Tartary Buckwheat Transcription Factor FtbZIP5, Regulated by FtSnRK2.6, Can Improve Salt/Drought Resistance in Transgenic Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. Cloning and Molecular Characterization of FtbZIP5
2.2. FtbZIP5 Expression is Involved in the Response to Abiotic Stress
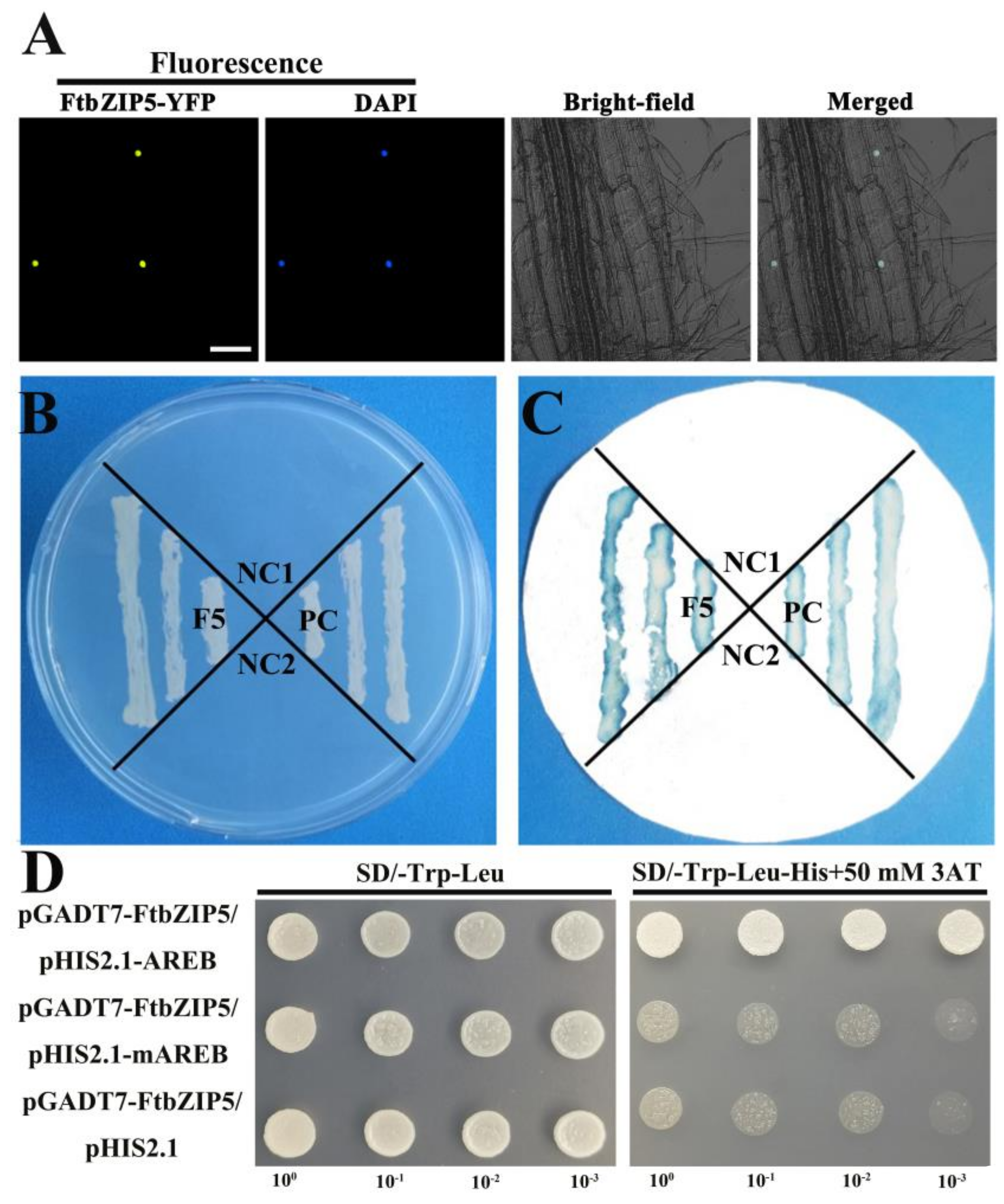
2.3. Characterization of FtbZIP5 as a Transcription Factor
2.4. Ectopic Expression of FtbZIP5 Increased the Sensitivity of Arabidopsis Seeds to Exogenous ABA at the Germination Stage
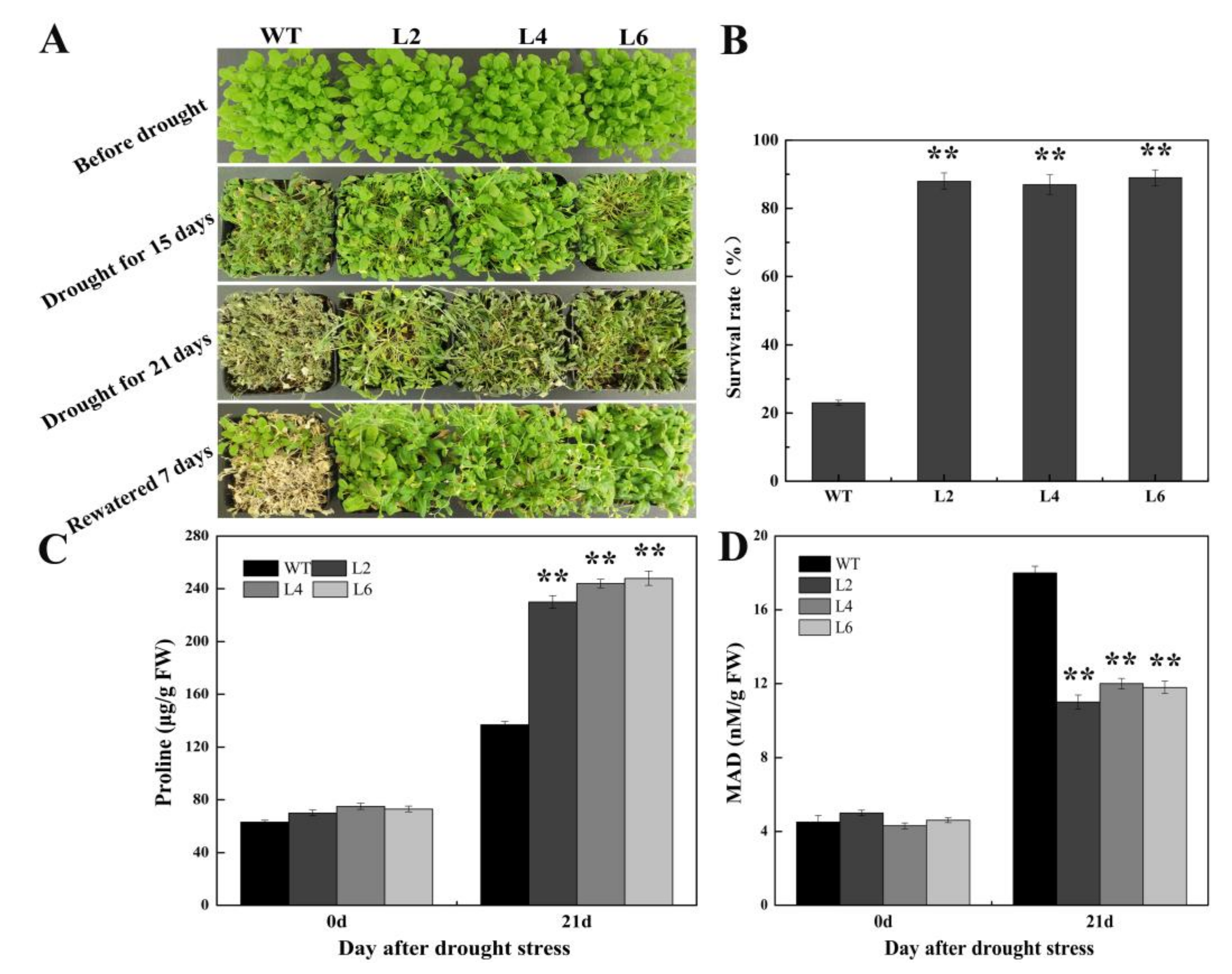
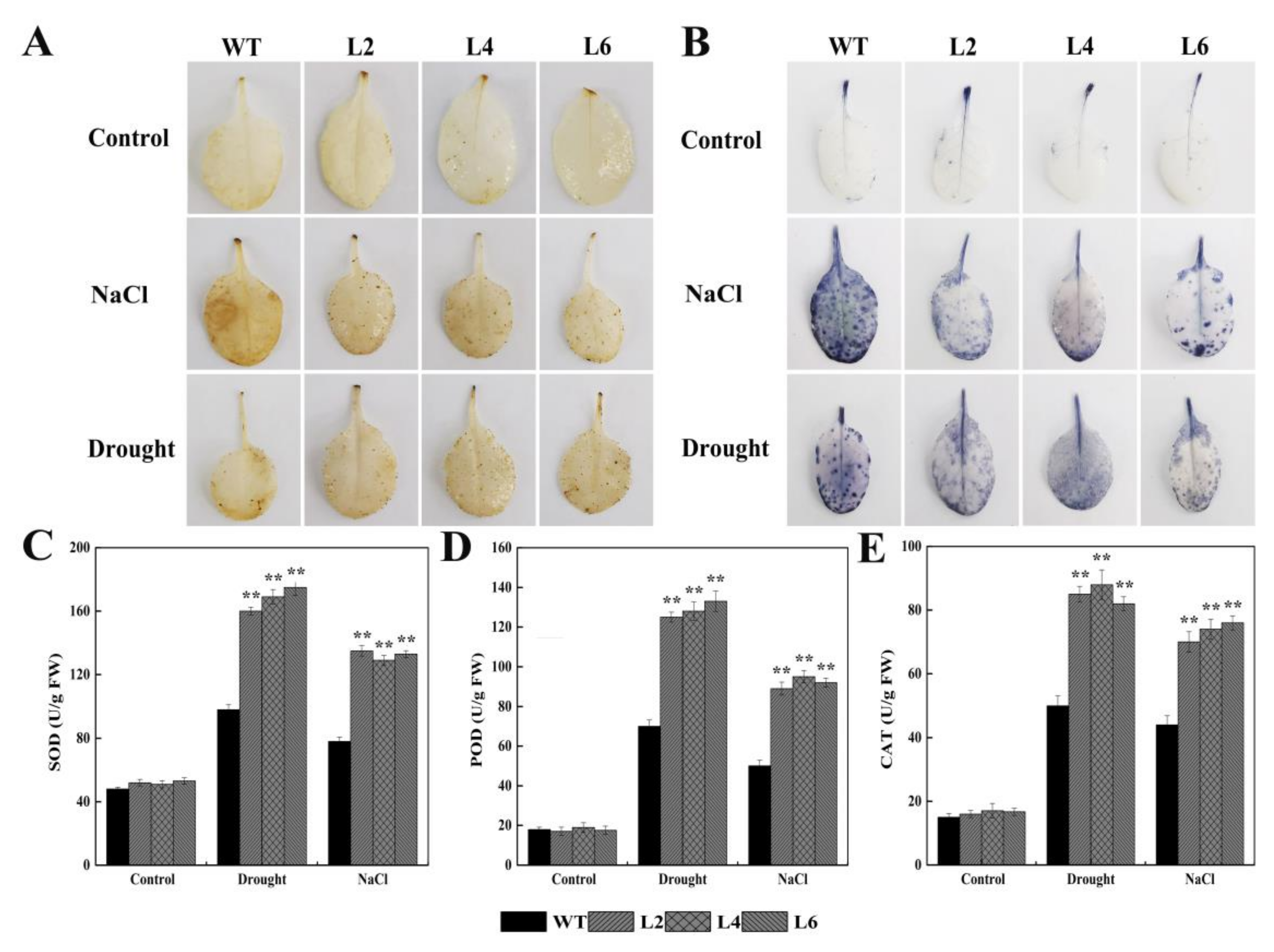
2.5. Ectopic Expression of FtbZIP5 Enhanced the Resistance of Transgenic Arabidopsis to Drought and Salt Stress
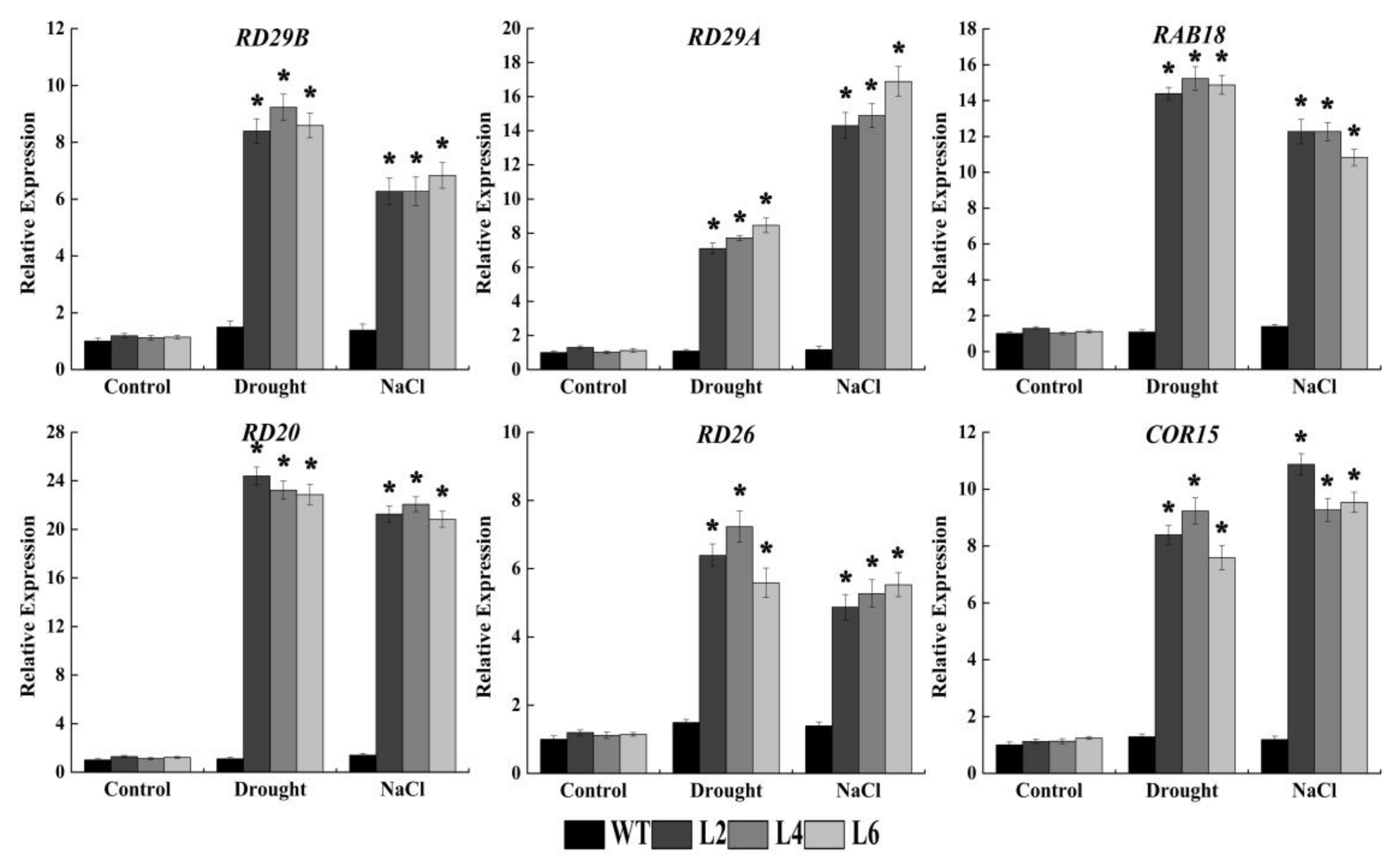
2.6. Ectopic Expression of FtbZIP5 Regulated Downstream Stress Response Genes
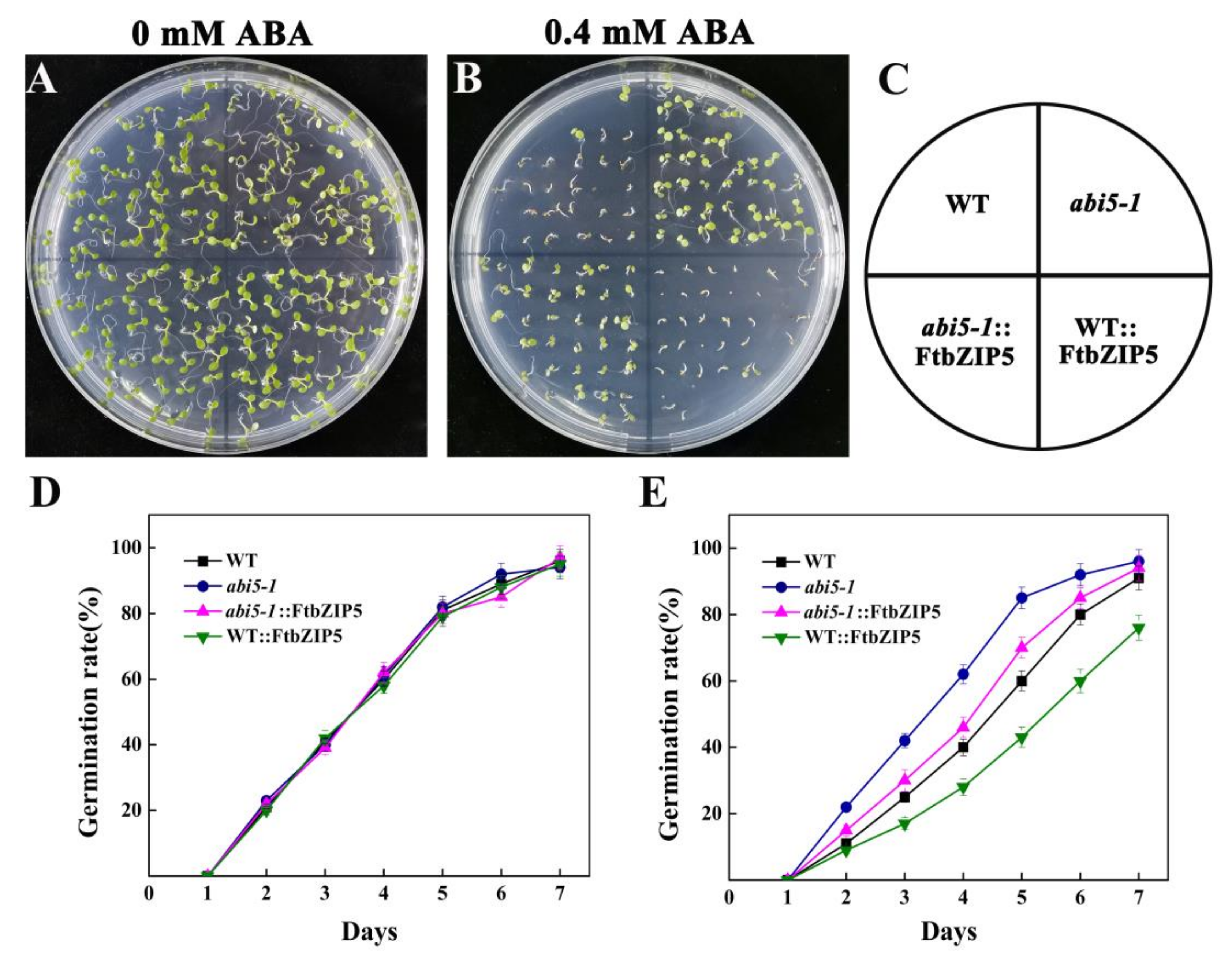
2.7. Ectopic Expression of FtbZIP5 Partially Recovered the Insensitivity of the Mutant abi5-1 to ABA
2.8. The Physical Interaction between FtbZIP5 and SnRK2.6 Proteins in Yeast
3. Discussion
4. Materials and Methods
4.1. Cloning of FtbZIP5 Gene, Construction of the Vector and Transformation of Arabidopsis Thaliana
4.2. Plant Materials and Stress Treatments
4.3. RNA Extraction and qRT-PCR Analysis
4.4. Subcellular Localization, Transcriptional Activation and Binding with ABRE Elements of FtbZIP5
4.5. Measurement of Antioxidant Enzymes, Proline and Malondialdehyde
