The Molecular Mechanism of Perillaldehyde Inducing Cell Death in Aspergillus flavus by Inhibiting Energy Metabolism Revealed by Transcriptome Sequencing
Abstract
:1. Introduction
2. Results
2.1. A Preliminary Analysis of Transcriptome Sequencing Data
2.2. Expression Patterns of DEGs
2.3. GO Enrichment Analysis of DEGs
2.4. KEGG Enrichment Analysis of DEGs
2.5. A Protein-Protein Interaction Network Analysis of DEGs
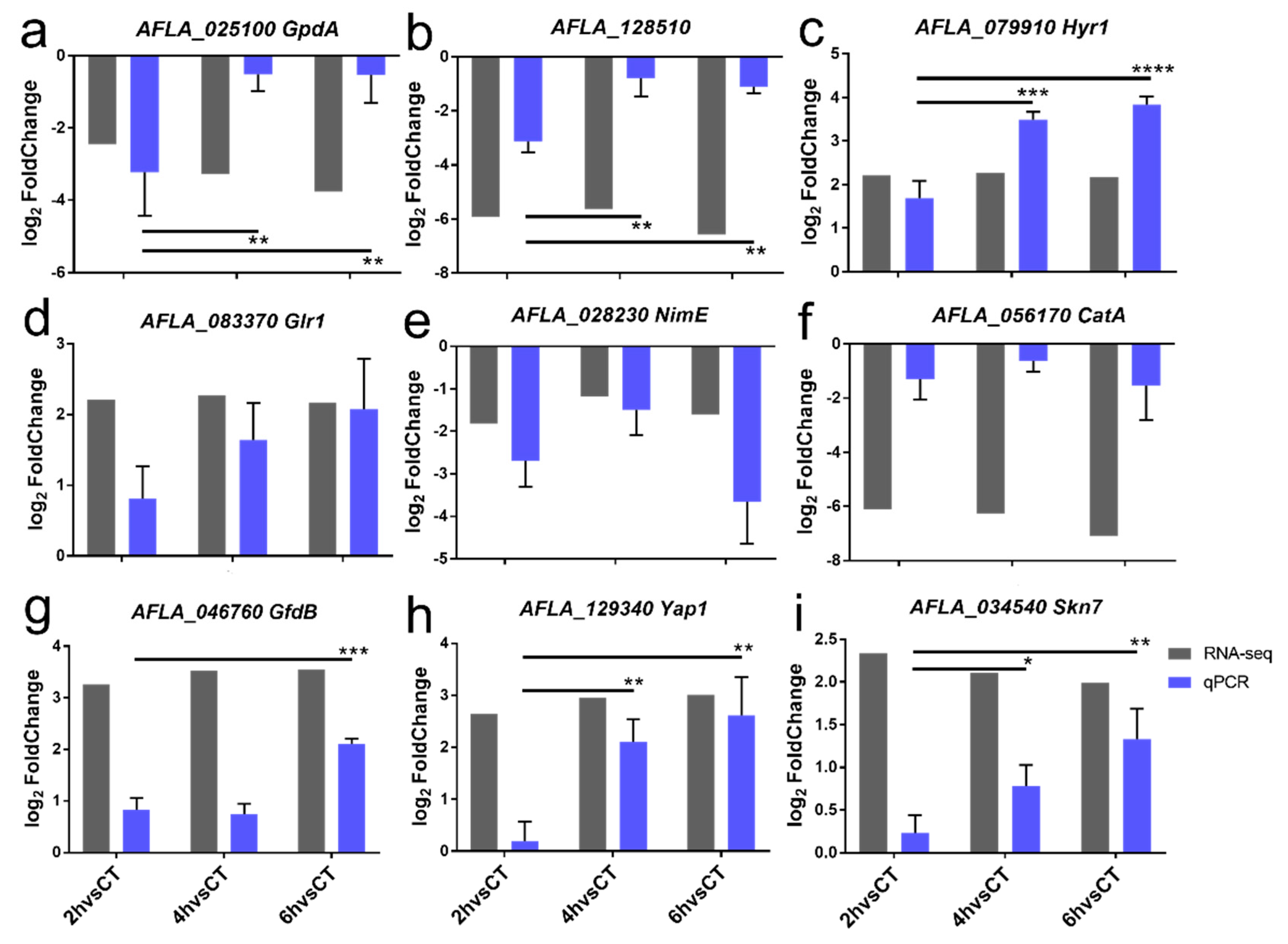
2.6. Confirmation of DEGs by RT-qPCR
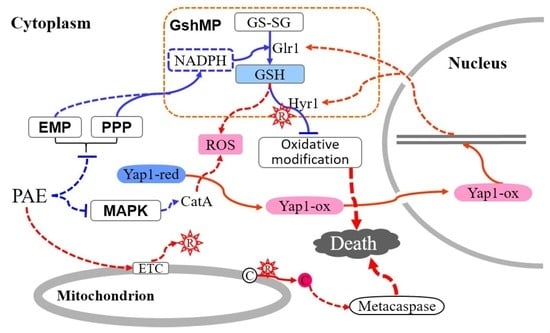
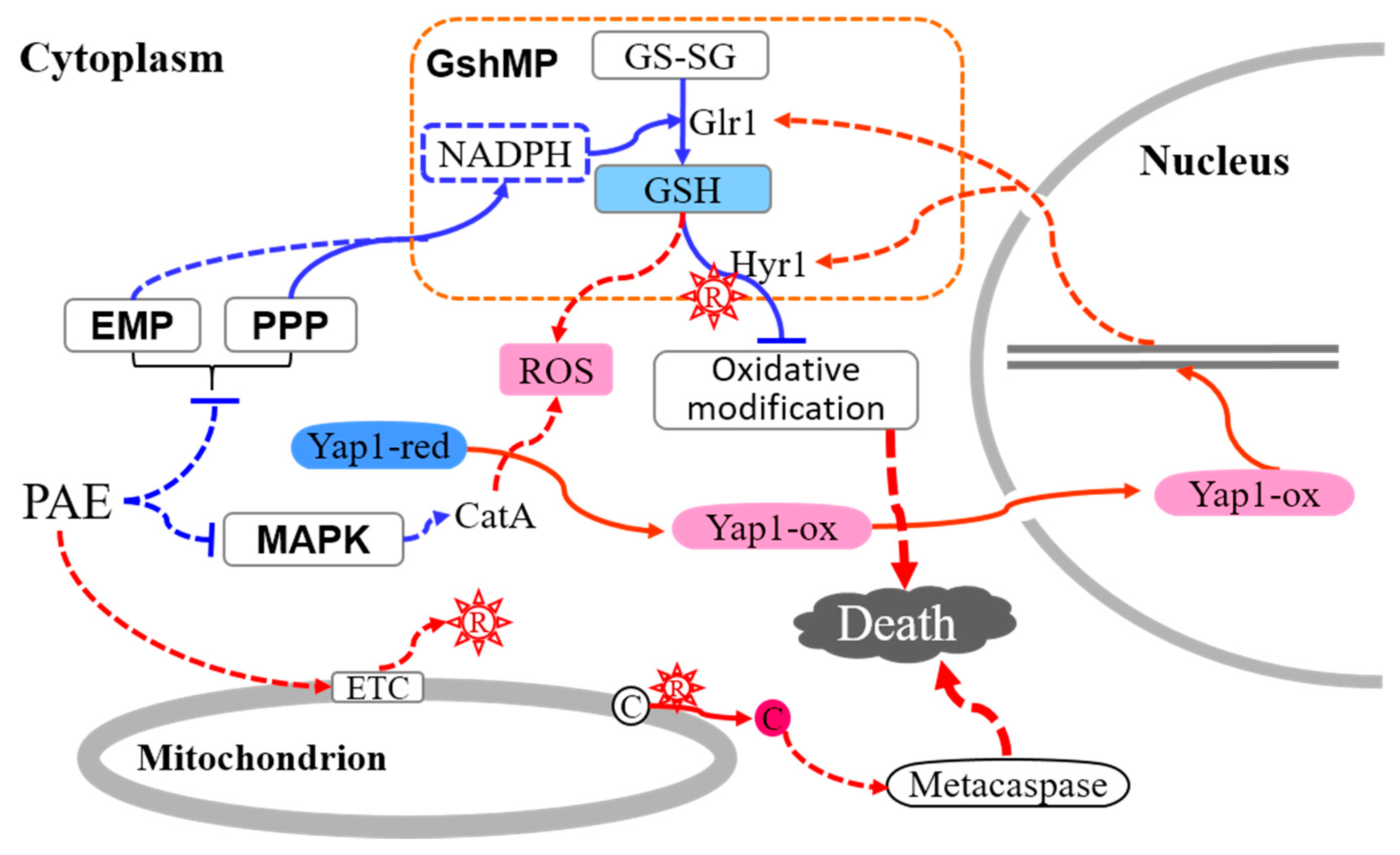
3. Discussion
4. Materials and Methods
4.1. Chemical and Fungus Strain
4.2. PAE Treatment and Sample Collection
4.3. RNA Isolation, Library Construction, and Sequencing
4.4. Data Analysis
4.5. Differential Expression Analysis
4.6. Validation of DEGs by RT-qPCR
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Cyt c | cytochrome c |
| DEGs | differentially expressed genes |
| EOs | essential oils |
| ETC | electron transport chain |
| FPKM | fragments per kilobase of transcript per million fragments mapped |
| GO | Gene Ontology |
| GSH | glutathione |
| GshMP | GSH metabolism pathway |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | mitogen activated protein kinase |
| MDS | mitogen activated protein kinase |
| MDS | multidimensional scaling plot |
| NADPH | reduced nicotinamide adenine dinucleotide phosphate |
| NOXs | NADPH oxidases |
| PAE | perillaldehyde |
| PPP | pentose phosphate pathway |
| RT-qPCR | real-time qPCR |
| ROS | reactive oxygen species |
| Trx | thioredoxin |
References
- Diener, U.L.; Cole, R.J.; Sanders, T.H.; Payne, G.A.; Lee, L.S.; Klich, M.A. Epidemiology of aflatoxin formation by Aspergillus flavus. Annu. Rev. Phytopathol. 1987, 25, 249–270. [Google Scholar] [CrossRef]
- Klich, M.A. Aspergillus flavus: The major producer of aflatoxin. Mol. Plant Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Lewis, L.; Onsongo, M.; Njapau, H.; Schurz-Rogers, H.; Luber, G.; Kieszak, S.; Nyamongo, J.; Backer, L.; Dahiye, A.M.; Misore, A.; et al. Aflatoxin contamination of commercial maize products during an outbreak of acute aflatoxicosis in eastern and central Kenya. Environ. Health Perspect. 2005, 113, 1763–1767. [Google Scholar] [CrossRef] [PubMed]
- Amaike, S.; Keller, N.P. Aspergillus flavus. Annu. Rev. Phytopathol. 2011, 49, 107–133. [Google Scholar] [CrossRef]
- Hobbs, C.A.; Taylor, S.V.; Beevers, C.; Lloyd, M.; Bowen, R.; Lillford, L.; Maronpot, R.; Hayashi, S.M. Genotoxicity assessment of the flavouring agent, perillaldehyde. Food Chem. Toxicol. 2016, 97, 232–242. [Google Scholar] [CrossRef]
- Sato, K.; Krist, S.; Buchbauer, G. Antimicrobial effect of trans-cinnamaldehyde, (-)-perillaldehyde, (-)-citronellal, citral, eugenol and carvacrol on airborne microbes using an airwasher. Biol. Pharm. Bull. 2006, 29, 2292–2294. [Google Scholar] [CrossRef] [Green Version]
- Mcgeady, P.; Wansley, D.L.; Logan, D.A. Carvone and perillaldehyde interfere with the serum-induced formation of filamentous structures in Candida albicans at substantially lower concentrations than those causing significant inhibition of growth. J. Nat. Prod. 2002, 65, 953–955. [Google Scholar] [CrossRef]
- Tian, J.; Zeng, X.; Lü, A.; Zhu, A.; Peng, X.; Wang, Y. Perillaldehyde, a potential preservative agent in foods: Assessment of antifungal activity against microbial spoilage of cherry tomatoes. LWT-Food Sci. Technol. 2015, 60, 63–70. [Google Scholar] [CrossRef]
