N-terminal Backbone Pairing Shifts in CCL5-12AAA14 Dimer Interface: Structural Significance of the FAY Sequence
Abstract
:1. Introduction
2. Results
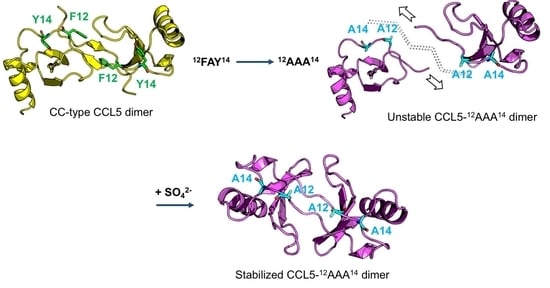
2.1. CCL5-12AAA14 Mutant
2.2. Single Mutations of F12A and Y14A
2.3. Sulfate Stabilized the CCL5-12AAA14 Dimer
2.4. Crystal Structure of CCL5-12AAA14
2.5. N-terminal Backbone Pairing of CCL5-12AAA14 Dimer
2.6. NMR Secondary Structural Prediction
2.7. NMR Backbone Dynamics
2.8. The Interface of the CCL5-12AAA14 Dimer
2.9. Sulfate Binding on CCL5
3. Discussion
4. Materials and Methods
4.1. Protein Preparation
4.2. X-ray Crystallization
4.3. NMR HSQC Spectroscopy for Titration
4.4. NMR Backbone Assignment
4.5. NMR Relaxation Experiments
5. Conclusions
6. Accession Number
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Raman, D.; Sobolik-Delmaire, T.; Richmond, A. Chemokines in health and disease. Exp. Cell Res. 2011, 317, 575–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appay, V.; Rowland-Jones, S.L. RANTES: A versatile and controversial chemokine. Trends Immunol. 2001, 22, 83–87. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. Chemokines: A new classification system and their role in immunity. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef] [Green Version]
- Schall, T.J.; Jongstra, J.; Dyer, B.J.; Jorgensen, J.; Clayberger, C.; Davis, M.M.; Krensky, A.M. A human T cell-specific molecule is a member of a new gene family. J. Immunol. 1988, 141, 1018–1025. [Google Scholar] [PubMed]
- Stone, M.J.; Hayward, J.A.; Huang, C.; E Huma, Z.; Sanchez, J. Mechanisms of Regulation of the Chemokine-Receptor Network. Int. J. Mol. Sci. 2017, 18, 342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proudfoot, A.E.I.; Johnson, Z.; Bonvin, P.; Handel, T.M. Glycosaminoglycan Interactions with Chemokines Add Complexity to a Complex System. Pharmaceuticals (Basel) 2017, 10, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyer, D.P.; Salanga, C.L.; Volkman, B.F.; Kawamura, T.; Handel, T.M. The dependence of chemokine-glycosaminoglycan interactions on chemokine oligomerization. Glycobiology 2016, 26, 312–326. [Google Scholar] [CrossRef]
- Wang, X.; Sharp, J.S.; Handel, T.M.; Prestegard, J.H. Chemokine oligomerization in cell signaling and migration. Prog. Mol. Biol. Transl. Sci. 2013, 117, 531–578. [Google Scholar]
- Proudfoot, A.E.; Handel, T.M.; Johnson, Z.; Lau, E.K.; LiWang, P.; Clark-Lewis, I.; Borlat, F.; Wells, T.N.; Kosco-Vilbois, M.H. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc. Natl. Acad. Sci. USA 2003, 100, 1885–1890. [Google Scholar] [CrossRef] [Green Version]
- Bacon, K.B.; Premack, B.A.; Gardner, P.; Schall, T.J. Activation of dual T cell signaling pathways by the chemokine RANTES. Science 1995, 269, 1727–1730. [Google Scholar] [CrossRef] [PubMed]
