Heterogeneity of Circulating Tumor Cells in Breast Cancer: Identifying Metastatic Seeds
Abstract
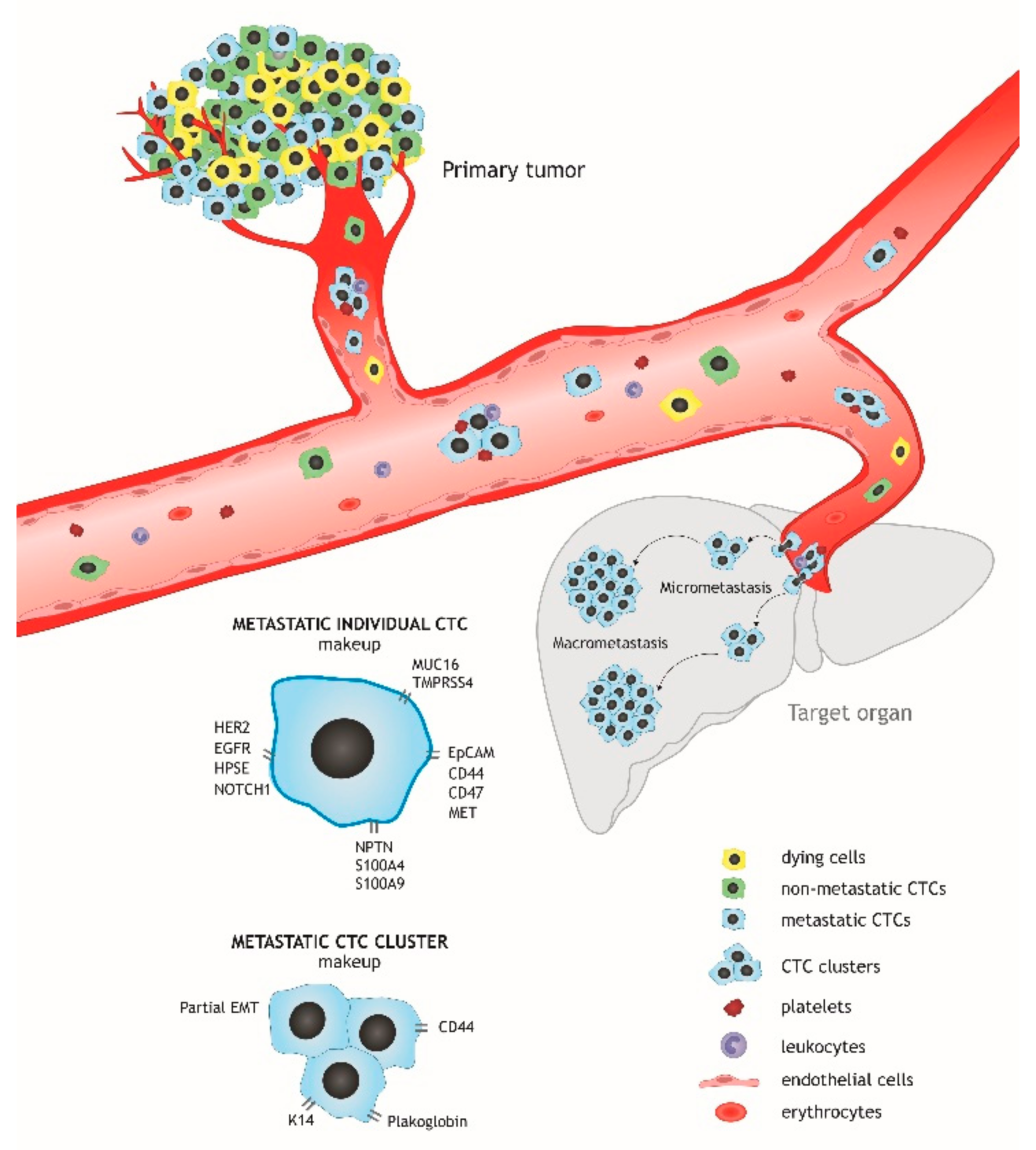
:1. Introduction
2. CTC Heterogeneity and the Molecular Makeup of Metastatic Cells
2.1. Morphological Heterogeneity. Circulating Clusters
2.2. Phenotypic Heterogeneity
2.3. Genetic Heterogeneity
3. In Vitro and in Vivo Study of the CTC Phenotype
4. Therapeutic Targeting of Metastatic CTCs
4.1. Chemo- and Targeted Therapy Based on Molecular Characteristics of CTCs
4.2. Impairment of CTC Adhesion to Hematopoietic Cells and Platelets
4.3. Elimination of CTCs
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lafourcade, A.; His, M.; Baglietto, L.; Boutron-Ruault, M.C.; Dossus, L.; Rondeau, V. Factors associated with breast cancer recurrences or mortality and dynamic prediction of death using history of cancer recurrences: The French E3N cohort. BMC Cancer 2018, 18, 171. [Google Scholar] [CrossRef] [PubMed]
- Dillekås, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christowitz, C.; Davis, T.; Isaacs, A.; van Niekerk, G.; Hattingh, S.; Engelbrecht, A.-M. Mechanisms of doxorubicin-induced drug resistance and drug resistant tumour growth in a murine breast tumour model. BMC Cancer 2019, 19, 757. [Google Scholar] [CrossRef] [Green Version]
- Jie, X.X.; Zhang, X.Y.; Xu, C.J. Epithelial-to-mesenchymal transition, circulating tumor cells and cancer metastasis: Mechanisms and clinical applications. Oncotarget 2017, 8, 81558–81571. [Google Scholar] [CrossRef] [Green Version]
- van de Vijver, M.J.; He, Y.D.; van’t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef] [Green Version]
- Vanharanta, S.; Massague, J. Origins of metastatic traits. Cancer Cell 2013, 24, 410–421. [Google Scholar] [CrossRef] [Green Version]
- Bulfoni, M.; Turetta, M.; Del Ben, F.; Di Loreto, C.; Beltrami, A.P.; Cesselli, D. Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack. Int. J. Mol. Sci. 2016, 17, 1775. [Google Scholar] [CrossRef]
- Paoletti, C.; Muñiz, M.C.; Thomas, D.G.; Griffith, K.A.; Kidwell, K.M.; Tokudome, N.; Brown, M.E.; Aung, K.; Miller, M.C.; Blossom, D.L.; et al. Development of circulating tumor cell-endocrine therapy index in patients with hormone receptor-positive breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 2487–2498. [Google Scholar] [CrossRef] [Green Version]
- Saxena, K.; Subbalakshmi, A.R.; Jolly, M.K. Phenotypic heterogeneity in circulating tumor cells and its prognostic value in metastasis and overall survival. EBioMedicine 2019, 46, 4–5. [Google Scholar] [CrossRef] [Green Version]
- Cherdyntseva, N.V.; Litviakov, N.V.; Denisov, E.V.; Gervas, P.A.; Cherdyntsev, E.S. Circulating tumor cells in breast cancer: Functional heterogeneity, pathogenetic and clinical aspects. Exp. Oncol. 2017, 39, 2–11. [Google Scholar] [CrossRef]
- Tellez-Gabriel, M.; Heymann, M.F.; Heymann, D. Circulating Tumor Cells as a Tool for Assessing Tumor Heterogeneity. Theranostics 2019, 9, 4580–4594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wu, T.; Peng, X.; Liu, J.; Liu, F.; Wu, S.; Liu, S.; Dong, Y.; Xie, S.; Ma, S. Mesenchymal phenotype of circulating tumor cells is associated with distant metastasis in breast cancer patients. Cancer Manag. Res. 2017, 9, 691–700. [Google Scholar] [CrossRef] [Green Version]
- Guan, X.; Ma, F.; Li, C.; Wu, S.; Hu, S.; Huang, J.; Sun, X.; Wang, J.; Luo, Y.; Cai, R.; et al. The prognostic and therapeutic implications of circulating tumor cell phenotype detection based on epithelial-mesenchymal transition markers in the first-line chemotherapy of HER2-negative metastatic breast cancer. Cancer Commun. 2019, 39, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Massague, J.; Obenauf, A.C. Metastatic colonization by circulating tumour cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Agnoletto, C.; Corrà, F.; Minotti, L.; Baldassari, F.; Crudele, F.; Cook, W.J.J.; Di Leva, G.; d’Adamo, A.P.; Gasparini, P.; Volinia, S. Heterogeneity in Circulating Tumor Cells: The Relevance of the Stem-Cell Subset. Cancers 2019, 11, 483. [Google Scholar] [CrossRef] [Green Version]