- Tian, J.; Wang, Y.; Zeng, H.; Li, Z.; Zhang, P.; Tessema, A.; Peng, X. Efficacy and possible mechanisms of perillaldehyde in control of Aspergillus niger causing grape decay. Int. J. Food Microbiol. 2015, 202, 27–34. [Google Scholar] [CrossRef]
- Tian, J.; Wang, Y.; Lu, Z.; Sun, C.; Zhang, M.; Zhu, A.; Peng, X. Perillaldehyde, a promising antifungal agent used in food preservation, triggers apoptosis through a metacaspase-dependent pathway in Aspergillus flavus. J. Agric. Food Chem. 2016, 64, 7404–7413. [Google Scholar] [CrossRef]
- Kagan, V.E.; Tyurin, V.A.; Jiang, J.F.; Tyurina, Y.Y.; Ritov, V.B.; Amoscato, A.A.; Osipov, A.N.; Belikova, N.A.; Kapralov, A.A.; Kini, V.; et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 2005, 1, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Pedrajas, J.R.; Miranda-Vizuete, A.; Javanmardy, N.; Gustafsson, J.A.; Spyrou, G. Mitochondria of Saccharomyces cerevisiae contain one-conserved cysteine type peroxiredoxin with thioredoxin peroxidase activity. J. Biol. Chem. 2000, 275, 16296–16301. [Google Scholar] [CrossRef] [Green Version]
- Circu, M.L.; Aw, T.Y. Glutathione and apoptosis. Free Radic. Res. Commun. 2008, 42, 689–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collet, J.F.; D’Souza, J.C.; Jakob, U.; Bardwell, J.C. Thioredoxin 2, an oxidative stress-induced protein, contains a high affinity zinc binding site. J. Biol. Chem. 2003, 278, 45325–45332. [Google Scholar] [CrossRef] [Green Version]
- Larochelle, M.; Drouin, S.; Robert, F.; Turcotte, B. Oxidative stress-activated zinc cluster protein Stb5 has dual activator/repressor functions required for pentose phosphate pathway regulation and NADPH production. Mol. Cell Biol. 2006, 26, 6690–6701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, F.; Kawai, S.; Mori, S.; Kono, E.; Murata, K. Identification of ATP-NADH kinase isozymes and their contribution to supply of NADP(H) in Saccharomyces cerevisiae. FEBS J. 2005, 272, 3337–3349. [Google Scholar] [CrossRef] [PubMed]
- Stanley, B.A.; Sivakumaran, V.; Shi, S.; McDonald, I.; Lloyd, D.; Watson, W.H.; Aon, M.A.; Paolocci, N. Thioredoxin reductase-2 is essential for keeping low levels of H2O2 emission from isolated heart mitochondria. J. Biol. Chem. 2011, 286, 33669–33677. [Google Scholar] [CrossRef] [Green Version]
- Maiorino, M.; Ursini, F.; Bosello, V.; Toppo, S.; Tosatto, S.C.; Mauri, P.; Becker, K.; Roveri, A.; Bulato, C.; Benazzi, L.; et al. The thioredoxin specificity of Drosophila GPx: A paradigm for a peroxiredoxin-like mechanism of many glutathione peroxidases. J. Mol. Biol. 2007, 365, 1033–1046. [Google Scholar] [CrossRef]
- Collinson, E.J.; Wheeler, G.L.; Garrido, E.O.; Avery, A.M.; Avery, S.V.; Grant, C.M. The yeast glutaredoxins are active as glutathione peroxidases. J. Biol. Chem. 2002, 277, 16712–16717. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [Green Version]
- Outten, C.E.; Culotta, V.C. Alternative start sites in the Saccharomyces cerevisiae GLR1 gene are responsible for mitochondrial and cytosolic isoforms of glutathione reductase. J. Biol. Chem. 2004, 279, 7785–7791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuzi, D.; Maeta, K.; Takatsume, Y.; Izawa, S.; Inoue, Y. Regulation of the yeast phospholipid hydroperoxide glutathione peroxidase GPX2 by oxidative stress is mediated by Yap1 and Skn7. FEBS Lett. 2004, 565, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.M.; Collinson, L.P.; Roe, J.H.; Dawes, I.W. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol. Microbiol. 1996, 21, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Latimer, H.R.; Veal, E.A. Peroxiredoxins in regulation of MAPK signalling pathways; sensors and barriers to signal transduction. Mol. Cells 2016, 39, 40–45. [Google Scholar]