- Bacon, K.B.; Szabo, M.C.; Yssel, H.; Bolen, J.B.; Schall, T.J. RANTES induces tyrosine kinase activity of stably complexed p125FAK and ZAP-70 in human T cells. J. Exp. Med. 1996, 184, 873–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szabo, M.C.; Butcher, E.C.; McIntyre, B.W.; Schall, T.J.; Bacon, K.B. RANTES stimulation of T lymphocyte adhesion and activation: Role for LFA-1 and ICAM-3. Eur. J. Immunol. 1997, 27, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Watson, C.; Sharp, J.S.; Handel, T.M.; Prestegard, J.H. Oligomeric structure of the chemokine CCL5/RANTES from NMR, MS, and SAXS data. Structure 2011, 19, 1138–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.C.; Chen, S.P.; Li, J.Y.; Chen, P.C.; Lee, Y.Z.; Li, K.M.; Zarivach, R.; Sun, Y.J.; Sue, S.C. Integrative model to coordinate the oligomerization and aggregation mechanisms of CCL5. J. Mol. Biol. 2020, 432, 1143–1157. [Google Scholar] [CrossRef] [PubMed]
- Appay, V.; Brown, A.; Cribbes, S.; Randle, E.; Czaplewski, L.G. Aggregation of RANTES is responsible for its inflammatory properties. Characterization of nonaggregating, noninflammatory RANTES mutants. J. Biol. Chem. 1999, 274, 27505–27512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czaplewski, L.G.; McKeating, J.; Craven, C.J.; Higgins, L.D.; Appay, V.; Brown, A.; Dudgeon, T.; Howard, L.A.; Meyers, T.; Owen, J.; et al. Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1a, MIP-1b, and RANTES. Characterization of active disaggregated chemokine variants. J. Biol. Chem. 1999, 274, 16077–16084. [Google Scholar] [CrossRef] [Green Version]
- Duma, L.; Haussinger, D.; Rogowski, M.; Lusso, P.; Grzesiek, S. Recognition of RANTES by extracellular parts of the CCR5 receptor. J. Mol. Biol. 2007, 365, 1063–1075. [Google Scholar] [CrossRef]
- Skelton, N.J.; Aspiras, F.; Ogez, J.; Schall, T.J. Proton NMR assignments and solution conformation of RANTES, a chemokine of the C-C type. Biochemistry 1995, 34, 5329–5342. [Google Scholar] [CrossRef]
- Chung, C.W.; Cooke, R.M.; Proudfoot, A.E.; Wells, T.N. The three-dimensional solution structure of RANTES. Biochemistry 1995, 34, 9307–9314. [Google Scholar] [CrossRef]
- Liang, W.G.; Triandafillou, C.G.; Huang, T.Y.; Zulueta, M.M.; Banerjee, S.; Dinner, A.R.; Hung, S.C.; Tang, W.J. Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3. Proc. Natl. Acad. Sci. USA 2016, 113, 5000–5005. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.C.; Li, K.M.; Zarivach, R.; Sun, Y.J.; Sue, S.C. Human CCL5 trimer: Expression, purification and initial crystallographic studies. Acta Crystallogr. Sect. F 2018, 74, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Pakianathan, D.R.; Kuta, E.G.; Artis, D.R.; Skelton, N.J.; Hebert, C.A. Distinct but overlapping epitopes for the interaction of a CC-chemokine with CCR1, CCR3 and CCR5. Biochemistry 1997, 36, 9642–9648. [Google Scholar] [CrossRef] [PubMed]
- Laurence, J.S.; Blanpain, C.; Burgner, J.W.; Parmentier, M.; LiWang, P.J. CC chemokine MIP-1 beta can function as a monomer and depends on Phe13 for receptor binding. Biochemistry 2000, 39, 3401–3409. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Wiktor, M.; Hartley, O.; Grzesiek, S. Characterization of structure, dynamics, and detergent interactions of the anti-HIV chemokine variant 5P12-RANTES. Biophys. J. 2013, 105, 2586–2597. [Google Scholar] [CrossRef] [Green Version]