4.6. Histochemical Staining with DAB and NBT
4.7. Y2H Assay
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nakashima, K.; Ito, Y.; Yamaguchishinozaki, K. Transcriptional Regulatory Networks in Response to Abiotic Stresses in Arabidopsis and Grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signaling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Gampala, S.S.L.; Rock, C.D. Abscisic Acid Signaling in Seeds and Seedlings. Plant Cell 2002, 14, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchishinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchishinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef]
- Drogelaser, W.; Snoek, L.B.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family—An update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Kim, S.Y.; Ma, J.; Perret, P.; Li, Z.; Thomas, T.L. Arabidopsis ABI5 Subfamily Members Have Distinct DNA-Binding and Transcriptional Activities. Plant Physiol. 2002, 130, 688–697. [Google Scholar] [CrossRef] [Green Version]
- Bensmihen, S.; Giraudat, J.; Parcy, F. Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J. Exp. Bot. 2005, 56, 597–603. [Google Scholar] [CrossRef] [Green Version]
- Bensmihen, S.; Rippa, S.; Lambert, G.; Jublot, D.; Pautot, V.; Granier, F.; Giraudat, J.; Parcy, F. The Homologous ABI5 and EEL Transcription Factors Function Antagonistically to Fine-Tune Gene Expression during Late Embryogenesis. Plant Cell 2002, 14, 1391–1403. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y. The role of ABF family bZIP class transcription factors in stress response. Physiol. Plant. 2005, 126, 519–527. [Google Scholar] [CrossRef]
- Lopezmolina, L.; Chua, N. A Null Mutation in a bZIP Factor Confers ABA-Insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2000, 41, 541–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, M.; Ketterling, M.G.; Li, Q.; Mccarty, D.R. Viviparous1 Alters Global Gene Expression Patterns through Regulation of Abscisic Acid Signaling. Plant Physiol. 2003, 132, 1664–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchishinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a Family of ABA-responsive Element Binding Factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchishinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis Abscisic Acid Response Gene ABI5 Encodes a Basic Leucine Zipper Transcription Factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Yu, X.; Wang, X.; Zhao, R.; Li, Y.; Fan, R.; Shang, Y.; Du, S.; Wang, X.; Wu, F. Two Calcium-Dependent Protein Kinases, CPK4 and CPK11, Regulate Abscisic Acid Signal Transduction in Arabidopsis. Plant Cell 2007, 19, 3019–3036. [Google Scholar] [CrossRef] [Green Version]
