A Celecoxib Derivative Eradicates Antibiotic-Resistant Staphylococcus aureus and Biofilms by Targeting YidC2 Translocase
Abstract
:1. Introduction
2. Results
2.1. S. aureus Isolates with Resistance to Celecoxib-Derived Antibacterial Agents All Have Missense Mutations at yidC2
2.2. YidC2-Overexpressing Staphylococci are Less Susceptible to Cpd36 and Cpd46
2.3. Cpd36 and Cpd46 Attenuate the Stress Tolerance of S. aureus
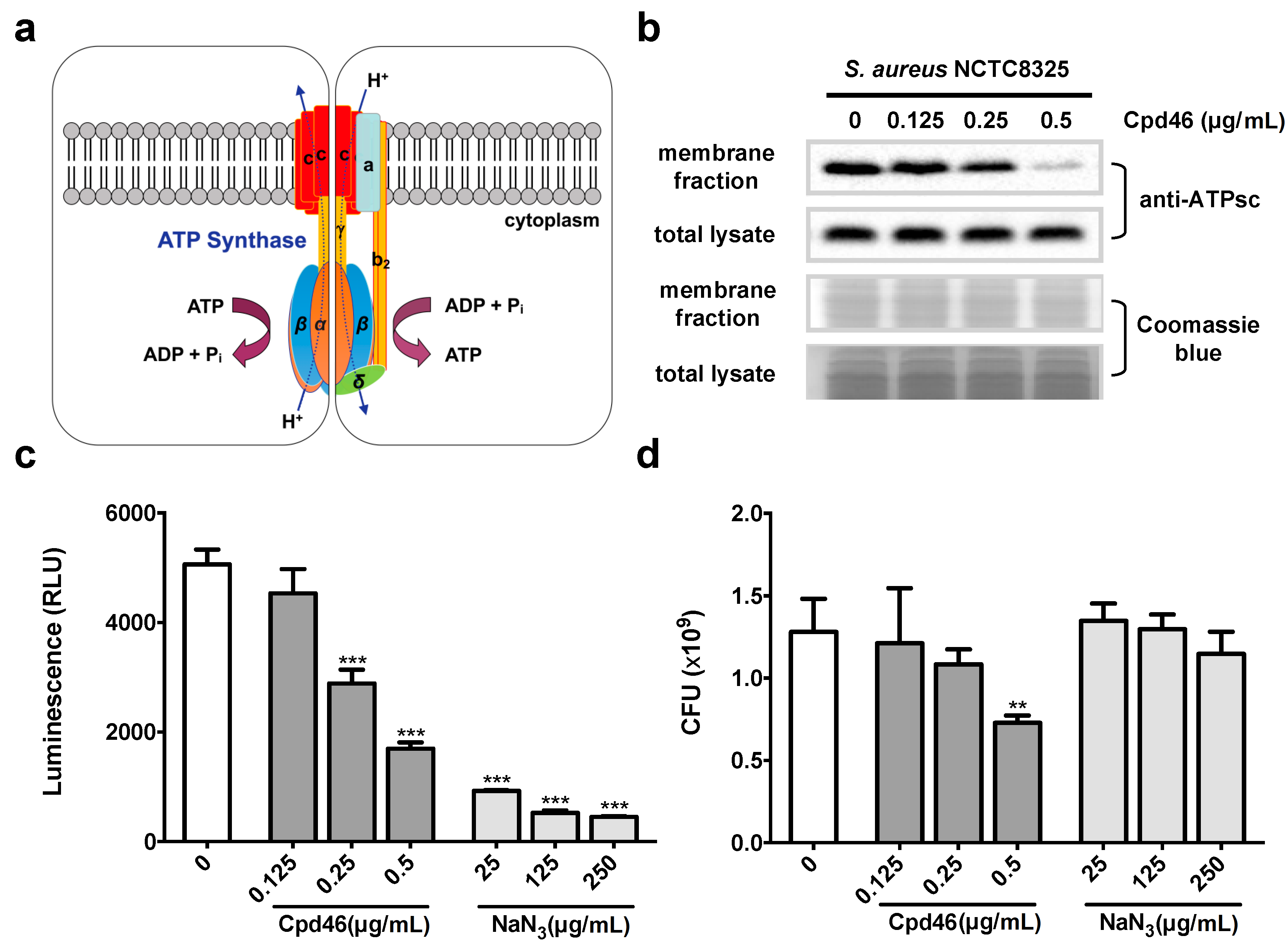
2.4. Cpd46 Blocks Membrane Insertion of FoF1 ATP Synthase Subunit C, Leading to Reduced ATP Production
2.5. Cpd36 and Cpd46 Enhance the Thermal Stability of YidC2
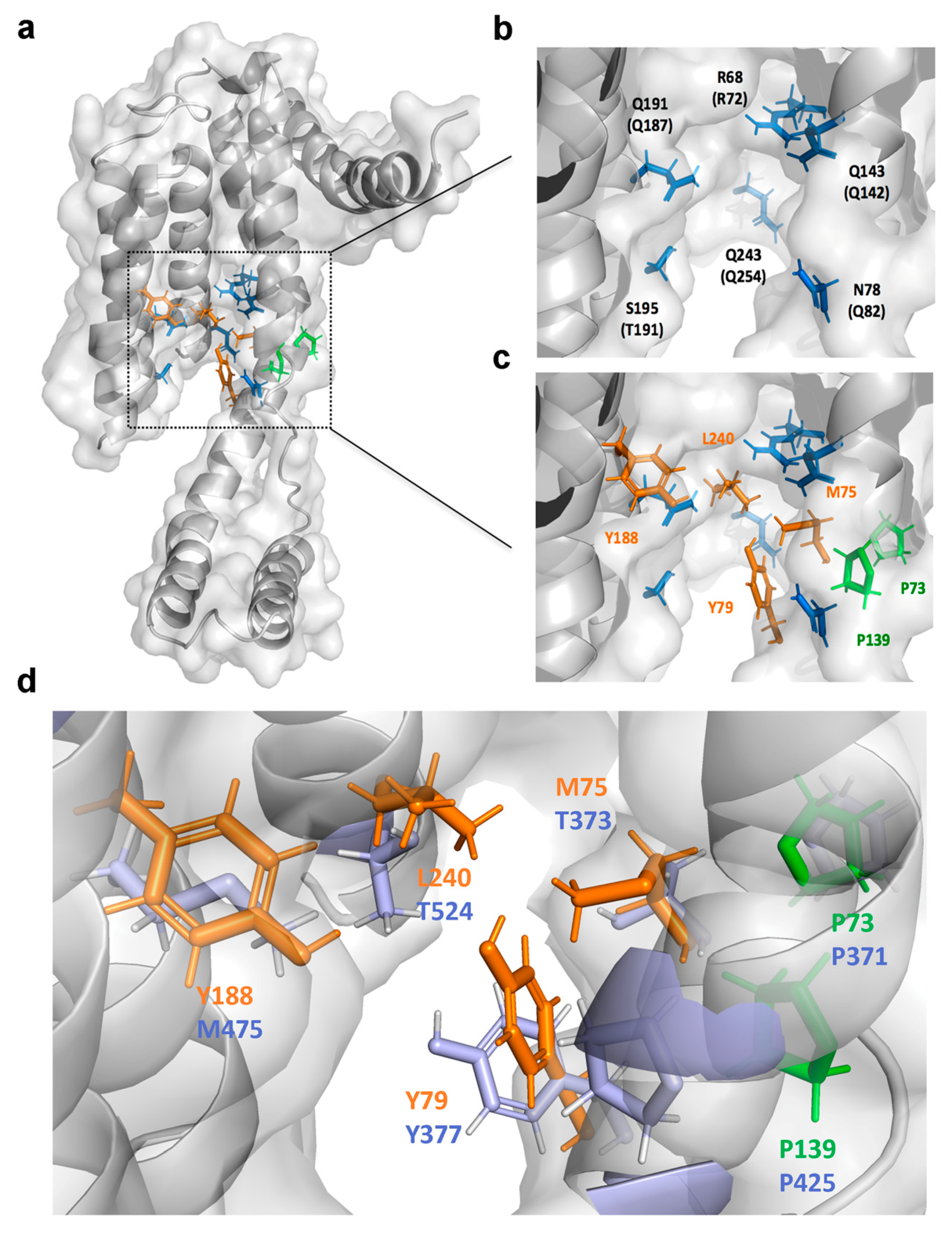
2.6. Identification of the Drug-Interacting Motif on YidC2 via Homology Modeling and Mutational Analyses
2.7. Cpd46 Effectively Eradicates MRSA Persisters and Biofilms
2.8. Cpd36 and Cpd46 are Active against Other Gram-Positive Bacteria
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Reagents and Antibodies
4.3. Antibacterial Assay
4.4. Isolation of Drug-Resistant S. aureus
4.5. Genomic Mutations Identification
4.6. Identification of YidC2 Orthologue
4.7. Thermal Shift Assay
4.8. Isolation of Bacterial Membrane Proteins
4.9. Intracellular ATP Measurement
4.10. Surface Plasmon Resonance (SPR) Analysis
4.11. Persisters Killing Assay
4.12. Biofilm Eradication Assay
4.13. Quantification and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MRSA | methicillin-resistant S. aureus |
| ATPsc | ATP synthase subunit c |
| Tm | melting temperature |
| MIC | minimal inhibitory concentration |
References
- Samuelson, J.C.; Chen, M.; Jiang, F.; Moller, I.; Wiedmann, M.; Kuhn, A.; Phillips, G.J.; Dalbey, R.E. YidC mediates membrane protein insertion in bacteria. Nature 2000, 406, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, A.; Koch, H.G.; Dalbey, R.E. Targeting and Insertion of Membrane Proteins. EcoSal Plus 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Jiang, F.; Chen, M.; Cain, B.; Bolhuis, A.; Dalbey, R.E. YidC is strictly required for membrane insertion of subunits a and c of the F(1)F(0)ATP synthase and SecE of the SecYEG translocase. Biochemistry 2003, 42, 10537–10544. [Google Scholar] [CrossRef] [PubMed]