- Pantel, K.; Brakenhoff, R.H. Dissecting the metastatic cascade. Nat. Rev. Cancer 2004, 4, 448–456. [Google Scholar] [CrossRef]
- Grosse-Wilde, A.; Fouquier d’Herouel, A.; McIntosh, E.; Ertaylan, G.; Skupin, A.; Kuestner, R.E.; del Sol, A.; Walters, K.A.; Huang, S. Stemness of the hybrid Epithelial/Mesenchymal State in Breast Cancer and Its Association with Poor Survival. PLoS ONE 2015, 10, e0126522. [Google Scholar] [CrossRef]
- Jia, D.; Jolly, M.K.; Boareto, M.; Parsana, P.; Mooney, S.M.; Pienta, K.J.; Levine, H.; Ben-Jacob, E. OVOL guides the epithelial-hybrid-mesenchymal transition. Oncotarget 2015, 6, 15436–15448. [Google Scholar] [CrossRef] [Green Version]
- Kaigorodova, E.; Tarabanovskaya, N.; Staheeva, M.; Savelieva, O.; Tashireva, L.; Denisov, E.; Perelmuter, V. Effect of small and radical surgical injury on the level of different populations of circulating tumor cells in the blood of breast cancer patients. Neoplasma 2017, 64, 437–443. [Google Scholar] [CrossRef]
- Aaltonen, K.E.; Novosadová, V.; Bendahl, P.O.; Graffman, C.; Larsson, A.M.; Rydén, L. Molecular characterization of circulating tumor cells from patients with metastatic breast cancer reflects evolutionary changes in gene expression under the pressure of systemic therapy. Oncotarget 2017, 8, 45544–45565. [Google Scholar] [CrossRef] [PubMed]
- Lianidou, E.S.; Mavroudis, D.; Sotiropoulou, G.; Agelaki, S.; Pantel, K. What’s new on circulating tumor cells? A meeting report. Breast Cancer Res. BCR 2010, 12, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.H.; Chen, Y.C.; Lin, E.; Brien, R.; Jung, S.; Chen, Y.T.; Lee, W.; Hao, Z.; Sahoo, S.; Min Kang, H.; et al. Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells. Nat. Commun. 2019, 10, 2163. [Google Scholar] [CrossRef] [PubMed]
- D’Avola, D.; Villacorta-Martin, C.; Martins-Filho, S.N.; Craig, A.; Labgaa, I.; von Felden, J.; Kimaada, A.; Bonaccorso, A.; Tabrizian, P.; Hartmann, B.M.; et al. High-density single cell mRNA sequencing to characterize circulating tumor cells in hepatocellular carcinoma. Sci. Rep. 2018, 8, 11570. [Google Scholar] [CrossRef]
- Garcia, J.; Monjaret, F.; Geiguer, F.; Aurel, J.-P.; Puisieux, A.; Morel, A.P.; Bardel, C.; Vilchez, G.; Brigitte, M.; Couraud, S.; et al. 46PWhole transcriptomics analyses of mimicking circulating tumor cells (CTCs) by single-cell RNA sequencing (scRNAseq). Ann. Oncol. 2019, 30. [Google Scholar] [CrossRef]
- Wu, C.P.; Wu, P.; Zhao, H.F.; Liu, W.L.; Li, W.P. Clinical Applications of and Challenges in Single-Cell Analysis of Circulating Tumor Cells. DNA Cell Biol. 2018, 37, 78–89. [Google Scholar] [CrossRef]
- Sharma, S.; Zhuang, R.; Long, M.; Pavlovic, M.; Kang, Y.; Ilyas, A.; Asghar, W. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol. Adv. 2018, 36, 1063–1078. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [Green Version]
- Yap, Y.S.; Leong, M.C.; Chua, Y.W.; Loh, K.W.J.; Lee, G.E.; Lim, E.H.; Dent, R.; Ng, R.C.H.; Lim, J.H.; Singh, G.; et al. Detection and prognostic relevance of circulating tumour cells (CTCs) in Asian breast cancers using a label-free microfluidic platform. PLoS ONE 2019, 14, e0221305. [Google Scholar] [CrossRef]
- Luzzi, K.J.; MacDonald, I.C.; Schmidt, E.E.; Kerkvliet, N.; Morris, V.L.; Chambers, A.F.; Groom, A.C. Multistep nature of metastatic inefficiency: Dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am. J. Pathol. 1998, 153, 865–873. [Google Scholar] [CrossRef]
