Acceleration of Carbon Fixation in Chilling-Sensitive Banana under Mild and Moderate Chilling Stresses
Abstract
:1. Introduction
2. Results
2.1. Morphological and Physiological Changes of Banana in Response to Mild and Moderate Chilling Stress
2.2. Transcriptomic Analysis of Banana Exposed to the Mild and Moderate Chilling Stress
2.2.1. Transcriptomic Analysis of Banana Exposed to 16 °C Chilling Stress
2.2.2. Transcriptomic Analysis of Banana Exposed to 10 °C Chilling Stress
2.2.3. Comparison of LT10 and LT16
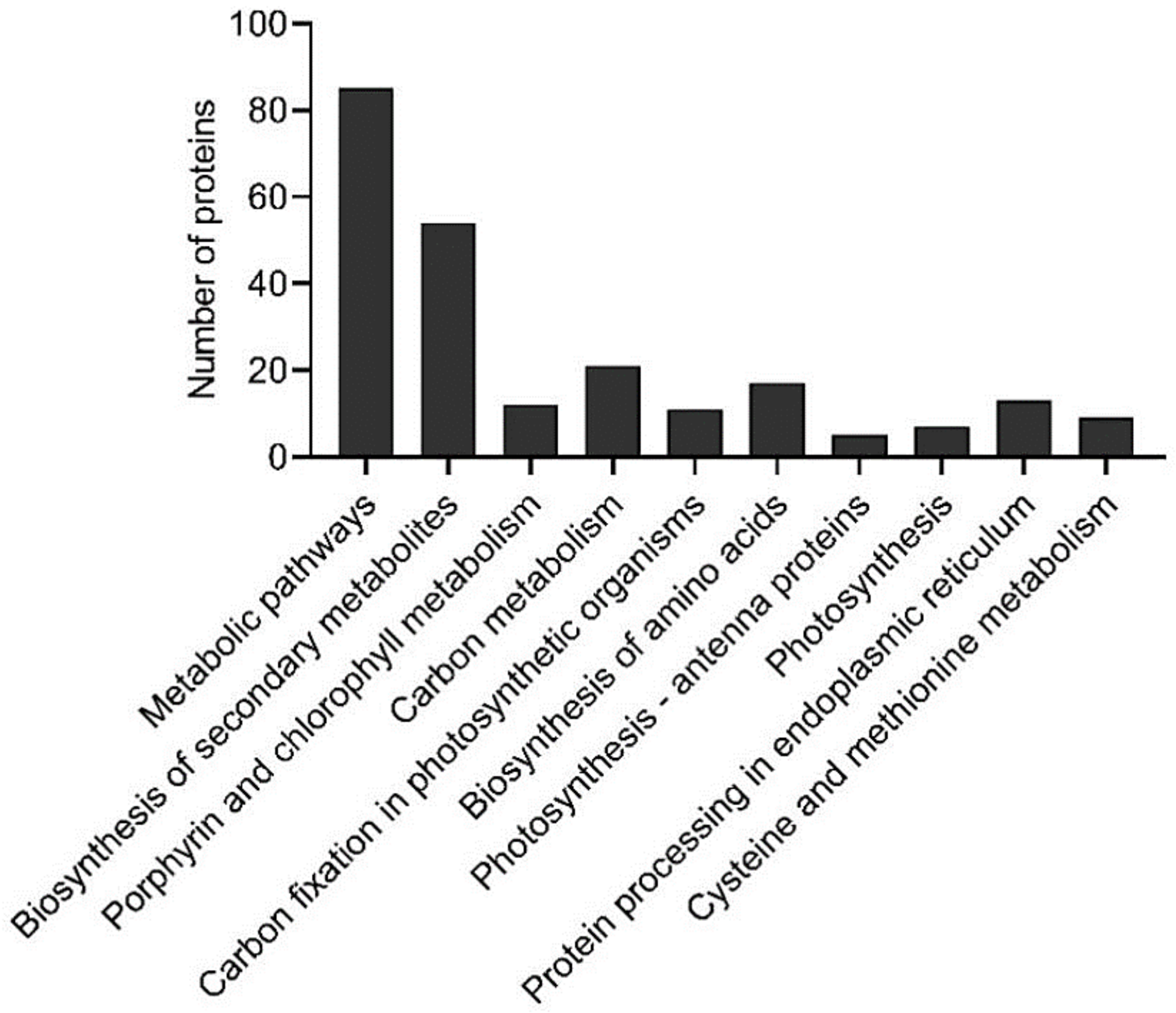
2.2.4. Evaluation of KEGG Pathways Enrichment
2.3. Proteomic Analysis of Banana Exposed to Mild Chilling Stress
2.4. DEGs and Proteins Involved in Photosynthesis upon LT Stress
2.4.1. DGE Analysis
2.4.2. Proteomics Analysis
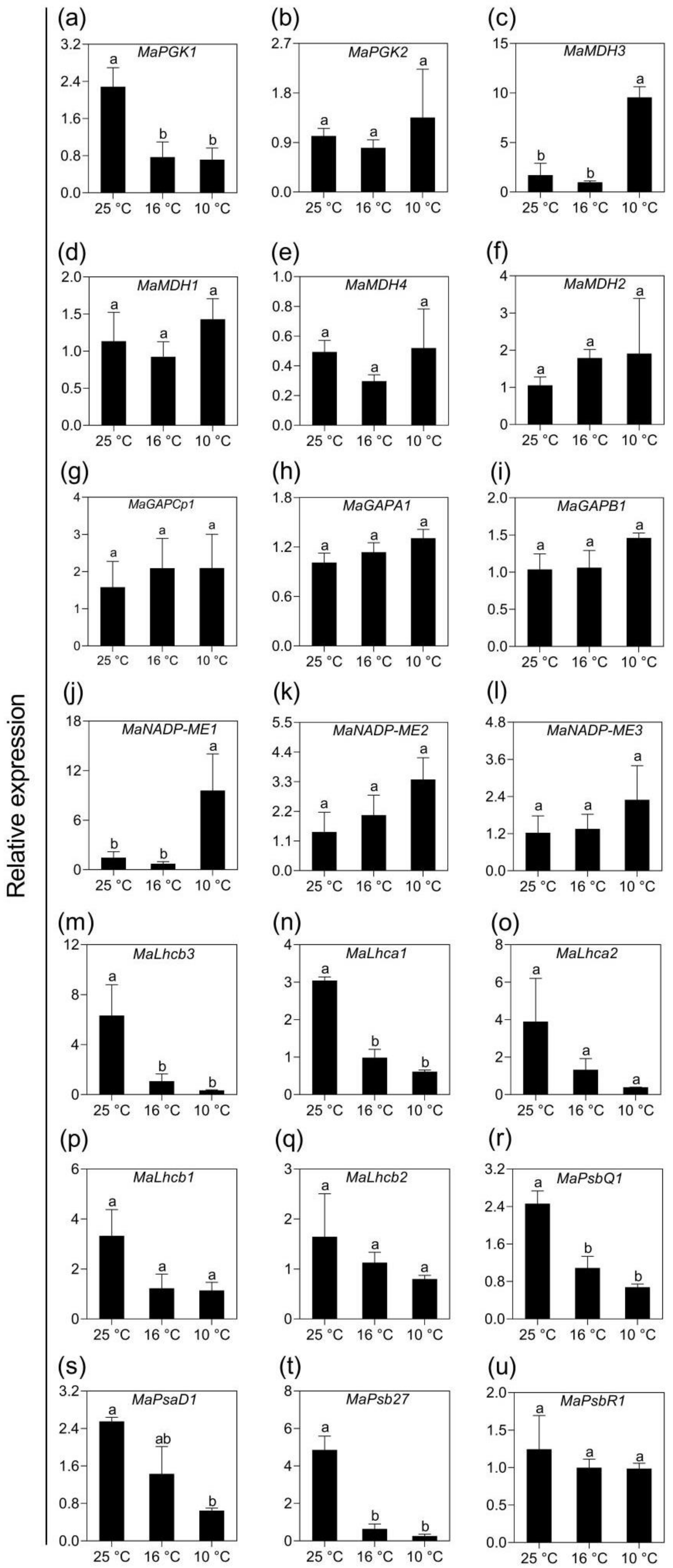
2.5. Validation of Transcriptomic Data Using qPCR Analysis
2.6. Immunofluorescence Labeling of Photosynthetic Proteins in Banana during Chilling Stress
3. Discussion
3.1. Mild and Moderate Chilling Results in Substantial Changes of Gene/Protein Expression in Banana
3.2. Upregulation of Carbon Fixation and Downregulation of the Electron Transport Chain in the Banana Response to Mild and Moderate Chilling Stresses
4. Materials and Methods
4.1. Plant Materials and LT Treatment
4.2. Measurement of Physiological Parameters
4.2.1. Chlorophyll Fluorescence Imaging
4.2.2. Determination of Enzyme Activities of PEPC and Rubisco
4.3. Immuno-labeling of Antibodies Related to Photosynthesis
4.4. DGE Analysis
4.5. Proteomic Analysis
4.5.1. Sample Preparation and Protein Digestion
4.5.2. Tandem Mass Tags Labeling and High-pH Reversed Phase Separation
4.5.3. LC-MS/MS Analysis
4.5.4. Protein Identification and Quantification
4.5.5. Bioinformatic Analysis
4.6. qPCR
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CK | the control |
| CS | chilling-sensitive |
| CT | chilling-tolerant |
| DEG | differentially expressed gene |
| DGE | digital gene expression |
| FDR | false discovery rate |
| kDa | kilodalton |
| KEGG | Kyoto encyclopedia of genes and genomes |
| LT | low temperature |
| LT10 | LT of 10 °C |
| LT16 | LT of 16 °C |
| NADH | nicotinamide adenine dinucleotide |
| PBS | phosphate-buffered saline |
| PEPC | phosphoenolpyruvate carboxylase |
| PPDK | pyruvate, phosphate dikinase |
| RNA-Seq | RNA sequencing |