- Du Plessis, D.J.; Nouwen, N.; Driessen, A.J. Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem. 2006, 281, 12248–12252. [Google Scholar] [CrossRef] [Green Version]
- Price, C.E.; Driessen, A.J. Conserved negative charges in the transmembrane segments of subunit K of the NADH:ubiquinone oxidoreductase determine its dependence on YidC for membrane insertion. J. Biol. Chem. 2010, 285, 3575–3581. [Google Scholar] [CrossRef] [Green Version]
- Van Bloois, E.; Nagamori, S.; Koningstein, G.; Ullers, R.S.; Preuss, M.; Oudega, B.; Harms, N.; Kaback, H.R.; Herrmann, J.M.; Luirink, J. The Sec-independent function of Escherichia coli YidC is evolutionary-conserved and essential. J. Biol. Chem. 2005, 280, 12996–13003. [Google Scholar] [CrossRef] [Green Version]
- Funes, S.; Kauff, F.; van der Sluis, E.O.; Ott, M.; Herrmann, J.M. Evolution of YidC/Oxa1/Alb3 insertases: Three independent gene duplications followed by functional specialization in bacteria, mitochondria and chloroplasts. Biol. Chem. 2011, 392, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Palmer, S.R.; Crowley, P.J.; Oli, M.W.; Ruelf, M.A.; Michalek, S.M.; Brady, L.J. YidC1 and YidC2 are functionally distinct proteins involved in protein secretion, biofilm formation and cariogenicity of Streptococcus mutans. Microbiology 2012, 158, 1702–1712. [Google Scholar] [CrossRef] [Green Version]
- Chiba, S.; Lamsa, A.; Pogliano, K. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 2009, 28, 3461–3475. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Palmer, S.R.; Hasona, A.; Nagamori, S.; Kaback, H.R.; Dalbey, R.E.; Brady, L.J. Functional overlap but lack of complete cross-complementation of Streptococcus mutans and Escherichia coli YidC orthologs. J. Bacteriol. 2008, 190, 2458–2469. [Google Scholar] [CrossRef] [Green Version]
- Hasona, A.; Crowley, P.J.; Levesque, C.M.; Mair, R.W.; Cvitkovitch, D.G.; Bleiweis, A.S.; Brady, L.J. Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc. Natl. Acad. Sci. USA 2005, 102, 17466–17471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, S.D.; Sharma, R.; Srivastava, S.; Navani, N.K.; Pathania, R. Downregulation of yidC in Escherichia coli by antisense RNA expression results in sensitization to antibacterial essential oils eugenol and carvacrol. PLoS ONE 2013, 