- Steeg, P.S. Tumor metastasis: Mechanistic insights and clinical challenges. Nat. Med. 2006, 12, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Tayoun, T.; Faugeroux, V.; Oulhen, M.; Aberlenc, A.; Pawlikowska, P.; Farace, F. CTC-Derived Models: A Window into the Seeding Capacity of Circulating Tumor Cells (CTCs). Cells 2019, 8, 1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bidard, F.C.; Peeters, D.J.; Fehm, T.; Nole, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Balasubramanian, P.; Chen, A.P.; Kummar, S.; Evrard, Y.A.; Kinders, R.J. Promise and limits of the CellSearch platform for evaluating pharmacodynamics in circulating tumor cells. Semin. Oncol. 2016, 43, 464–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cristofanilli, M.; Hayes, D.F.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Reuben, J.M.; Doyle, G.V.; Matera, J.; Allard, W.J.; Miller, M.C.; et al. Circulating tumor cells: A novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 1420–1430. [Google Scholar] [CrossRef] [PubMed]
- Dolfus, C.; Piton, N.; Toure, E.; Sabourin, J.C. Circulating tumor cell isolation: The assets of filtration methods with polycarbonate track-etched filters. Chin. J. Cancer Res. Chung-Kuo Yen Cheng Yen Chiu 2015, 27, 479–487. [Google Scholar] [CrossRef]
- Riahi, R.; Gogoi, P.; Sepehri, S.; Zhou, Y.; Handique, K.; Godsey, J.; Wang, Y. A novel microchannel-based device to capture and analyze circulating tumor cells (CTCs) of breast cancer. Int. J. Oncol. 2014, 44, 1870–1878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hattori, M.; Nakanishi, H.; Yoshimura, M.; Iwase, M.; Yoshimura, A.; Adachi, Y.; Gondo, N.; Kotani, H.; Sawaki, M.; Fujita, N. Circulating tumor cells detection in tumor draining vein of breast cancer patients. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef] [Green Version]
- Krebs, M.G.; Hou, J.M.; Ward, T.H.; Blackhall, F.H.; Dive, C. Circulating tumour cells: Their utility in cancer management and predicting outcomes. Ther. Adv. Med Oncol. 2010, 2, 351–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danila, D.C.; Pantel, K.; Fleisher, M.; Scher, H.I. Circulating tumors cells as biomarkers: Progress toward biomarker qualification. Cancer J. (Sudbury Mass.) 2011, 17, 438–450. [Google Scholar] [CrossRef] [Green Version]
- Molnar, B.; Ladanyi, A.; Tanko, L.; Sreter, L.; Tulassay, Z. Circulating tumor cell clusters in the peripheral blood of colorectal cancer patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2001, 7, 4080–4085. [Google Scholar]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuliano, M.; Shaikh, A.; Lo, H.C.; Arpino, G.; De Placido, S.; Zhang, X.H.; Cristofanilli, M.; Schiff, R.; Trivedi, M.V. Perspective on Circulating Tumor Cell Clusters: Why It Takes a Village to Metastasize. Cancer Res. 2018, 78, 845–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, K.J.; Padmanaban, V.; Silvestri, V.; Schipper, K.; Cohen, J.D.; Fairchild, A.N.; Gorin, M.A.; Verdone, J.E.; Pienta, K.J.; Bader, J.S.; et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci. USA 2016, 113, E854–E863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Taftaf, R.; Kawaguchi, M.; Chang, Y.F.; Chen, W.; Entenberg, D.; Zhang, Y.; Gerratana, L.; Huang, S.; Patel, D.B.; et al. Homophilic CD44 Interactions Mediate Tumor Cell Aggregation and Polyclonal Metastasis in Patient-Derived Breast Cancer Models. Cancer Discov. 2019, 9, 96–113. [Google Scholar] [CrossRef] [Green Version]
- Paoletti, C.; Miao, J.; Dolce, E.M.; Darga, E.P.; Repollet, M.I.; Doyle, G.V.; Gralow, J.R.; Hortobagyi, G.N.; Smerage, J.B.; Barlow, W.E.; et al. Circulating Tumor Cell Clusters in Patients with Metastatic Breast Cancer: A SWOG S0500 Translational Medicine Study. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 6089–6097. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [Green Version]
- Li, C.F.; Chen, J.Y.; Ho, Y.H.; Hsu, W.H.; Wu, L.C.; Lan, H.Y.; Hsu, D.S.; Tai, S.K.; Chang, Y.C.; Yang, M.H. Snail-induced claudin-11 prompts collective migration for tumour progression. Nat. Cell Biol. 2019, 21, 251–262. [Google Scholar] [CrossRef]