| SPAD | Soil Plant Analysis Development |
References
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, S.A.; Men, S.N.; Wang, L.C. Chilling and drought stresses in crop plants: Implications, cross talk, and potential management opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.S.; Gao, J.; He, W.D.; Dou, T.X.; Ding, L.J.; Wu, J.H.; Li, C.Y.; Peng, X.X.; Zhang, S.; Yi, G.J. Comparative transcriptomics analysis reveals difference of key gene expression between banana and plantain in response to cold stress. BMC Genom. 2015, 16, 446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.J.; Zhang, Y.F.; Xu, Z.Q.; Luan, A.P.; Mao, Q.; Feng, J.J.; Xie, T.; Gong, X.; Wang, X.S.; Chen, X.; et al. Transcriptome profiling of the pineapple under low temperature to facilitate its breeding for cold tolerance. PLoS ONE 2016, 11, e0163315. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.S.; Zhang, Q.; Cui, F.; Hou, L.; Zhao, S.Z.; Xia, H.; Qiu, J.J.; Li, T.T.; Zhang, Y.; Wang, X.J.; et al. Genome-wide analysis of gene expression provides new insights into cold responses in Thellungiella salsuginea. Front. Plant Sci. 2017, 8, 713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Wu, T.K.; Li, S.H.; He, Q.; Deng, H.F. The methylation patterns and transcriptional responses to chilling stress at the seedling stage in rice. Int. J. Mol. Sci. 2019, 20, 5089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.L.; Li, Y.H.; He, C.; Zhou, J.T.; Chen, Y.Q.; Yu, Z.; Wang, P.; Ni, D.J. Nonvolatile metabolism in postharvest tea (Camellia sinensis L.) leaves: Effects of different withering treatments on nonvolatile metabolites, gene expression levels, and enzyme activity. Food Chem. 2020, 327, 126992. [Google Scholar] [CrossRef]
- Chai, F.M.; Liu, W.W.; Xiang, Y.; Meng, X.B.; Sun, X.M.; Cheng, C.; Liu, G.T.; Duan, L.X.; Xin, H.P.; Li, S.H. Comparative metabolic profiling of Vitis amurensis and Vitis vinifera during cold acclimation. Hortic. Res. 2019, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.L.; Khan, R.; Li, Q.Y.; Liu, G.L.; Xu, N.; Yang, Y.J.; Wang, Y.; Wang, S.S.; Chen, A.G. Transcriptomic analyses of chilling stress responsiveness in leaves of tobacco (Nicotiana tabacum) seedlings. Plant Mol. Biol. Rep. 2019, 38, 1–13. [Google Scholar] [CrossRef]