8, e57370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofbauer, B.; Vomacka, J.; Stahl, M.; Korotkov, V.S.; Jennings, M.C.; Wuest, W.M.; Sieber, S.A. Dual Inhibitor of Staphylococcus aureus Virulence and Biofilm Attenuates Expression of Major Toxins and Adhesins. Biochemistry 2018, 57, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.C.; Lee, S.L.; Kapuriya, N.; Wang, D.; Chen, Y.R.; Yu, S.L.; Kulp, S.K.; Teng, L.J.; Chen, C.S. Development of novel antibacterial agents against methicillin-resistant Staphylococcus aureus. Bioorg. Med. Chem. 2012, 20, 4653–4660. [Google Scholar] [CrossRef] [Green Version]
- Corrigan, R.M.; Foster, T.J. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 2009, 61, 126–129. [Google Scholar] [CrossRef]
- Guo, H.; Suzuki, T.; Rubinstein, J.L. Structure of a bacterial ATP synthase. Elife 2019, 8, e43128. [Google Scholar] [CrossRef]
- Bald, D.; Amano, T.; Muneyuki, E.; Pitard, B.; Rigaud, J.L.; Kruip, J.; Hisabori, T.; Yoshida, M.; Shibata, M. ATP synthesis by F0F1-ATP synthase independent of noncatalytic nucleotide binding sites and insensitive to azide inhibition. J. Biol. Chem. 1998, 273, 865–870. [Google Scholar] [CrossRef] [Green Version]
- Jafari, R.; Almqvist, H.; Axelsson, H.; Ignatushchenko, M.; Lundback, T.; Nordlund, P.; Martinez Molina, D. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014, 9, 2100–2122. [Google Scholar] [CrossRef]
- Kumazaki, K.; Chiba, S.; Takemoto, M.; Furukawa, A.; Nishiyama, K.; Sugano, Y.; Mori, T.; Dohmae, N.; Hirata, K.; Nakada-Nakura, Y.; et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature 2014, 509, 516–520. [Google Scholar] [CrossRef] [Green Version]
- Kumazaki, K.; Kishimoto, T.; Furukawa, A.; Mori, H.; Tanaka, Y.; Dohmae, N.; Ishitani, R.; Tsukazaki, T.; Nureki, O. Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci. Rep. 2014, 4, 7299. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klenner, C.; Kuhn, A. Dynamic disulfide scanning of the membrane-inserting Pf3 coat protein reveals multiple YidC substrate contacts. J. Biol. Chem. 2012, 287, 3769–3776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Koningstein, G.; Pop, A.; Luirink, J. The conserved third transmembrane segment of YidC contacts nascent Escherichia coli inner membrane proteins. J. Biol. Chem. 2008, 283, 34635–34642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.C.; Huang, Y.T.; Chen, C.S.; Chen, Y.W.; Huang, Y.T.; Su, J.C.; Teng, L.J.; Shiau, C.W.; Chiu, H.C. In vitro and in vivo activity of a novel sorafenib derivative SC5005 against MRSA. J. Antimicrob. Chemother. 2016, 71, 449–459. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Tian, H.F.; Wen, J.F. The evolution of YidC/Oxa/Alb3 family in the three domains of life: A phylogenomic analysis. BMC Evol. Biol. 2009, 9, 137. [Google Scholar] [CrossRef] [Green Version]
- Su, J.C.; Huang, Y.T.; Chen, C.S.; Chiu, H.C.; Shiau, C.W. Design and Synthesis of Malonamide Derivatives as Antibiotics against Methicillin-Resistant Staphylococcus aureus. Molecules 2017, 23, 27. [Google Scholar] [CrossRef] [Green Version]



| MIC of Cpd36 (μg/mL) | MIC of Cpd46 (μg/mL) | yidC1 | yidC2 | |
|---|---|---|---|---|
| S. aureus (ATCC12598) | 1 | 0.5 | − | + |
| S. epidermidis (ATCC12228) | 1 | 0.5 | − | + |
| S. haemolyticus (ATCC29970) | 2 | 2 | + | + |
| S. hominis (ATCC27844) | 2 | 2 | + | + |
| S. intermedius (ATCC29663) | 1 | 0.5 | − a | + |
| S. saprophyticus (ATCC15305) | 2 | 2 | + | + |
| S. lugdunensis (NTUH isolate) | 2 | 2 | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tzeng, S.-R.; Huang, Y.-W.; Zhang, Y.-Q.; Yang, C.-Y.; Chien, H.-S.; Chen, Y.-R.; Yu, S.-L.; Chen, C.S.; Chiu, H.-C. A Celecoxib Derivative Eradicates Antibiotic-Resistant Staphylococcus aureus and Biofilms by Targeting YidC2 Translocase. Int. J. Mol. Sci. 2020, 21, 9312. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21239312
Tzeng S-R, Huang Y-W, Zhang Y-Q, Yang C-Y, Chien H-S, Chen Y-R, Yu S-L, Chen CS, Chiu H-C. A Celecoxib Derivative Eradicates Antibiotic-Resistant Staphylococcus aureus and Biofilms by Targeting YidC2 Translocase. International Journal of Molecular Sciences. 2020; 21(23):9312. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21239312
Chicago/Turabian StyleTzeng, Shiou-Ru, Yi-Wei Huang, Yao-Qing Zhang, Ching-Yi Yang, Han-Sheng Chien, Yi-Ru Chen, Sung-Liang Yu, Ching S. Chen, and Hao-Chieh Chiu. 2020. "A Celecoxib Derivative Eradicates Antibiotic-Resistant Staphylococcus aureus and Biofilms by Targeting YidC2 Translocase" International Journal of Molecular Sciences 21, no. 23: 9312. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21239312