- Gkountela, S.; Castro-Giner, F.; Szczerba, B.M.; Vetter, M.; Landin, J.; Scherrer, R.; Krol, I.; Scheidmann, M.C.; Beisel, C.; Stirnimann, C.U.; et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 2019, 176, 98–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thangavel, H.; Angelis, C.; Vasaikar, S.; Bhat, R.; Jolly, M.K.; Nagi, C.; Creighton, C.J.; Chen, F.; Dobrolecki, L.E.; George, J.T.; et al. A CTC-Cluster-Specific Signature Derived from OMICS Analysis of Patient-Derived Xenograft Tumors Predicts Outcomes in Basal-Like Breast Cancer. J. Clin. Med. 2019, 8, 1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Zeinali, M.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; Miyamoto, D.T.; et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 2015, 12, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Tripathi, S.C.; Jia, D.; Mooney, S.M.; Celiktas, M.; Hanash, S.M.; Mani, S.A.; Pienta, K.J.; Ben-Jacob, E.; Levine, H. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget 2016, 7, 27067–27084. [Google Scholar] [CrossRef] [Green Version]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef]
- Fehm, T.; Hoffmann, O.; Aktas, B.; Becker, S.; Solomayer, E.F.; Wallwiener, D.; Kimmig, R.; Kasimir-Bauer, S. Detection and characterization of circulating tumor cells in blood of primary breast cancer patients by RT-PCR and comparison to status of bone marrow disseminated cells. Breast Cancer Res. BCR 2009, 11, R59. [Google Scholar] [CrossRef] [Green Version]
- Pantel, K.; Schlimok, G.; Braun, S.; Kutter, D.; Lindemann, F.; Schaller, G.; Funke, I.; Izbicki, J.R.; Riethmuller, G. Differential expression of proliferation-associated molecules in individual micrometastatic carcinoma cells. J. Natl. Cancer Inst. 1993, 85, 1419–1424. [Google Scholar] [CrossRef]
- Muller, V.; Stahmann, N.; Riethdorf, S.; Rau, T.; Zabel, T.; Goetz, A.; Janicke, F.; Pantel, K. Circulating tumor cells in breast cancer: Correlation to bone marrow micrometastases, heterogeneous response to systemic therapy and low proliferative activity. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 3678–3685. [Google Scholar] [CrossRef] [Green Version]
- Spiliotaki, M.; Mavroudis, D.; Kapranou, K.; Markomanolaki, H.; Kallergi, G.; Koinis, F.; Kalbakis, K.; Georgoulias, V.; Agelaki, S. Evaluation of proliferation and apoptosis markers in circulating tumor cells of women with early breast cancer who are candidates for tumor dormancy. Breast Cancer Res. BCR 2014, 16, 485. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.B.; Di Lee, W.; Vasudevan, J.; Lim, W.T.; Lim, C.T. Liquid biopsy: One cell at a time. NPJ Precis. Oncol. 2019, 3, 23. [Google Scholar] [CrossRef]
- Powell, A.A.; Talasaz, A.H.; Zhang, H.; Coram, M.A.; Reddy, A.; Deng, G.; Telli, M.L.; Advani, R.H.; Carlson, R.W.; Mollick, J.A.; et al. Single cell profiling of circulating tumor cells: Transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS ONE 2012, 7, e33788. [Google Scholar] [CrossRef] [Green Version]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Jakabova, A.; Bielcikova, Z.; Pospisilova, E.; Matkowski, R.; Szynglarewicz, B.; Staszek-Szewczyk, U.; Zemanova, M.; Petruzelka, L.; Eliasova, P.; Kolostova, K.; et al. Molecular characterization and heterogeneity of circulating tumor cells in breast cancer. Breast Cancer Res. Treat. 2017, 166, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Kaigorodova, E.V.; Savelieva, O.E.; Tashireva, L.A.; Tarabanovskaya, N.A.; Simolina, E.I.; Denisov, E.V.; Slonimskaya, E.M.; Choynzonov, E.L.; Perelmuter, V.M. Heterogeneity of Circulating Tumor Cells in Neoadjuvant Chemotherapy of Breast Cancer. Molecules 2018, 23, 727. [Google Scholar] [CrossRef] [Green Version]