- Blain, K.Y.; Kwiatkowski, W.; Zhao, Q.; La Fleur, D.; Naik, C.; Chun, T.W.; Tsareva, T.; Kanakaraj, P.; Laird, M.W.; Shah, R.; et al. Structural and functional characterization of CC chemokine CCL14. Biochemistry 2007, 46, 10008–10015. [Google Scholar] [CrossRef]
- Shinkai, A.; Yoshisue, H.; Koike, M.; Shoji, E.; Nakagawa, S.; Saito, A.; Takeda, T.; Imabeppu, S.; Kato, Y.; Hanai, N.; et al. A novel human CC chemokine, eotaxin-3, which is expressed in IL-4-stimulated vascular endothelial cells, exhibits potent activity toward eosinophils. J. Immunol. 1999, 163, 1602–1610. [Google Scholar]
- Zhang, X.; Chen, L.; Bancroft, D.P.; Lai, C.K.; Maione, T.E. Crystal structure of recombinant human platelet factor 4. Biochemistry 1994, 33, 8361–8366. [Google Scholar] [CrossRef]
- Chen, Y.P.; Wu, H.L.; Boye, K.; Pan, C.Y.; Chen, Y.C.; Pujol, N.; Lin, C.W.; Chiu, L.Y.; Billottet, C.; Alves, I.D.; et al. Oligomerization State of CXCL4 Chemokines Regulates G Protein-Coupled Receptor Activation. ACS Chem. Biol. 2017, 12, 2767–2778. [Google Scholar] [CrossRef]
- Malkowski, M.G.; Wu, J.Y.; Lazar, J.B.; Johnson, P.H.; Edwards, B.F. The crystal structure of recombinant human neutrophil-activating peptide-2 (M6L) at 1.9-Å resolution. J. Biol. Chem. 1995, 270, 7077–7087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swaminathan, G.J.; Holloway, D.E.; Colvin, R.A.; Campanella, G.K.; Papageorgiou, A.C.; Luster, A.D.; Acharya, K.R. Crystal structures of oligomeric forms of the IP-10/CXCL10 chemokine. Structure 2003, 11, 521–532. [Google Scholar] [CrossRef]
- Asojo, O.A.; Boulegue, C.; Hoover, D.M.; Lu, W.; Lubkowski, J. Structures of thymus and activation-regulated chemokine (TARC). Acta Crystallogr. Sect. D Biol. Crystallogr. 2003, 59, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, A.E.; Fritchley, S.; Borlat, F.; Shaw, J.P.; Vilbois, F.; Zwahlen, C.; Trkola, A.; Marchant, D.; Clapham, P.R.; Wells, T.N. The BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. J. Biol. Chem. 2001, 276, 10620–10626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, J.P.; Johnson, Z.; Borlat, F.; Zwahlen, C.; Kungl, A.; Roulin, K.; Harrenga, A.; Wells, T.N.; Proudfoot, A.E. The X-ray structure of RANTES: Heparin-derived disaccharides allows the rational design of chemokine inhibitors. Structure 2004, 12, 2081–2093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Kett, W.C.; Severin, I.C.; Agyekum, I.; Duan, J.; Amster, I.J.; Proudfoot, A.E.; Coombe, D.R.; Woods, R.J. The Interaction of Heparin Tetrasaccharides with Chemokine CCL5 Is Modulated by Sulfation Pattern and pH. J. Biol. Chem. 2015, 290, 15421–15436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshauer, C.; Morgan, A.M.; Ryan, E.O.; Handel, T.M.; Prestegard, J.H.; Wang, X. Interactions of the Chemokine CCL5/RANTES with Medium-Sized Chondroitin Sulfate Ligands. Structure 2015, 23, 1066–1077. [Google Scholar] [CrossRef] [Green Version]
- Abayev, M.; Rodrigues, J.; Srivastava, G.; Arshava, B.; Jaremko, L.; Jaremko, M.; Naider, F.; Levitt, M.; Anglister, J. The solution structure of monomeric CCL5 in complex with a doubly sulfated N-terminal segment of CCR5. FEBS J. 2018, 285, 1988–2003. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delaglio, F.; Walker, G.S.; Farley, K.A.; Sharma, R.; Hoch, J.C.; Arbogast, L.W.; Brinson, R.G.; Marino, J.P. Non-Uniform Sampling for All: More NMR Spectral Quality, Less Measurement Time. Am. Pharm. Rev. 2017, 20, 339681. [Google Scholar] [PubMed]
- Leopold, M.F.; Urbauer, J.L.; Wand, A.J. Resonance assignment strategies for the analysis of NMR spectra of proteins. Mol. Biotechnol. 1994, 2, 61–93. [Google Scholar] [CrossRef] [PubMed]
- Ishima, R.; Torchia, D.A. Protein dynamics from NMR. Nat. Struct. Biol. 2000, 7, 740–743. [Google Scholar] [CrossRef] [PubMed]









| Data Collection | |
|---|---|
| Molecule | CCL5-12AAA14 |
| Beamline | NSRRC TPS 05A1 |
| Wavelength (Å) | 0.99984 |
| Space Group | P3121 |
| Cell Dimensions | |
| a, b, c (Å) | 48.3, 48.3, 60.4 |
| α, β, γ (o) | 90, 90, 120 |
| Resolution (Å) a | 25.0–2.55 (2.64–2.55) |
| Rmergeb | 0.041 (0.359) |
| Rmeasc | 0.049 (0.319) |
| Rpimd | 0.026 (0.212) |
| CC1/2 e | 0.978 (0.906) |
| I/σI | 31.3 (2.2) |
| Completeness (%) | 99.4 (100.0) |
| Total no. Unique Reflections | 2866 (269) |
| Multiplicity | 3.4 (3.4) |
| Refinement | |
| Resolution (Å) | 18.88–2.55 (2.61–2.55) |
| Unique Reflections | 2749 (103) |
| Rwork (%) | 21.4 (32.3) |
| Rfree (%) | 24.9 (44.7) |
| No. Atoms | |
| Protein | 501 |
| Water | 7 |
| B-Factors | |
| Protein | 83.06 |
| Water | 80.47 |
| RMSD | |
| Bond Lengths (Å) | 0.011 |
| Angles (°) | 2.23 |
| Ramachandran | |
| Most Favored (%) | 93.5 |
| Additional Allowed (%) | 4.9 |
| Generously Allowed (%) | 1.6 |
| Disallowed (%) | 0.0 |
| Clash Score | 9.02 |
| PDB Accession | 6LOG |
| pH | Oligomeric State | R1 (s−1) | R2 (s−1) | NOE | |
|---|---|---|---|---|---|
| Mouse CCL5 a | 3.8 | Dimer | 1.27 ± 0.05 | 11.21 ± 0.43 | 0.69 ± 0.03 |
| 5P12-E66S CCL5 b | 3.2 | Monomer | 2.26 ± 0.13 | 6.08 ± 0.78 | 0.71 ± 0.04 |
| CCL5-12AAA14 | 3.2 | Dimer | 1.38 ± 0.013 | 10.31 ± 0.073 | 0.68 ± 0.027 |
| CCL5-12AAA14 (with SO42-) | 3.2 | Dimer | 1.26 ± 0.015 | 11.47 ± 0.097 | 0.70 ± 0.032 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.-Y.; Chen, Y.-C.; Lee, Y.-Z.; Huang, C.-H.; Sue, S.-C. N-terminal Backbone Pairing Shifts in CCL5-12AAA14 Dimer Interface: Structural Significance of the FAY Sequence. Int. J. Mol. Sci. 2020, 21, 1689. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051689
Li J-Y, Chen Y-C, Lee Y-Z, Huang C-H, Sue S-C. N-terminal Backbone Pairing Shifts in CCL5-12AAA14 Dimer Interface: Structural Significance of the FAY Sequence. International Journal of Molecular Sciences. 2020; 21(5):1689. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051689
Chicago/Turabian StyleLi, Jin-Ye, Yi-Chen Chen, Yi-Zong Lee, Chun-Hsiang Huang, and Shih-Che Sue. 2020. "N-terminal Backbone Pairing Shifts in CCL5-12AAA14 Dimer Interface: Structural Significance of the FAY Sequence" International Journal of Molecular Sciences 21, no. 5: 1689. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051689