- Kagaya, Y.; Hobo, T.; Murata, M.; Ban, A.; Hattori, T. Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 2002, 14, 3177–3189. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchishinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K. Three SnRK2 Protein Kinases are the Main Positive Regulators of Abscisic Acid Signaling in Response to Water Stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correa, L.G.G.; Rianopachon, D.M.; Schrago, C.G.; Santos, R.V.D.; Muellerroeber, B.; Vincentz, M. The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS ONE 2008, 3, e2944. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, Q.; Wang, A.; Lv, B.; Dong, Q.; Yao, Y.; Wu, Q.; Zhao, H.; Li, C.; Chen, H.; et al. Tartary buckwheat transcription factor FtbZIP83 improves the drought/salt tolerance of Arabidopsis via an ABA-mediated pathway. Plant Physiol. Biochem. PPB 2019, 144, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yue, J.; Wu, X.; Xu, C.; Yu, J. An ABA-responsive DRE-binding protein gene from Setaria italica, SiARDP, the target gene of SiAREB, plays a critical role under drought stress. J. Exp. Bot. 2014, 65, 5415–5427. [Google Scholar] [CrossRef] [Green Version]
- Lu, G.; Gao, C.; Zheng, X.; Han, B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 2009, 229, 605–615. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Li, Q.; Mao, X.; Li, A.; Jing, R. Wheat Transcription Factor TaAREB3 Participates in Drought and Freezing Tolerances in Arabidopsis. Int. J. Biol. Sci. 2016, 12, 257–269. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Meng, Z.; Meng, Z.; Malik, W.; Yan, R.; Lwin, K.M.; Lin, F.; Wang, Y.X.; Sun, G.; Zhou, T. GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.). Sci. Rep. 2016, 6, 35040. [Google Scholar] [CrossRef] [Green Version]
- Stone, S.L.; Williams, L.A.; Farmer, L.M.; Vierstra, R.D.; Callis, J. KEEP ON GOING, a RING E3 Ligase Essential for Arabidopsis Growth and Development, Is Involved in Abscisic Acid Signaling. Plant Cell 2006, 18, 3415–3428. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, L.; Ye, T.; Lu, Y.; Chen, X.; Wu, Y. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 2013, 64, 675–684. [Google Scholar] [CrossRef]
- Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and Role of the Arabidopsis Abscisic Acid-Insensitive 5 Gene in Abscisic Acid, Sugar, and Stress Response. Plant Physiol. 2002, 129, 1533–1543. [Google Scholar] [CrossRef] [Green Version]
- Skubacz, A.; Daszkowskagolec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkelstein, R.R.; Gampala, S.S.L.; Lynch, T.J.; Thomas, T.L.; Rock, C.D. Redundant and distinct functions of the ABA response loci ABA- INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 2005, 59, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ye, N.; Liu, R.; Chen, M.; Zhang, J. H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J. Exp. Bot. 2010, 61, 2979–2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Wang, L.; Meng, H.; Wen, H.; Fan, Y.; Zhao, J. Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 2011, 75, 365–378. [Google Scholar] [CrossRef] [Green Version]
- Environment and plant metabolism: Flexibility and acclimation. In Environment & Plant Metabolism Flexibility & Acclimation XVI + P; Smirnoff, N. (Ed.) Bios Scientific Publishers: Oxford, UK, 1995; Volume 7, pp. 405–410. [Google Scholar]
- Tu, M.; Wang, X.; Huang, L.; Guo, R.; Zhang, H.; Cai, J.; Wang, X. Expression of a grape bZIP transcription factor, VqbZIP39, in transgenic Arabidopsis thaliana confers tolerance of multiple abiotic stresses. Plant Cell Tissue Organ Cult. 2016, 125, 537–551. [Google Scholar] [CrossRef]
- Rejeb, K.B.; Abdelly, C.; Savoure, A. How reactive oxygen species and proline face stress together. Plant Physiol. Biochem. 2014, 80, 278–284. [Google Scholar] [CrossRef]
- Liu, J.; Chu, J.; Ma, C.; Jiang, Y.; Ma, Y.; Xiong, J.; Cheng, Z. Overexpression of an ABA-dependent grapevine bZIP transcription factor, VvABF2, enhances osmotic stress in Arabidopsis. Plant Cell Rep. 2019, 38, 587–596. [Google Scholar] [CrossRef]
- Wang, C.; Lu, G.; Hao, Y.; Guo, H.; Guo, Y.; Zhao, J.; Cheng, H. ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 2017, 246, 453–469. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchishinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef]