- Pizon, M.; Zimon, D.; Carl, S.; Pachmann, U.; Pachmann, K.; Camara, O. Heterogeneity of circulating epithelial tumour cells from individual patients with respect to expression profiles and clonal growth (sphere formation) in breast cancer. Ecancermedicalscience 2013, 7, 343. [Google Scholar] [CrossRef] [PubMed]
- Kwan, T.T.; Bardia, A.; Spring, L.M.; Giobbie-Hurder, A.; Kalinich, M.; Dubash, T.; Sundaresan, T.; Hong, X.; LiCausi, J.A.; Ho, U.; et al. A Digital RNA Signature of Circulating Tumor Cells Predicting Early Therapeutic Response in Localized and Metastatic Breast Cancer. Cancer Discov. 2018, 8, 1286–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vega, S.; Morales, A.V.; Ocaña, O.H.; Valdés, F.; Fabregat, I.; Nieto, M.A. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004, 18, 1131–1143. [Google Scholar] [CrossRef] [Green Version]
- Krawczyk, N.; Meier-Stiegen, F.; Banys, M.; Neubauer, H.; Ruckhaeberle, E.; Fehm, T. Expression of stem cell and epithelial-mesenchymal transition markers in circulating tumor cells of breast cancer patients. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- Zavyalova, M.V.; Denisov, E.V.; Tashireva, L.A.; Savelieva, O.E.; Kaigorodova, E.V.; Krakhmal, N.V.; Perelmuter, V.M. Intravasation as a Key Step in Cancer Metastasis. Biochem. Biokhimiia 2019, 84, 762–772. [Google Scholar] [CrossRef]
- Dongre, A.; Rashidian, M.; Reinhardt, F.; Bagnato, A.; Keckesova, Z.; Ploegh, H.L.; Weinberg, R.A. Epithelial-to-Mesenchymal Transition Contributes to Immunosuppression in Breast Carcinomas. Cancer Res. 2017, 77, 3982–3989. [Google Scholar] [CrossRef] [Green Version]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jolly, M.K.; Somarelli, J.A.; Sheth, M.; Biddle, A.; Tripathi, S.C.; Armstrong, A.J.; Hanash, S.M.; Bapat, S.A.; Rangarajan, A.; Levine, H. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol. Ther. 2019, 194, 161–184. [Google Scholar] [CrossRef] [PubMed]
- Kröger, C.; Afeyan, A.; Mraz, J.; Eaton, E.N.; Reinhardt, F.; Khodor, Y.L.; Thiru, P.; Bierie, B.; Ye, X.; Burge, C.B.; et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7353–7362. [Google Scholar] [CrossRef] [Green Version]
- Bocci, F.; Gearhart-Serna, L.; Boareto, M.; Ribeiro, M.; Ben-Jacob, E.; Devi, G.R.; Levine, H.; Onuchic, J.N.; Jolly, M.K. Toward understanding cancer stem cell heterogeneity in the tumor microenvironment. Proc. Natl. Acad. Sci. USA 2019, 116, 148–157. [Google Scholar] [CrossRef] [Green Version]
- Aktas, B.; Tewes, M.; Fehm, T.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. BCR 2009, 11, R46. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 2013, 5, 180ra48. [Google Scholar] [CrossRef] [Green Version]
- Theodoropoulos, P.A.; Polioudaki, H.; Agelaki, S.; Kallergi, G.; Saridaki, Z.; Mavroudis, D.; Georgoulias, V. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2010, 288, 99–106. [Google Scholar] [CrossRef]
- Armstrong, A.J.; Marengo, M.S.; Oltean, S.; Kemeny, G.; Bitting, R.L.; Turnbull, J.D.; Herold, C.I.; Marcom, P.K.; George, D.J.; Garcia-Blanco, M.A. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol. Cancer Res. MCR 2011, 9, 997–1007. [Google Scholar] [CrossRef] [Green Version]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Abreu, M.; Cabezas-Sainz, P.; Pereira-Veiga, T.; Falo, C.; Abalo, A.; Morilla, I.; Curiel, T.; Cueva, J.; Rodriguez, C.; Varela-Pose, V.; et al. Looking for a Better Characterization of Triple-Negative Breast Cancer by Means of Circulating Tumor Cells. J. Clin. Med. 2020, 9, 353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimondi, C.; Gradilone, A.; Naso, G.; Vincenzi, B.; Petracca, A.; Nicolazzo, C.; Palazzo, A.; Saltarelli, R.; Spremberg, F.; Cortesi, E.; et al. Epithelial-mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Cancer Res. Treat. 2011, 130, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bauerle, T.; Wallwiener, M.; et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013, 31, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Pestrin, M.; Salvianti, F.; Galardi, F.; De Luca, F.; Turner, N.; Malorni, L.; Pazzagli, M.; Di Leo, A.; Pinzani, P. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol. Oncol. 2015, 9, 749–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, S.V.; Bingham, C.; Fittipaldi, P.; Austin, L.; Palazzo, J.; Palmer, G.; Alpaugh, K.; Cristofanilli, M. TP53 mutations detected in circulating tumor cells present in the blood of metastatic triple negative breast cancer patients. Breast Cancer Res. BCR 2014, 16, 445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, J.A.; Guttery, D.S.; Hills, A.; Fernandez-Garcia, D.; Page, K.; Rosales, B.M.; Goddard, K.S.; Hastings, R.K.; Luo, J.; Ogle, O.; et al. Mutation Analysis of Cell-Free DNA and Single Circulating Tumor Cells in Metastatic Breast Cancer Patients with High Circulating Tumor Cell Counts. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 88–96. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Guo, L.; Feng, L.; Zhang, W.; Xiao, T.; Di, X.; Chen, G.; Zhang, K. Single nucleotide variant profiles of viable single circulating tumour cells reveal CTC behaviours in breast cancer. Oncol. Rep. 2018, 39, 2147–2159. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.B.; Lim, C.T.; Lim, W.T. Single-Cell Analysis of Circulating Tumor Cells: Why Heterogeneity Matters. Cancers 2019, 11, 1595. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Zhao, L.; Han, L.; Tuo, X.; Ma, S.; Wang, Y.; Feng, X.; Liang, D.; Sun, C.; Wang, Q.; et al. The Discordance of Gene Mutations between Circulating Tumor Cells and Primary/Metastatic Tumor. Mol. Ther. Oncolytics 2019, 15, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Paoletti, C.; Cani, A.K.; Larios, J.M.; Hovelson, D.H.; Aung, K.; Darga, E.P.; Cannell, E.M.; Baratta, P.J.; Liu, C.J.; Chu, D.; et al. Comprehensive Mutation and Copy Number Profiling in Archived Circulating Breast Cancer Tumor Cells Documents Heterogeneous Resistance Mechanisms. Cancer Res. 2018, 78, 1110–1122. [Google Scholar] [CrossRef] [Green Version]
- Joosse, S.A.; Brandwijk, K.I.; Mulder, L.; Wesseling, J.; Hannemann, J.; Nederlof, P.M. Genomic signature of BRCA1 deficiency in sporadic basal-like breast tumors. Genes Chromosomes Cancer 2011, 50, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Joosse, S.A.; van Beers, E.H.; Tielen, I.H.; Horlings, H.; Peterse, J.L.; Hoogerbrugge, N.; Ligtenberg, M.J.; Wessels, L.F.; Axwijk, P.; Verhoef, S.; et al. Prediction of BRCA1-association in hereditary non-BRCA1/2 breast carcinomas with array-CGH. Breast Cancer Res. Treat. 2009, 116, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Kawazu, M.; Kojima, S.; Ueno, T.; Totoki, Y.; Nakamura, H.; Kunita, A.; Qu, W.; Yoshimura, J.; Soda, M.; Yasuda, T.; et al. Integrative analysis of genomic alterations in triple-negative breast cancer in association with homologous recombination deficiency. PLoS Genet. 2017, 13, e1006853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klotz, R.; Thomas, A.; Teng, T.; Han, S.M.; Iriondo, O.; Li, L.; Restrepo-Vassalli, S.; Wang, A.; Izadian, N.; MacKay, M.; et al. Circulating Tumor Cells Exhibit Metastatic Tropism and Reveal Brain Metastasis Drivers. Cancer Discov. 2020, 10, 86–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamińska, A.; Szymborski, T.; Witkowska, E.; Kijeńska-Gawrońska, E.; Świeszkowski, W.; Niciński, K.; Trzcińska-Danielewicz, J.; Girstun, A. Detection of Circulating Tumor Cells Using Membrane-Based SERS Platform: A New Diagnostic Approach for ‘Liquid Biopsy’. Nanomaterials 2019, 9, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millner, L.M.; Linder, M.W.; Valdes, R., Jr. Circulating tumor cells: A review of present methods and the need to identify heterogeneous phenotypes. Ann. Clin. Lab. Sci. 2013, 43, 295–304. [Google Scholar]
- Tellez-Gabriel, M.; Cochonneau, D.; Cadé, M.; Jubellin, C.; Heymann, M.F.; Heymann, D. Circulating Tumor Cell-Derived Pre-Clinical Models for Personalized Medicine. Cancers 2018, 11, 19. [Google Scholar] [CrossRef] [Green Version]
- Cayrefourcq, L.; Mazard, T.; Joosse, S.; Solassol, J.; Ramos, J.; Assenat, E.; Schumacher, U.; Costes, V.; Maudelonde, T.; Pantel, K.; et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res. 2015, 75, 892–901. [Google Scholar] [CrossRef] [Green Version]
- Bobek, V.; Matkowski, R.; Gurlich, R.; Grabowski, K.; Szelachowska, J.; Lischke, R.; Schutzner, J.; Harustiak, T.; Pazdro, A.; Rzechonek, A.; et al. Cultivation of circulating tumor cells in esophageal cancer. Folia Histochem. Cytobiol. 2014, 52, 171–177. [Google Scholar] [CrossRef] [Green Version]
- Cegan, M.; Kolostova, K.; Matkowski, R.; Broul, M.; Schraml, J.; Fiutowski, M.; Bobek, V. In vitro culturing of viable circulating tumor cells of urinary bladder cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 7164–7171. [Google Scholar]
- Bobek, V.; Gurlich, R.; Eliasova, P.; Kolostova, K. Circulating tumor cells in pancreatic cancer patients: Enrichment and cultivation. World J. Gastroenterol. 2014, 20, 17163–17170. [Google Scholar] [CrossRef] [PubMed]
- Kolostova, K.; Matkowski, R.; Gürlich, R.; Grabowski, K.; Soter, K.; Lischke, R.; Schützner, J.; Bobek, V. Detection and cultivation of circulating tumor cells in gastric cancer. Cytotechnology 2016, 68, 1095–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thoma, C.R.; Zimmermann, M.; Agarkova, I.; Kelm, J.M.; Krek, W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Deliv. Rev. 2014, 69, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Weiswald, L.B.; Bellet, D.; Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nath, S.; Devi, G.R. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 2016, 163, 94–108. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Shiratsuchi, H.; Lin, J.; Chen, G.; Reddy, R.M.; Azizi, E.; Fouladdel, S.; Chang, A.C.; Lin, L.; Jiang, H.; et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget 2014, 5, 12383–12397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, Y.R.; Leung, A.W. Next generation organoids for biomedical research and applications. Biotechnol. Adv. 2018, 36, 132–149. [Google Scholar] [CrossRef]
- Weeber, F.; van de Wetering, M.; Hoogstraat, M.; Dijkstra, K.K.; Krijgsman, O.; Kuilman, T.; Gadellaa-van Hooijdonk, C.G.; van der Velden, D.L.; Peeper, D.S.; Cuppen, E.P.; et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl. Acad. Sci. USA 2015, 112, 13308–13311. [Google Scholar] [CrossRef] [Green Version]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef]
- Hodgkinson, C.L.; Morrow, C.J.; Li, Y.; Metcalf, R.L.; Rothwell, D.G.; Trapani, F.; Polanski, R.; Burt, D.J.; Simpson, K.L.; Morris, K.; et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 2014, 20, 897–903. [Google Scholar] [CrossRef]
- Girotti, M.R.; Gremel, G.; Lee, R.; Galvani, E.; Rothwell, D.; Viros, A.; Mandal, A.K.; Lim, K.H.; Saturno, G.; Furney, S.J.; et al. Application of Sequencing, Liquid Biopsies, and Patient-Derived Xenografts for Personalized Medicine in Melanoma. Cancer Discov. 2016, 6, 286–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, D.A.; Kessenbrock, K.; Davis, R.T.; Pervolarakis, N.; Werb, Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 2018, 20, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Q.; Wang, T.; Bian, L.; Zhang, S.; Hu, H.; Li, S.; Hu, Z.; Wu, S.; Liu, B.; et al. Circulating tumor cells in HER2-positive metastatic breast cancer patients: A valuable prognostic and predictive biomarker. BMC Cancer 2013, 13, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalykaki, A.; Agelaki, S.; Kallergi, G.; Xyrafas, A.; Mavroudis, D.; Georgoulias, V. Elimination of EGFR-expressing circulating tumor cells in patients with metastatic breast cancer treated with gefitinib. Cancer Chemother. Pharmacol. 2014, 73, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Rugo, H.S.; Cortes, J.; Awada, A.; O’Shaughnessy, J.; Twelves, C.; Im, S.A.; Hannah, A.; Lu, L.; Sy, S.; Caygill, K.; et al. Change in Topoisomerase 1-Positive Circulating Tumor Cells Affects Overall Survival in Patients with Advanced Breast Cancer after Treatment with Etirinotecan Pegol. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 3348–3357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandalovičová, A.; Rosel, D.; Fernandes, M.; Veselý, P.; Heneberg, P.; Čermák, V.; Petruželka, L.; Kumar, S.; Sanz-Moreno, V.; Brábek, J. Migrastatics-Anti-metastatic and Anti-invasion Drugs: Promises and Challenges. Trends Cancer 2017, 3, 391–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosette, C.; Roth, R.B.; Oeth, P.; Braun, A.; Kammerer, S.; Ekblom, J.; Denissenko, M.F. Role of ICAM1 in invasion of human breast cancer cells. Carcinogenesis 2005, 26, 943–950. [Google Scholar] [CrossRef] [Green Version]
- Nieswandt, B.; Hafner, M.; Echtenacher, B.; Mannel, D.N. Lysis of tumor cells by natural killer cells in mice is impeded by platelets. Cancer Res. 1999, 59, 1295–1300. [Google Scholar] [PubMed]
- Amirkhosravi, A.; Meyer, T.; Chang, J.Y.; Amaya, M.; Siddiqui, F.; Desai, H.; Francis, J.L. Tissue factor pathway inhibitor reduces experimental lung metastasis of B16 melanoma. Thromb. Haemost. 2002, 87, 930–936. [Google Scholar] [PubMed]
- Amirkhosravi, A.; Meyer, T.; Amaya, M.; Davila, M.; Mousa, S.A.; Robson, T.; Francis, J.L. The role of tissue factor pathway inhibitor in tumor growth and metastasis. Semin. Thromb. Hemost. 2007, 33, 643–652. [Google Scholar] [CrossRef] [Green Version]
- Wojciechowski, J.C.; Narasipura, S.D.; Charles, N.; Mickelsen, D.; Rana, K.; Blair, M.L.; King, M.R. Capture and enrichment of CD34-positive haematopoietic stem and progenitor cells from blood circulation using P-selectin in an implantable device. Br. J. Haematol. 2008, 140, 673–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faltas, B. Cornering metastases: Therapeutic targeting of circulating tumor cells and stem cells. Front. Oncol. 2012, 2, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, M.J.; Chen, C.S.; Ponmudi, V.; Hughes, A.D.; King, M.R. E-selectin liposomal and nanotube-targeted delivery of doxorubicin to circulating tumor cells. J. Control. Release Off. J. Control. Release Soc. 2012, 160, 609–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rana, K.; Liesveld, J.L.; King, M.R. Delivery of apoptotic signal to rolling cancer cells: A novel biomimetic technique using immobilized TRAIL and E-selectin. Biotechnol. Bioeng. 2009, 102, 1692–1702. [Google Scholar] [CrossRef]
- Scarberry, K.E.; Dickerson, E.B.; McDonald, J.F.; Zhang, Z.J. Magnetic nanoparticle-peptide conjugates for in vitro and in vivo targeting and extraction of cancer cells. J. Am. Chem. Soc. 2008, 130, 10258–10262. [Google Scholar] [CrossRef]
- Scarberry, K.E.; Dickerson, E.B.; Zhang, Z.J.; Benigno, B.B.; McDonald, J.F. Selective removal of ovarian cancer cells from human ascites fluid using magnetic nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 399–408. [Google Scholar] [CrossRef]
- Scarberry, K.E.; Mezencev, R.; McDonald, J.F. Targeted removal of migratory tumor cells by functionalized magnetic nanoparticles impedes metastasis and tumor progression. Nanomedicine 2011, 6, 69–78. [Google Scholar] [CrossRef]
- Weight, R.M.; Viator, J.A.; Dale, P.S.; Caldwell, C.W.; Lisle, A.E. Photoacoustic detection of metastatic melanoma cells in the human circulatory system. Opt. Lett. 2006, 31, 2998–3000. [Google Scholar] [CrossRef]
- Hughes, A.D.; King, M.R. Nanobiotechnology for the capture and manipulation of circulating tumor cells. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2012, 4, 291–309. [Google Scholar] [CrossRef]
- Galanzha, E.I.; Menyaev, Y.A.; Yadem, A.C.; Sarimollaoglu, M.; