive NK Cells Effectively Kill Primary Acute Lymphoblastic Leukemia Cells. Cancer Immunol. Res. 2017, 5, 654–665. [Google Scholar] [CrossRef] [Green Version]
- Torelli, G.F.; Peragine, N.; Raponi, S.; Pagliara, D.; De Propris, M.S.; Vitale, A.; Bertaina, A.; Barberi, W.; Moretta, L.; Basso, G.; et al. Recognition of adult and pediatric acute lymphoblastic leukemia blasts by natural killer cells. Haematologica 2014, 99, 1248–1254. [Google Scholar] [CrossRef] [Green Version]
- Foley, B.; Ta, C.; Barnes, S.; de Jong, E.; Nguyen, M.; Cheung, L.C.; Buzzai, A.; Wagner, T.; Wylie, B.; Fernandez, S.; et al. Identifying the optimal donor for natural killer cell adoptive therapy to treat pediatric B- and T-cell acute lymphoblastic leukemia. Clin. Transl. Immunol. 2020, 9, e1151. [Google Scholar] [CrossRef]
- Cichocki, F.; Valamehr, B.; Bjordahl, R.; Zhang, B.; Rezner, B.; Rogers, P.; Gaidarova, S.; Moreno, S.; Tuininga, K.; Dougherty, P.; et al. GSK3 Inhibition Drives Maturation of NK Cells and Enhances Their Antitumor Activity. Cancer Res. 2017, 77, 5664–5675. [Google Scholar] [CrossRef] [Green Version]
- Augello, G.; Emma, M.R.; Cusimano, A.; Azzolina, A.; Montalto, G.; McCubrey, J.A.; Cervello, M. The Role of GSK-3 in Cancer Immunotherapy: GSK-3 Inhibitors as a New Frontier in Cancer Treatment. Cells 2020, 9, 1427. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barnes, S.; Schilizzi, O.; Audsley, K.M.; Newnes, H.V.; Foley, B. Deciphering the Immunological Phenomenon of Adaptive Natural Killer (NK) Cells and Cytomegalovirus (CMV). Int. J. Mol. Sci. 2020, 21, 8864. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228864
Barnes S, Schilizzi O, Audsley KM, Newnes HV, Foley B. Deciphering the Immunological Phenomenon of Adaptive Natural Killer (NK) Cells and Cytomegalovirus (CMV). International Journal of Molecular Sciences. 2020; 21(22):8864. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228864
Chicago/Turabian StyleBarnes, Samantha, Ophelia Schilizzi, Katherine M. Audsley, Hannah V. Newnes, and Bree Foley. 2020. "Deciphering the Immunological Phenomenon of Adaptive Natural Killer (NK) Cells and Cytomegalovirus (CMV)" International Journal of Molecular Sciences 21, no. 22: 8864. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228864