- Hobo, T.; Kowyama, Y.; Hattori, T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 1999, 96, 15348–15353. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, S.; Katagiri, T.; Yamaguchishinozaki, K.; Shinozaki, K. An Arabidopsis Gene Encoding a Ca2+-Binding Protein is Induced by Abscisic Acid during Dehydration. Plant Cell Physiol. 2000, 41, 898–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.N.M.; Giammaria, V.; Grandellis, C.; Tellezinon, M.T.; Ulloa, R.M.; Capiati, D.A. Characterization of StABF1, a stress-responsive bZIP transcription factor from Solanum tuberosum L. that is phosphorylated by StCDPK2 in vitro. Planta 2012, 235, 761–778. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchishinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Sun, M.; Lu, J.; Liu, Y.; You, C.; Hao, Y. An apple CIPK protein kinase targets a novel residue of AREB transcription factor for ABA-dependent phosphorylation. Plant Cell Environ. 2017, 40, 2207–2219. [Google Scholar] [CrossRef]
- Bechtold, N.; Pelletier, G. In planta Agrobacterium mediated transformation of adult Arabidopsis thaliana plants by vacuum infifiltration. Methods Mol. Biol. 1998, 82, 259–266. [Google Scholar]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, W.; Zhao, Y.; Gong, X.; Guo, L.; Zhu, G.; Wang, X.; Gong, Z.; Schumaker, K.S.; Guo, Y. SAD2, an Importin β-Like Protein, Is Required for UV-B Response in Arabidopsis by Mediating MYB4 Nuclear Trafficking. Plant Cell 2007, 19, 3805–3818. [Google Scholar] [CrossRef] [Green Version]
- Jaffar, M.A.; Song, A.; Faheem, M.; Chen, S.; Jiang, J.; Liu, C.; Fan, Q.; Chen, F. Involvement of CmWRKY10 in Drought Tolerance of Chrysanthemum through the ABA-Signaling Pathway. Int. J. Mol. Sci. 2016, 17, 693. [Google Scholar] [CrossRef]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Zhao, H.; Wang, X.; Kang, J.; Lv, B.; Dong, Q.; Li, C.; Chen, H.; Wu, Q. Tartary Buckwheat Transcription Factor FtbZIP5, Regulated by FtSnRK2.6, Can Improve Salt/Drought Resistance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1123. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21031123
Li Q, Zhao H, Wang X, Kang J, Lv B, Dong Q, Li C, Chen H, Wu Q. Tartary Buckwheat Transcription Factor FtbZIP5, Regulated by FtSnRK2.6, Can Improve Salt/Drought Resistance in Transgenic Arabidopsis. International Journal of Molecular Sciences. 2020; 21(3):1123. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21031123
Chicago/Turabian StyleLi, Qi, Haixia Zhao, Xiaoli Wang, Jingyue Kang, Bingbing Lv, Qixin Dong, Chenglei Li, Hui Chen, and Qi Wu. 2020. "Tartary Buckwheat Transcription Factor FtbZIP5, Regulated by FtSnRK2.6, Can Improve Salt/Drought Resistance in Transgenic Arabidopsis" International Journal of Molecular Sciences 21, no. 3: 1123. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21031123