- Lee, J.; Godon, C.; Lagniel, G.; Spector, D.; Garin, J.; Labarre, J.; Toledano, M.B. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999, 274, 16040–16046. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Rodriguez, P.; Fernandez, E.; Almeida, A.; Bolanos, J.P. Excitotoxic stimulus stabilizes PFKFB3 causing pentose-phosphate pathway to glycolysis switch and neurodegeneration. Cell Death. Differ. 2012, 19, 1582–1589. [Google Scholar] [CrossRef] [Green Version]
- Patra, K.C.; Hay, N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef] [Green Version]
- Fraser, J.A.; Davis, M.A.; Hynes, M.J. A gene from Aspergillus nidulans with similarity to URE2 of Saccharomyces cerevisiae encodes a glutathione S-transferase which contributes to heavy metal and xenobiotic resistance. Appl. Environ. Microbiol. 2002, 68, 2802–2808. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.D.; Wang, J.; Stubbe, J.; Elledge, S.J. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol. Cell 2008, 32, 70–80. [Google Scholar] [CrossRef] [Green Version]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef]
- Kabeya, Y.; Kamada, Y.; Baba, M.; Takikawa, H.; Sasaki, M.; Ohsumi, Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 2005, 16, 2544–2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, D.K.; Carton, J.; Shah, A.K.; Meredith, F.; Uhlinger, D.J.; Hannun, Y.A. Serine palmitoyltransferase regulates de novoceramide generation during etoposide-induced apoptosis. J. Biol. Chem. 2000, 275, 9078–9084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vries, R.P.; Flitter, S.J.; Van De Vondervoort, P.J.; Chaveroche, M.K.; Fontaine, T.; Fillinger, S.; Ruijter, G.J.; D’Enfert, C.; Visser, J. Glycerol dehydrogenase, encoded by gldB is essential for osmotolerance in Aspergillus nidulans. Mol. Microbiol. 2003, 49, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Caceres, I.; El Khoury, R.; Bailly, S.; Oswald, I.P.; Puel, O.; Bailly, J.D. Piperine inhibits aflatoxin B1 production in Aspergillus flavus by modulating fungal oxidative stress response. Fungal Genet. Biol. 2017, 107, 77–85. [Google Scholar] [CrossRef]
- Boyce, K.J.; Cao, C.; Andrianopoulos, A. Two-component signaling regulates osmotic stress adaptation via SskA and the high-osmolarity glycerol MAPK pathway in the human pathogen talaromyces marneffei. MSphere 2016, 1, e00086-15. [Google Scholar] [CrossRef] [Green Version]
- Balázs, A.; Pócsi, I.; Hamari, Z.; Leiter, É.; Emri, T.; Miskei, M.; Oláh, J.; Tóth, V.; Hegedűs, N.; Prade, R.A. AtfA bZIP-type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans. Mol. Genet. Genom. 2010, 283, 289–303. [Google Scholar] [CrossRef]
- Ren, S.; Yang, M.; Li, Y.; Zhang, F.; Chen, Z.; Zhang, J.; Yang, G.; Yue, Y.; Li, S.; Ge, F.; et al. Global phosphoproteomic analysis reveals the involvement of phosphorylation in aflatoxins biosynthesis in the pathogenic fungus Aspergillus flavus. Sci. Rep. 2016, 6, 34078. [Google Scholar] [CrossRef]
