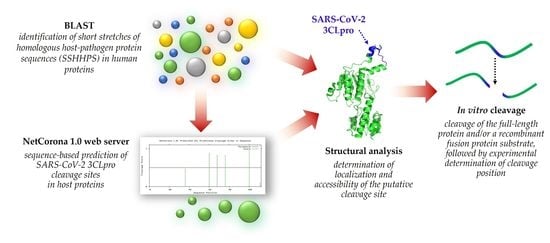
Identification of Host Cellular Protein Substrates of SARS-COV-2 Main Protease
Abstract
:1. Introduction
2. Results
2.1. Comparison of SARS-CoV and SARS-CoV-2 Protease Cleavage Sites
2.2. Testing NetCorona 1.0 Webserver for Prediction of SARS-CoV-2 3CLpro Cleavage Sites
2.3. Identification of Host Targets by SSHHPS Analysis and NetCorona Prediction
2.4. Selection of Targets for In Vitro Investigation
2.5. In Vitro Cleavage of Recombinant Proteins by SARS-CoV-2 3CLpro
2.6. Identification of Cleavage Position in CTBP1 and in His6-MBP-mEYFP Substrates
2.7. Comparison of Cleavage Efficiencies of SARS-CoV-2 and CTBP Cleavage Sites
3. Discussion
4. Materials and Methods
4.1. In Silico Analyses
4.1.1. BLAST Analysis
4.1.2. NetCorona Prediction
4.1.3. Structures
4.2. In Vitro Analyses
4.2.1. Materials
4.2.2. Expression and Purification of SARS-CoV-2 3CLpro
4.2.3. Vector Construction for the Expression of a His6-MBP-mEYFP Substrates
4.2.4. Expression and Purification of the His6-MBP-mEYFP Substrates
4.2.5. Cleavage Reactions by SARS-CoV-2 3CLpro
4.2.6. Cleavage Site Identification by MALDI-TOF MS
4.2.7. Proteinase Assay with His6-MBP-mEYFP Substrates
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 3CLpro | 3-chymotrypsin-like protease (main protease) |
| COVID-19 | coronavirus disease-19 |
| CTBP1 | C-terminal-binding protein 1 |
| CTBP2 | C-terminal-binding protein 2 |
| MBP | Maltose-binding protein |
| mEYFP | Monomeric enhanced yellow fluorescent protein |
| PLpro | Papain-like protease |
| PR | Protease |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SSHHPS | Short stretches of homologous host-pathogen protein sequences |
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef] [Green Version]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nyodu, R.; Maurya, V.K.; Saxena, S.K. Host Immune Response and Immunobiology of Human SARS-CoV-2 Infection. In Coronavirus Disease 2019 (COVID-19); Springer: Singapore, 2020; pp. 43–53. [Google Scholar]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Wunderink, R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology 2018, 23, 130–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannis, D.; Ziogas, I.; Gianni, P. Coagulation disorders in coronavirus infected patients: COVID-19, SARS-CoV-1, MERS-CoV and lessons from the past. J. Clin. Virol. 2020, 127, 104362. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, L.; Zhang, D.; Xu, J.; Dai, H.; Tang, N.; Su, X.; Cao, B. SARS-CoV-2 and viral sepsis: Observations and hypotheses. Lancet 2020, 395, 1517–1520. [Google Scholar] [CrossRef]
- Lin, L.; Jiang, X.; Zhang, Z.; Huang, S.; Zhang, Z.; Fang, Z.; Gu, Z.; Gao, L.; Shi, H.; Mai, L.; et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 2020, 69, 997–1001. [Google Scholar] [CrossRef]
- Baig, A.M. Neurological manifestations in COVID-19 caused by SARS-CoV-2. CNS Neurosci. Ther. 2020, 26, 499–501. [Google Scholar] [CrossRef] [Green Version]
- Xue, X.; Yu, H.; Yang, H.; Xue, F.; Wu, Z.; Shen, W.; Li, J.; Zhou, Z.; Ding, Y.; Zhao, Q.; et al. Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. J. Virol. 2008, 82, 2515–2527. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Xie, W.; Xue, X.; Yang, K.; Ma, J.; Liang, W.; Zhao, Q.; Zhou, Z.; Pei, D.; Ziebuhr, J.; et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005, 3, e324. [Google Scholar]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schechter, I.; Berger, A. On the size of the active site in proteases. I. papain. Biochem. Biophys Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Stoddard, S.V.; Stoddard, S.D.; Oelkers, B.K.; Fitts, K.; Whalum, K.; Whalum, K.; Hemphill, A.D.; Manikonda, J.; Martinez, L.M.; Riley, E.G.; et al. Optimization Rules for SARS-CoV-2 Mpro Antivirals: Ensemble Docking and Exploration of the Coronavirus Protease Active Site. Viruses 2020, 12, 942. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, T.; Takemoto, C.; Kim, Y.T.; Wang, H.; Nishii, W.; Terada, T.; Shirouzu, M.; Yokoyama, S. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. USA 2016, 113, 12997–13002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.W.; Tsai, F.J.; Wan, L.; Lai, C.C.; Lin, K.H.; Hsieh, T.H.; Shiu, S.Y.; Li, J.Y. Binding interaction of SARS coronavirus 3CL(pro) protease with vacuolar-H+ ATPase G1 subunit. FEBS Lett. 2005, 579, 6089–6094. [Google Scholar] [CrossRef] [Green Version]
- Liao, H.H.; Wang, Y.C.; Chen, M.C.; Tsai, H.Y.; Lin, J.; Chen, S.T.; Tsay, G.J.; Cheng, S.L. Down-regulation of granulocyte-macrophage colony-stimulating factor by 3C-like proteinase in transfected A549 human lung carcinoma cells. BMC Immunol. 2011, 12, 16. [Google Scholar] [CrossRef] [Green Version]
- Koudelka, T.; Boger, J.; Henkel, A.; Schönherr, R.; Krantz, S.; Fuchs, S.; Rodríguez, E.; Redecke, L.; Tholey, A. N-Terminomics for the Identification of in vitro Substrates and Cleavage Site Specificity of the SARS-CoV-2 Main Protease. Proteomics 2020, e2000246. [Google Scholar] [CrossRef]
- Shen, H.B.; Chou, K.C. HIVcleave: A web-server for predicting human immunodeficiency virus protease cleavage sites in proteins. Anal. Biochem. 2008, 375, 388–390. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Hansen, J.; Blaas, D.; Brunak, S. Cleavage site analysis in picornaviral polyproteins: Discovering cellular targets by neural networks. Protein Sci. 1996, 5, 2203–2216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morazzani, E.M.; Compton, J.R.; Leary, D.H.; Berry, A.V.; Hu, X.; Marugan, J.J.; Glass, P.J.; Legler, P.M. Proteolytic cleavage of host proteins by the Group IV viral proteases of Venezuelan equine encephalitis virus and Zika virus. Antivir. Res. 2019, 164, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Compton, J.R.; Legler, P.M. Analysis of Group IV Viral SSHHPS Using In Vitro and In Silico Methods. J. Vis. Exp. 2019, 154, e60421. [Google Scholar] [CrossRef] [PubMed]
- Kiemer, L.; Lund, O.; Brunak, S.; Blom, N. Coronavirus 3CLpro proteinase cleavage sites: Possible relevance to SARS virus pathology. BMC Bioinform. 2004, 5, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaru-Ampornpan, P.; Jengarn, J.; Wanitchang, A.; Jongkaewwattana, A. Porcine Epidemic Diarrhea Virus 3C-Like Protease-Mediated Nucleocapsid Processing: Possible Link to Viral Cell Culture Adaptability. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Guy, J.S.; Snijder, E.J.; Denniston, D.A.; Timoney, P.J.; Balasuriya, U.B. Genomic characterization of equine coronavirus. Virology 2007, 369, 92–104. [Google Scholar] [CrossRef] [Green Version]
- Taylor, E.W.; Radding, W. Understanding Selenium and Glutathione as Antiviral Factors in COVID-19: Does the Viral Mpro Protease Target Host Selenoproteins and Glutathione Synthesis? Front. Nutr. 2020, 7, 143. [Google Scholar] [CrossRef]
- Fan, K.; Ma, L.; Han, X.; Liang, H.; Wei, P.; Liu, Y.; Lai, L. The substrate specificity of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 2005, 329, 934–940. [Google Scholar] [CrossRef]
- Zhang, C.; Zheng, W.; Huang, X.; Bell, E.W.; Zhou, X.; Zhang, Y. Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. J. Proteome Res. 2020, 19, 1351–1360. [Google Scholar] [CrossRef] [Green Version]
- Grum-Tokars, V.; Ratia, K.; Begaye, A.; Baker, S.C.; Mesecar, A.D. Evaluating the 3C-like protease activity of SARS-Coronavirus: Recommendations for standardized assays for drug discovery. Virus Res. 2008, 133, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Bozóki, B.; Gazda, L.; Tóth, F.; Miczi, M.; Mótyán, J.A.; Tőzsér, J. A recombinant fusion protein-based, fluorescent protease assay for high throughput-compatible substrate screening. Anal. Biochem. 2018, 540, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Bozóki, B.; Mótyán, J.A.; Miczi, M.; Gazda, L.D.; Tőzsér, J. Use of Recombinant Fusion Proteins in a Fluorescent Protease Assay Platform and Their In-gel Renaturation. J. Vis. Exp. 2019, 16, e58824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mótyán, J.A.; Miczi, M.; Bozóki, B.; Tőzsér, J. Data supporting Ni-NTA magnetic bead-based fluorescent protease assay using recombinant fusion prostein substrates. Data Brief. 2018, 18, 203–208. [Google Scholar] [CrossRef]
- Gazda, L.D.; Joóné, M.K.; Nagy, T.; Mótyán, J.A.; Tőzsér, J. Biochemical characterization of Ty1 retrotransposon protease. PLoS ONE 2020, 15, e0227062. [Google Scholar] [CrossRef]
- Bozóki, B.; Mótyán, J.A.; Hoffka, G.; Waugh, D.S.; Tőzsér, J. Specificity Studies of the Venezuelan Equine Encephalitis Virus Non-Structural Protein 2 Protease Using Recombinant Fluorescent Substrates. Int. J. Mol. Sci. 2020, 21, 7686. [Google Scholar]
- Golda, M.; Mótyán, J.A.; Mahdi, M.; Tőzsér, J. Functional Study of the Retrotransposon-Derived Human PEG10 Protease. Int. J. Mol. Sci. 2020, 21, 2424. [Google Scholar] [CrossRef] [Green Version]
- Stankiewicz, T.R.; Gray, J.J.; Winter, A.N.; Linseman, D.A. C-terminal binding proteins: Central players in development and disease. Biomol. Concepts 2014, 5, 489–511. [Google Scholar] [CrossRef]
- Subramanian, T.; Zhao, L.J.; Chinnadurai, G. Interaction of CtBP with adenovirus E1A suppresses immortalization of primary epithelial cells and enhances virus replication during productive infection. Virology 2013, 443, 313–320. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Schäffer, A.A.; Aravind, L.; Madden, T.L.; Shavirin, S.; Spouge, J.L.; Wolf, Y.I.; Koonin, E.V.; Altschul, S.F. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001, 29, 2994–3005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilbert, B.J.; Morris, B.L.; Ellis, K.C.; Paulsen, J.L.; Schiffer, C.A.; Grossman, S.R.; Royer, W.E., Jr. Structure-guided design of a high affinity inhibitor to human CtBP. ACS Chem. Biol. 2015, 10, 1118–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Terzyan, S.; Tang, J.; Loy, J.A.; Lin, X.; Zhang, X.C. Human plasminogen catalytic domain undergoes an unusual conformational change upon activation. J. Mol. Biol. 2000, 295, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Kryger, G.; Harel, M.; Giles, K.; Toker, L.; Velan, B.; Lazar, A.; Kronman, C.; Barak, D.; Ariel, N.; Shafferman, A.; et al. Structures of recombinant native and E202Q mutant human acetylcholinesterase complexed with the snake-venom toxin fasciculin-II. Acta Crystallogr. D Biol. Crystallogr. 2000, 56 Pt 11, 1385–1394. [Google Scholar] [CrossRef] [Green Version]
- Hong, E.; Shin, J.; Kim, H.I.; Lee, S.T.; Lee, W. Solution structure and backbone dynamics of the non-receptor protein-tyrosine kinase-6 Src homology 2 domain. J. Biol. Chem. 2004, 279, 29700–29708. [Google Scholar] [CrossRef] [Green Version]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Qiao, Q.; Ferrao, R.; Shen, C.; Hatcher, J.M.; Buhrlage, S.J.; Gray, N.S.; Wu, H. Crystal structure of human IRAK1. Proc. Natl. Acad. Sci. USA 2017, 114, 13507–13512. [Google Scholar] [CrossRef] [Green Version]
- Mitternacht, S. FreeSASA: An open source C library for solvent accessible surface area calculations. F1000Res 2016, 5, 189. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]










| SARS-CoV-2 Cleavage Site | Protease | Sequence | NetCorona 1.0 Score |
|---|---|---|---|
| nsp1 | PLpro | ELNGG*AYTRY | no score given |
| nsp2 | PLpro | TLKGG*APTKV | no score given |
| nsp3 | PLpro | ALKGG*KIVNN | no score given |
| nsp4 | 3CLpro | SAVLQ*SGFRK | 0.891 |
| nsp5 | 3CLpro | GVTFQ*SAVKR | 0.458 |
| nsp6 | 3CLpro | VATVQ*SKMSD | 0.783 |
| nsp7 | 3CLpro | RATLQ*AIASE | 0.838 |
| nsp8 | 3CLpro | AVKLQ*NNELS | 0.860 |
| nsp9 | 3CLpro | TVRLQ*AGNAT | 0.904 |
| nsp10 | 3CLpro | EPMLQ*SADAQ | 0.865 |
| nsp12 | 3CLpro | HTVLQ*AVGAC | 0.905 |
| nsp13 | 3CLpro | VATLQ*AENVT | 0.680 |
| nsp14 | 3CLpro | FTRLQ*SLENV | 0.964 |
| nsp15 | 3CLpro | YPKLQ*SSQAW | 0.899 |
| Cleavage Site | KM (µM) | kcat (s−1) | kcat/KM (µM−1s−1) |
|---|---|---|---|
| TSAVLQ*SGFRKM | 5.086 ± 2.046 | 1.349 ± 0.370 | 0.2652 ± 0.1291 |
| REGTRVQ*SVEQIRE | 0.860 ± 0.303 | 0.0033 ± 0.0004 | 0.0038 ± 0.0014 |
| Cleavage Site | Sequence | Oligonucleotide Primer Sequence |
|---|---|---|
| SARS-CoV-2 nsp4 | TSAVLQ* SGFRKM | FW: 5’-TAAAACCTCTGCGGTGCTGCAGTCTGGCTTTCGTAAAATGG-3’ RV: 5’-CTAGCCATTTTACGAAAGCCAGACTGCAGCACCGCAGAGGTTTTAAT-3’ |
| CTBP1/2 (151–164) | REGTRV* SVEQIRE | FW: 5’-TAAACGTGAAGGCACCCGTGTGCAGTCTGTGGAACAGATCCGTGAAG-3’ RV: 5’-CTAGCTTCACGGATCTGTTCCACAGACTGCACACGGGTGCCTTCACGTTTAAT-3’ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miczi, M.; Golda, M.; Kunkli, B.; Nagy, T.; Tőzsér, J.; Mótyán, J.A. Identification of Host Cellular Protein Substrates of SARS-COV-2 Main Protease. Int. J. Mol. Sci. 2020, 21, 9523. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21249523
Miczi M, Golda M, Kunkli B, Nagy T, Tőzsér J, Mótyán JA. Identification of Host Cellular Protein Substrates of SARS-COV-2 Main Protease. International Journal of Molecular Sciences. 2020; 21(24):9523. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21249523
Chicago/Turabian StyleMiczi, Márió, Mária Golda, Balázs Kunkli, Tibor Nagy, József Tőzsér, and János András Mótyán. 2020. "Identification of Host Cellular Protein Substrates of SARS-COV-2 Main Protease" International Journal of Molecular Sciences 21, no. 24: 9523. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21249523