- Yang, Q.S.; Wu, J.H.; Li, C.Y.; Wei, Y.R.; Shen, O.; Hu, C.H.; Kuang, R.B. Quantitative proteomic analysis reveals that antioxidation mechanisms contribute to cold tolerance in plantain (Musa paradisiaca L.; ABB group) seedlings. Mol. Cell. Proteom. 2012, 11, 1853–1869. [Google Scholar] [CrossRef] [Green Version]
- Trzcinska-Danielewicz, J.; Bilska, A.; Fronk, J.; Zielenkiewicz, P.; Jarochowska, E.; Roszczyk, M.; Jonczyk, M.; Axentowicz, E.; Skoneczny, M.; Sowinski, P. Global analysis of gene expression in maize leaves treated with low temperature. I. Moderate chilling (14 °C). Plant Sci. 2009, 177, 648–658. [Google Scholar] [CrossRef]
- Spence, A.K.; Boddu, J.; Wang, D.; James, B.; Swaminathan, K.; Moose, S.P.; Long, S.P. Transcriptional responses indicate maintenance of photosynthetic proteins as key to the exceptional chilling tolerance of C4 photosynthesis in Miscanthus × giganteus. J. Exp. Bot. 2014, 65, 3737–3747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FAOSTAT. 2020. Available online: https://www.fao.org/faostat/en/#data/QC/visualize (accessed on 15 February 2020).
- Turner, D.W.; Lahav, E. The growth of banana plants in relation to temperature. Funct. Plant Biol. 1983, 10, 43–53. [Google Scholar] [CrossRef]
- Liu, H.; Ouyang, B.; Zhang, J.H.; Wang, T.T.; Li, H.X.; Zhang, Y.Y.; Yu, C.Y.; Ye, Z.B. Differential modulation of photosynthesis, signaling, and transcriptional regulation between tolerant and sensitive tomato genotypes under cold stress. PLoS ONE 2012, 7, e50785. [Google Scholar] [CrossRef]
- Xu, C.X. Fruit Cultivation in South China; China Agriculture Press: Beijing, China, 2015; pp. 85–86. ISBN 7-109-07750-0. [Google Scholar]
- Xu, C.X.; Chen, J.Z.; Liang, L.F. Effects of low temperature on the contents of glycerol, starch and sugars in banana leaves [in Chinese with English abstract]. J. Fruit Sci. 2000, 17, 105–109. [Google Scholar]
- Feng, R.J.; Zhang, L.L.; Wang, J.Y.; Luo, J.M.; Peng, M. Proteomic analysis of cold stress responses in banana leaves. J. Ameri. Soc. Hort. Sci. 2015, 140, 214–222. [Google Scholar] [CrossRef] [Green Version]
- Lai, Z.X.; Chen, Y.; Lin, Y.L.; Zhao, Q.Y.; Chen, Y.T. Discovery and taxonomy of wild banana (Musa spp.) in Fuzhou. [in Chinese with an English abstract]. Subtrop. Agric. Res. 2007, 3, 1–5. [Google Scholar]
- Liu, W.H.; Cheng, C.Z.; Lin, Y.L.; Xuhan, X.; Lai, Z.X. Genome-wide identification and characterization of mRNAs and lncRNAs involved in cold stress in the wild banana (Musa itinerans). PLoS ONE 2018, 13, e0200002. [Google Scholar] [CrossRef]
- Yamori, W.; Hikosaka, K.; Way, D.A. Temperature response of photosynthesis in C3, C4, and CAM plants: Temperature acclimation and temperature adaptation. Photosynth. Res. 2014, 119, 101–117. [Google Scholar] [CrossRef]
- Sobkowiak, A.; Jończyk, M.; Adamczyk, J.; Szczepanik, J.; Solecka, D.; Kuciara, I.; Hetmańczyk, K.; Trzcinska-Danielewicz, J.; Grzybowski, M.; Skoneczny, M.; et al. Molecular foundations of chilling-tolerance of modern maize. BMC Genom. 2016, 17, 125. [Google Scholar] [CrossRef] [Green Version]
- Friesen, P.C.; Sage, R.F. Photosynthetic responses to chilling in a chilling-tolerant and chilling-sensitive Miscanthus hybrid. Plant Cell Environ. 2016, 39, 1420–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajihashemi, S.; Noedoost, F.; Geuns, J.M.C.; Djalovic, I.; Siddique, K.H.M. Effect of cold stress on photosynthetic traits, carbohydrates, morphology, and anatomy in nine cultivars of Stevia rebaudiana. Front. Plant Sci. 2018, 9, 1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nouri, M.Z.; Moumeni, A.; Komatsu, S. Abiotic stresses: Insight into gene regulation and protein expression in photosynthetic pathways of plants. Int. J. Mol. Sci. 2015, 16, 20392–20416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.Y.; Nawaz, M.A.; Wei, N.N.; Cheng, F.; Bie, Z.L. Suboptimal temperature acclimation enhances chilling tolerance by improving photosynthetic adaptability and osmoregulation ability in watermelon. Hortic. Plant J. 2020, 6, 49–60. [Google Scholar] [CrossRef]
- Naidu, S.L.; Moose, S.P.; AL-Shoaibi, A.K.; Raines, C.A.; Long, S.P. Cold tolerance of C4 photosynthesis in Miscanthus × giganteus: Adaptation in amounts and sequence of C4 photosynthetic enzymes. Plant Physiol. 2003, 132, 1688–1697. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.F.; Naidu, S.L.; Portis, A.R., Jr.; Moose, S.P.; Long, S.P. Can the cold tolerance of C4 photosynthesis in Miscanthus × giganteus relative to Zea mays be explained by differences in activities and thermal properties of Rubisco? J. Exp. Bot. 2008, 59, 1779–1787. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.F.; Portis, A.R.; Moose, S.P.; Long, S.P. Cool C4 photosynthesis: Pyruvate Pi dikinase expression and activity corresponds to the exceptional cold tolerance of carbon assimilation in Miscanthus × giganteus. Plant Physiol. 2008, 148, 557–567. [Google Scholar] [CrossRef] [Green Version]
- Long, S.P.; Spence, A.K. Toward cool C4 crops. Annu. Rev. Plant Biol. 2013, 64, 701–722. [Google Scholar] [CrossRef]
- Bilska-Kos, A.; Panek, P.; Szulc-Głaz, A.; Ochodzki, P.; Cisło, A.; Zebrowski, J. Chilling-induced physiological, anatomical and biochemical responses in the leaves of Miscanthus × giganteus and maize (Zea mays L.). J. Plant Physiol. 2018, 228, 178–188. [Google Scholar] [CrossRef]
- Caffarri, S.; Frigerio, S.; Olivieri, E.; Righetti, P.G.; Bassi, R. Differential accumulation of Lhcb gene products in thylakoid membranes of Zea mays plants grown under contrasting light and temperature conditions. Proteomics 2005, 5, 758–768. [Google Scholar] [CrossRef]
- Cavaco, A.M.; De Silva, A.B.; Arrabaca, M.C. Effects of long-term chilling on growth and photosynthesis of the C4 gramineae Paspalum dilatatum. Physiol. Plant. 2003, 119, 87–96. [Google Scholar] [CrossRef]
- Soares-Cordeiro, A.S.; Driscoll, S.P.; Arrabaca, M.C.; Foyer, C.H. Dorsoventral variations in dark chilling effects on photosynthesis and stomatal function in Paspalum dilatatum leaves. J. Exp. Bot. 2010, 62, 687–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.; Sharma, N.; Deswal, R. The molecular biology of the low-temperature response in plants. Bioessays 2005, 27, 1048–1059. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.L.; Takác, T.; Li, X.Q.; Chen, H.B.; Wang, Y.Y.; Xu, E.F.; Xie, L.; Su, Z.H.; Šamaj, J.; Xu, C.X. Variable content and distribution of arabinogalactan proteins in banana (Musa spp.) under low temperature stress. Front. Plant Sci. 2015, 6, 353. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.; Hu, B.; Yi, G.J.; Li, X.Q.; Chen, H.B.; Wang, Y.Y.; Yuan, W.N.; Xing, Y.Q.; Sheng, Q.M.; Su, Z.X.; et al. Genome-wide analyses of banana fasciclin-like AGP genes and their differential expression under low-temperature stress in chilling sensitive and tolerant cultivars. Plant Cell Rep. 2020, 39, 693–708. [Google Scholar] [CrossRef]
- Xia, X.J.; Wang, Y.J.; Zhou, Y.H.; Tao, Y.; Mao, W.H.; Shi, K.; Asami, T.; Chen, Z.X.; Yu, J.Q. Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 2009, 150, 801–814. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, X.; Ma, Y.H.; Qing, Y.H.; Wang, H.C.; Huang, X.M. The highly drought-tolerant pitaya (Hylocereus undatus) is a non-facultative CAM plant under both well-watered and drought conditions. J. Horticult. Sci. Biotechnol. 2019, 94, 643–652. [Google Scholar] [CrossRef]
- Xu, C.X.; Takáč, T.; Burbach, C.; Menzel, D.; Samaj, J. Developmental localization and the role of hydroxyproline rich glycoproteins during somatic embryogenesis of banana (Musa spp. AAA). BMC Plant Biol. 2011, 11, 38. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.Q.; Hu, B.; Li, X.Q.; Chen, H.B.; Takáč, T.; Šamaj, J.; Xu, C.X. Comparative digital gene expression analysis of tissue-cultured plantlets of highly resistant and susceptible banana cultivars in response to Fusarium oxysporum. Int. J. Mol. Sci. 2018, 19, 350. [Google Scholar] [CrossRef] [Green Version]
- Fang, D.L.; Yang, W.J.; Deng, Z.L.; An, X.X.; Zhao, L.Y.; Hu, Q.H. Proteomic investigation of metabolic changes of mushroom (Flammulina velutipes) packaged with nanocomposite material during cold storage. J. Agric. Food Chem. 2017, 65, 10368–10381. [Google Scholar] [CrossRef]
- Oberg, A.L.; Mahoney, D.W.; Eckel-Passow, J.E.; Malone, C.J.; Wolfinger, R.D.; Hill, E.G.; Cooper, L.T.; Onuma, O.K.; Spiro, C.; Therneau, T.M.; et al. Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. J. Proteome Res. 2008, 7, 225–233. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Takáč, T.; Yi, G.; Chen, H.; Wang, Y.; Meng, J.; Yuan, W.; Tan, Y.; Ning, T.; He, Z.; et al. Acceleration of Carbon Fixation in Chilling-Sensitive Banana under Mild and Moderate Chilling Stresses. Int. J. Mol. Sci. 2020, 21, 9326. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21239326
Liu J, Takáč T, Yi G, Chen H, Wang Y, Meng J, Yuan W, Tan Y, Ning T, He Z, et al. Acceleration of Carbon Fixation in Chilling-Sensitive Banana under Mild and Moderate Chilling Stresses. International Journal of Molecular Sciences. 2020; 21(23):9326. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21239326
Chicago/Turabian StyleLiu, Jing, Tomáš Takáč, Ganjun Yi, Houbin Chen, Yingying Wang, Jian Meng, Weina Yuan, Yehuan Tan, Tong Ning, Zhenting He, and et al. 2020. "Acceleration of Carbon Fixation in Chilling-Sensitive Banana under Mild and Moderate Chilling Stresses" International Journal of Molecular Sciences 21, no. 23: 9326. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21239326