- Alves de Castro, P.; dos Reis, T.F.; Dolan, S.K.; Oliveira Manfiolli, A.; Brown, N.A.; Jones, G.W.; Doyle, S.; Riaño-Pachón, D.M.; Squina, F.M.; Caldana, C. The Aspergillus fumigatus SchASCH9 kinase modulates SakAHOG1 MAP kinase activity and it is essential for virulence. Mol. Microbiol. 2016, 102, 642–671. [Google Scholar] [CrossRef] [Green Version]
- Pramila, T.; Miles, S.; GuhaThakurta, D.; Jemiolo, D.; Breeden, L.L. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 2002, 16, 3034–3045. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, N.; Zhang, M.Q. Identifying cooperativity among transcription factors controlling the cell cycle in yeast. Nucleic Acids Res. 2003, 31, 7024–7031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omari-Siaw, E.; Zhu, Y.; Wang, H.; Peng, W.; Firempong, C.K.; Wang, Y.W.; Cao, X.; Deng, W.; Yu, J.; Xu, X. Hypolipidemic potential of perillaldehyde-loaded self-nanoemulsifying delivery system in high-fat diet induced hyperlipidemic mice: Formulation, in vitro and in vivo evaluation. Eur. J. Pharm. Sci. 2016, 85, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Uemura, T.; Yashiro, T.; Oda, R.; Shioya, N.; Nakajima, T.; Hachisu, M.; Kobayashi, S.; Nishiyama, C.; Arimura, G.I. Intestinal anti-inflammatory activity of perillaldehyde. J. Agric. Food. Chem. 2018, 66, 3443–3448. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Chen, L.; Tian, H.; Wang, Z.; Wang, F.; Wang, L.; Li, J.; Ji, H.; Xi, L.; Feng, Z.; et al. Effect of perillaldehyde on prophylaxis and treatment of vaginal candidiasis in a murine model. Front. Microbiol. 2019, 10, 1466. [Google Scholar] [CrossRef]
- Ito, N.; Nagai, T.; Oikawa, T.; Yamada, H.; Hanawa, T. Antidepressant-like effect of l-perillaldehyde in stress-induced depression-like model mice through regulation of the olfactory nervous system. Evid. Based Complement. Altern. Med. 2011, 2011, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, J.; Pan, C.; Zhang, M.; Gan, Y.; Pan, S.; Liu, M.; Li, Y.; Zeng, X. Induced cell death in Ceratocystis fimbriata by pro-apoptotic activity of a natural organic compound, perillaldehyde, through Ca2+ overload and accumulation of reactive oxygen species. Plant Pathol. 2019, 68, 344–357. [Google Scholar] [CrossRef]
- Tian, H.; Qu, S.; Wang, Y.; Lu, Z.; Zhang, M.; Gan, Y.; Zhang, P.; Tian, J. Calcium and oxidative stress mediate perillaldehyde-induced apoptosis in Candida albicans. Appl. Microbiol. Biotechnol. 2017, 101, 3335–3345. [Google Scholar] [CrossRef]
- Knobloch, K.; Pauli, A.; Iberl, B.; Weigand, H.; Weis, N. Antibacterial and antifungal properties of essential oil components. J. Essent. Oil Res. 1989, 1, 119–128. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, X.; Zhou, Z.; Xing, K.; Tessema, A.; Zeng, H.; Tian, J. Inhibitory effect of nerol against Aspergillus niger on grapes through a membrane lesion mechanism. Food Control 2015, 55, 54–61. [Google Scholar] [CrossRef]
- Cha, M.K.; Kim, H.K.; Kim, I.H. Thioredoxin-linked “thiol peroxidase” from periplasmic space of Escherichia coli. J. Biol. Chem. 1995, 270, 28635–28641. [Google Scholar] [CrossRef] [Green Version]
- Bakkali, F.; Averbeck, S.; Averbeck, D.; Idaomar, M. Biological effects of essential oils--a review. Food Chem. Toxicol. 2008, 46, 446–475. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valko, M.; Morris, H.; Cronin, M.T. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005, 12, 1161–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reczek, C.R.; Chandel, N.S. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015, 33, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Toledano, M.B.; Kumar, C.; Le Moan, N.; Spector, D.; Tacnet, F. The system biology of thiol redox system in Escherichia coli and yeast: Differential functions in oxidative stress, iron metabolism and DNA synthesis. FEBS Lett. 2007, 581, 3598–3607. [Google Scholar] [CrossRef] [Green Version]
- Le, M.N.; Clement, G.; Le, M.S.; Tacnet, F.; Toledano, M.B. The Saccharomyces cerevisiae proteome of oxidized protein thiols: Contrasted functions for the thioredoxin and glutathione pathways. J. Biol. Chem. 2006, 281, 10420–10430. [Google Scholar]
- Bauer, H.; Kanzok, S.M.; Schirmer, R.H. Thioredoxin-2 but not thioredoxin-1 is a substrate of thioredoxin peroxidase-1 from Drosophila melanogaster: Isolation and characterization of a second thioredoxin in D. Melanogaster and evidence for distinct biological functions of Trx-1 and Trx-2. J. Biol. Chem. 2002, 277, 17457–17463. [Google Scholar] [CrossRef] [Green Version]
- Caccuri, A.M.; Antonini, G.; Allocati, N.; Di Ilio, C.; De Maria, F.; Innocenti, F.; Parker, M.W.; Masulli, M.; Lo Bello, M.; Turella, P.; et al. GSTB1-1 from Proteus mirabilis: A snapshot of an enzyme in the evolutionary pathway from a redox enzyme to a conjugating enzyme. J. Biol. Chem. 2002, 277, 18777–18784. [Google Scholar] [CrossRef] [Green Version]
- Golla, U.; Singh, V.; Azad, G.K.; Singh, P.; Verma, N.; Mandal, P.; Chauhan, S.; Tomar, R.S. Sen1p contributes to genomic integrity by regulating expression of ribonucleotide reductase 1 (RNR1) in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e64798. [Google Scholar] [CrossRef]
- Rodriguez, A.; de la Cera, T.; Herrero, P.; Moreno, F. The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem. J. 2001, 355, 625–631. [Google Scholar] [CrossRef] [Green Version]
- Reeves, R.E.; Serrano, R.; South, D.J. 6-phosphofructokinase (pyrophosphate). Properties of the enzyme from Entamoeba histolytica and its reaction mechanism. J. Biol. Chem. 1976, 251, 2958–2962. [Google Scholar] [PubMed]
- Outten, C.E.; Culotta, V.C. A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J. 2014, 22, 2015–2024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izawa, S.; Inoue, Y.; Kimura, A. Importance of catalase in the adaptive response to hydrogen peroxide: Analysis of acatalasaemic Saccharomyces cerevisiae. Biochem. J. 1996, 320, 61–67. [Google Scholar] [CrossRef] [Green Version]
- Raspelli, E.; Facchinetti, S.; Fraschini, R. Swe1 and Mih1 regulate mitotic spindle dynamics in budding yeast via Bik1. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nierman, W.C.; Yu, J.; Fedorova-Abrams, N.D.; Losada, L.; Cleveland, T.E.; Bhatnagar, D.; Bennett, J.W.; Dean, R.; Payne, G.A. Genome sequence of Aspergillus flavus NRRL 3357, a strain that causes aflatoxin contamination of food and feed. Genome Announc. 2015, 3, e00168-15. [Google Scholar] [CrossRef] [Green Version]
- Dhar, R.; Sagesser, R.; Weikert, C.; Wagner, A. Yeast adapts to a changing stressful environment by evolving cross-protection and anticipatory gene regulation. Mol. Biol. Evol. 2013, 30, 573–588. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [Green Version]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Liaw, W.H.A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; Schwartz, M. Gplots: Various R Programming Tools for Plotting Data. R package version 3.0. 1. Compr. R Arch. Netw. 2016. Available online: https://rdrr.io/cran/gplots/ (accessed on 11 January 2020).
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seip, E.R.; Woloshuk, C.P.; Payne, G.A.; Curtis, S.E. Isolation and sequence analysis of a beta-tubulin gene from Aspergillus flavus and its use as a selectable marker. Appl. Environ. Microbiol. 1990, 56, 3686–3692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


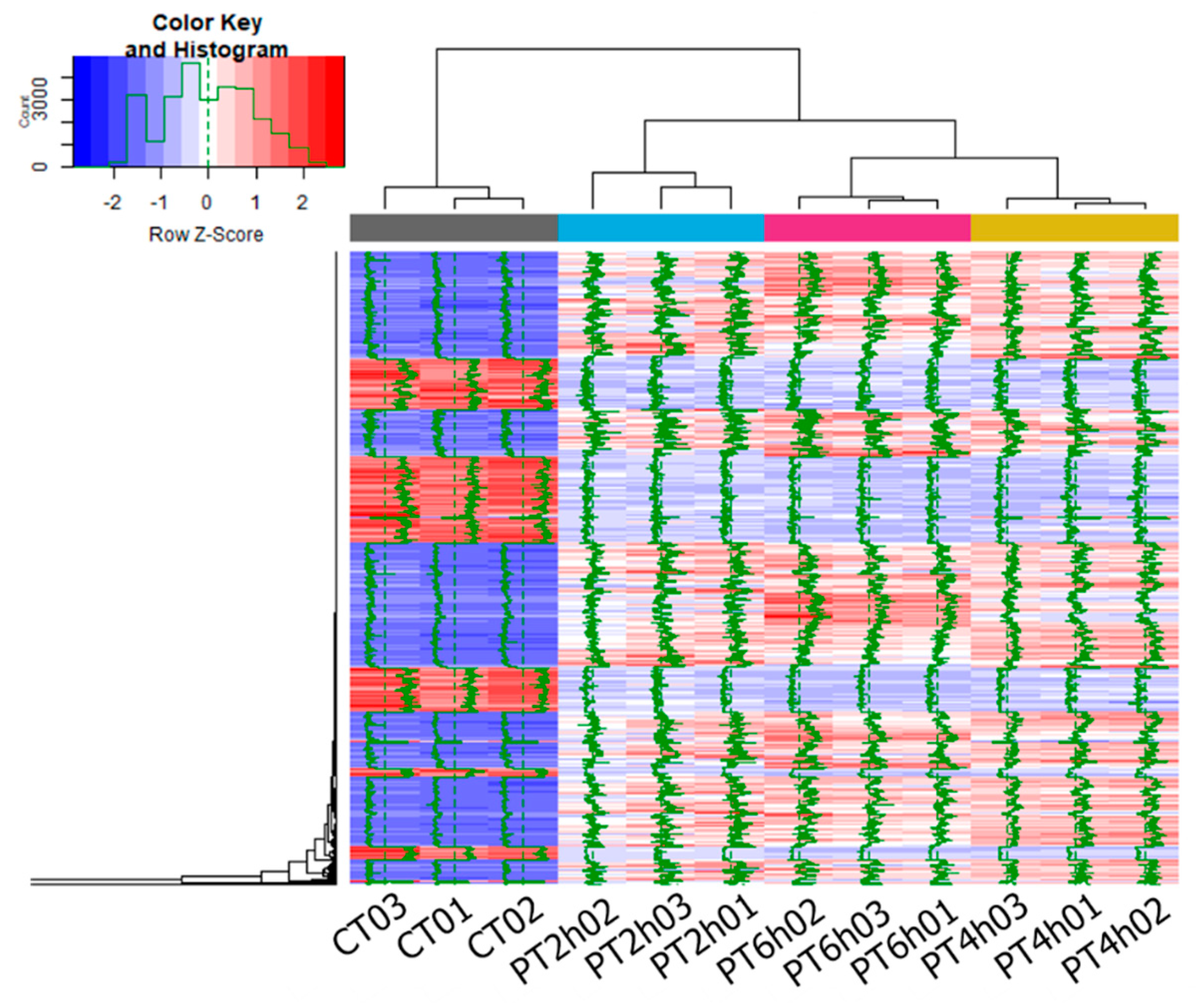
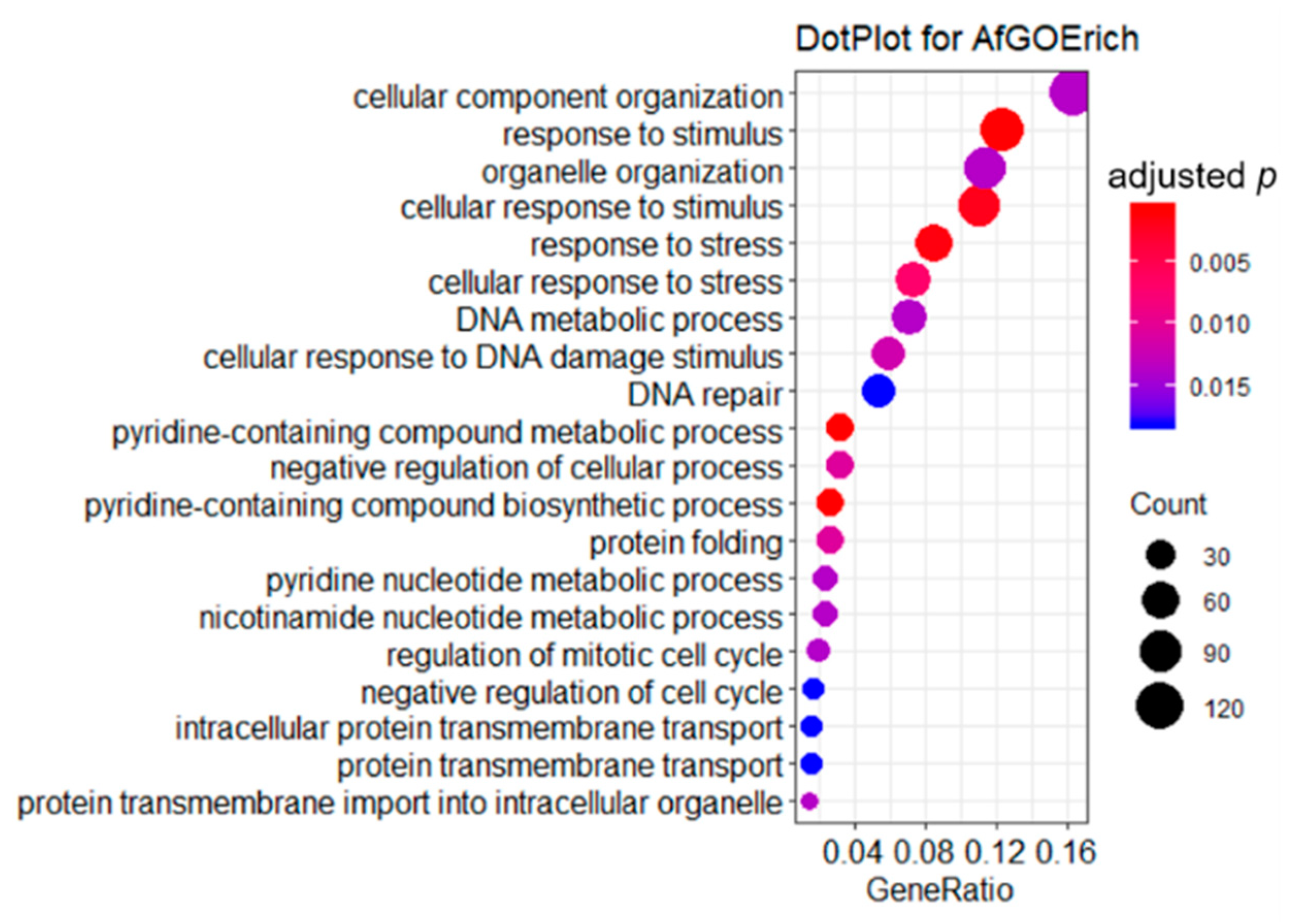
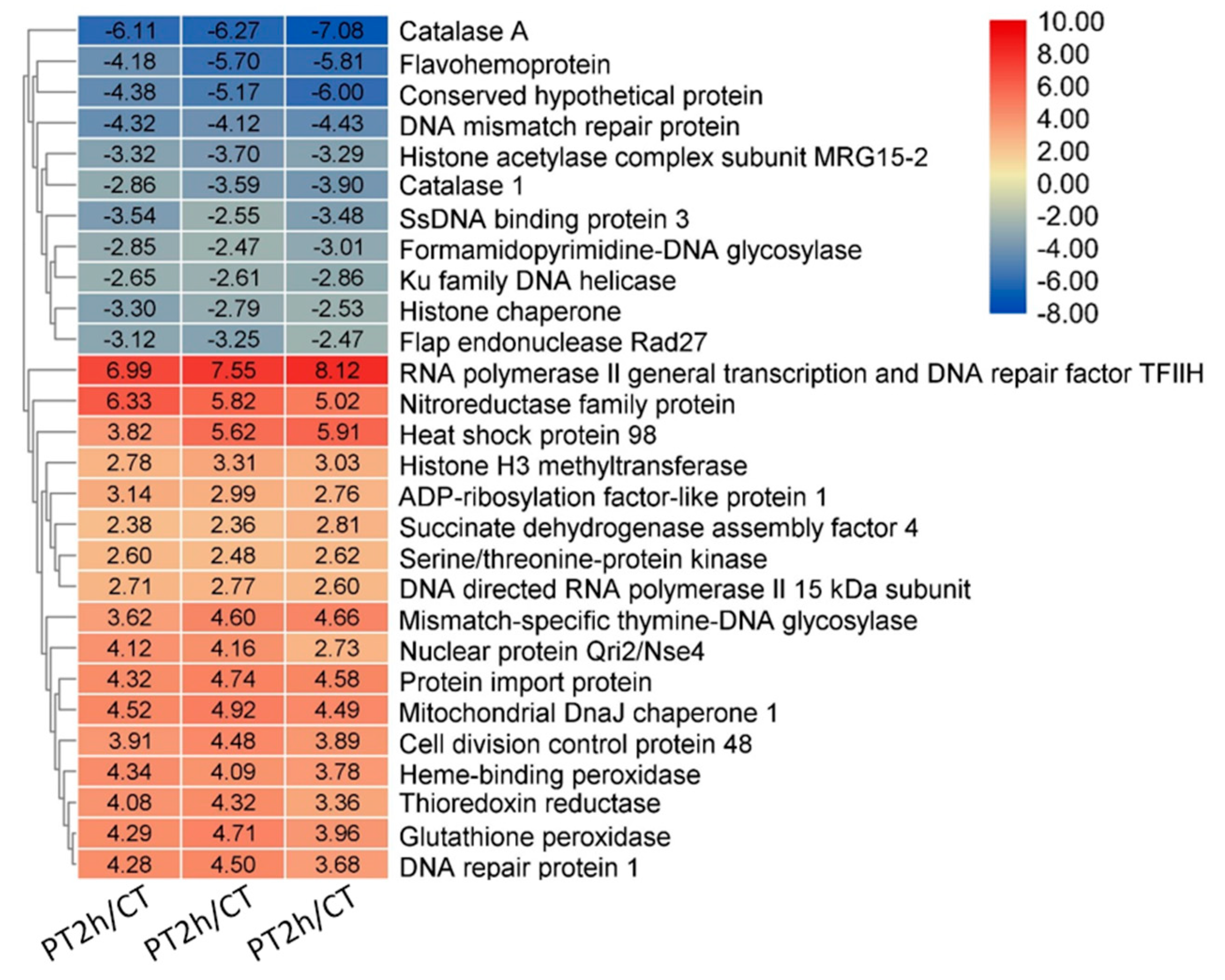


| Samples | Duplicates | Raw Reads | Clean Reads | Clean Bases/Gb | Q30/% | Mapped/% |
|---|---|---|---|---|---|---|
| 1 | 14,463,254 | 14,010,225 | 3,528,805,818 | 88.10 | 86.06 | |
| CT | 2 | 12,973,364 | 12,519,649 | 3,153,153,160 | 87.79 | 85.49 |
| 3 | 12,891,043 | 12,502,271 | 3,148,706,828 | 88.17 | 86.23 | |
| 1 | 16,474,698 | 14,912,152 | 3,754,934,036 | 88.70 | 90.18 | |
| PT2h | 2 | 15,322,010 | 13,912,871 | 3,503,972,558 | 88.41 | 89.78 |
| 3 | 16,769,520 | 15,215,838 | 3,832,001,194 | 89.98 | 90.31 | |
| 1 | 18,805,595 | 17,107,102 | 4,308,390,052 | 89.92 | 90.27 | |
| PT4h | 2 | 17,976,647 | 16,481,808 | 4,150,736,108 | 90.18 | 90.55 |
| 3 | 18,935,548 | 17,335,243 | 4,365,533,832 | 89.94 | 89.73 | |
| 1 | 16,979,324 | 13,183,354 | 3,319,958,436 | 85.43 | 84.33 | |
| PT6h | 2 | 17,624,059 | 15,768,269 | 3,971,226,468 | 90.00 | 89.71 |
| 3 | 21,925,695 | 19,631,355 | 4,942,988,542 | 89.96 | 89.82 |
| Comparisons | Up | Down | Total |
|---|---|---|---|
| PT2h/CT | 1848 | 950 | 2798 |
| PT4h/CT | 1928 | 1024 | 2952 |
| PT6h/CT | 1872 | 1082 | 2954 |
| Pathway | PT vs. CT log2 Fold Change | Name | Product | ||
|---|---|---|---|---|---|
| 2 h | 4 h | 6 h | |||
| Glycolysis | |||||
| AFLA_073260 | −1.09 | −1.88 | −1.08 | hexokinase | |
| AFLA_024480 | −1.49 | −1.76 | −2.75 | 6-phosphofructokinase alpha subunit | |
| AFLA_025100 | −2.45 | −3.28 | −3.76 | GpdA | glyceraldehyde 3-phosphate dehydrogenase |
| AFLA_027070 | −2.35 | −2.43 | −2.06 | acetate-CoA ligase | |
| AFLA_037480 | −2.52 | −3.22 | −1.96 | AspF22 | enolase |
| AFLA_069370 | −1.26 | −1.70 | −1.77 | PgkA | phosphoglycerate kinase |
| AFLA_094630 | −1.15 | −1.68 | −1.68 | triosephosphate isomerase | |
| PPP | |||||
| AFLA_080390 | −2.12 | −1.96 | −3.27 | 6-phosphogluconolactonase | |
| AFLA_030710 | −1.65 | −2.02 | −2.65 | PgmA | phosphoglucomutase |
| AFLA_015700 | −1.31 | −1.01 | −1.78 | ribulose-phosphate 3-epimerase | |
| AFLA_044820 | −1.83 | −2.13 | −2.10 | glucose-6-phosphate isomerase | |
| AFLA_123760 | 2.84 | 4.39 | 2.76 | deoxyribose-phosphate aldolase | |
| AFLA_125700 | −2.70 | −2.24 | −2.02 | ribokinase | |
| AFLA_128510 | −5.91 | −5.62 | −6.57 | 6-phosphogluconate dehydrogenase | |
| GSH metabolism | |||||
| AFLA_076480 | 3.45 | 3.27 | 2.73 | glutathione S-transferase | |
| AFLA_079910 | 4.29 | 4.71 | 3.96 | Hyr1 | glutathione peroxidase |
| AFLA_082280 | −1.22 | −1.80 | −1.69 | glutathione S-transferase | |
| AFLA_083370 | 2.21 | 2.27 | 2.17 | Glr1 | glutathione oxidoreductase |
| AFLA_029890 | 3.06 | 3.62 | 3.81 | glutathione S-transferase | |
| AFLA_017920 | −1.75 | −1.55 | −1.82 | spermidine synthase | |
| AFLA_113250 | 2.43 | 2.98 | 2.44 | Rnr-1 | ribonucleotide reductase large subunit |
| AFLA_056570 | 2.75 | 2.61 | 2.44 | glutathione synthetase | |
| AFLA_123650 | −2.95 | −1.12 | −2.95 | glutathione synthetase | |
| MAPK | |||||
| AFLA_028230 | −1.82 | −1.18 | −1.60 | NimE | G2/M-specific cyclin |
| AFLA_031340 | 1.98 | 2.27 | 2.01 | AtfA | bZIP transcription factor |
| AFLA_034380 | −2.86 | −3.59 | −3.90 | catalase | |
| AFLA_037000 | −3.04 | −3.9 | −3.88 | Mih1 | cell cycle control protein tyrosine phosphatase |
| AFLA_037350 | 2.05 | 2.07 | 2.31 | casein kinase I | |
| AFLA_039130 | 2.58 | 3.49 | 3.60 | TcsB/Sln1 | sensor histidine kinase/response regulator |
| AFLA_046760 | 3.27 | 3.54 | 3.55 | GfdB | glycerol 3-phosphate dehydrogenase |
| AFLA_056170 | −6.11 | −6.27 | −7.08 | CatA | spore-specific catalase |
| AFLA_062210 | 1.75 | 1.65 | 1.20 | response regulator | |
| AFLA_068590 | 1.71 | 1.97 | 1.57 | SskB | MAP kinase kinase kinase |
| AFLA_090910 | 2.81 | 3.61 | 3.57 | Mcm1 | MADS box transcription factor |
| AFLA_129340 | 2.65 | 2.95 | 3.01 | Yap1 | AP-1-like transcription factor |
| AFLA_034540 | 2.34 | 2.11 | 1.99 | Skn7 | stress response transcription factor |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, C.; Li, Y.-X.; Yang, K.; Famous, E.; Ma, Y.; He, X.; Geng, Q.; Liu, M.; Tian, J. The Molecular Mechanism of Perillaldehyde Inducing Cell Death in Aspergillus flavus by Inhibiting Energy Metabolism Revealed by Transcriptome Sequencing. Int. J. Mol. Sci. 2020, 21, 1518. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21041518
Pan C, Li Y-X, Yang K, Famous E, Ma Y, He X, Geng Q, Liu M, Tian J. The Molecular Mechanism of Perillaldehyde Inducing Cell Death in Aspergillus flavus by Inhibiting Energy Metabolism Revealed by Transcriptome Sequencing. International Journal of Molecular Sciences. 2020; 21(4):1518. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21041518
Chicago/Turabian StylePan, Chao, Yong-Xin Li, Kunlong Yang, Erhunmwunsee Famous, Yan Ma, Xiaona He, Qingru Geng, Man Liu, and Jun Tian. 2020. "The Molecular Mechanism of Perillaldehyde Inducing Cell Death in Aspergillus flavus by Inhibiting Energy Metabolism Revealed by Transcriptome Sequencing" International Journal of Molecular Sciences 21, no. 4: 1518. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21041518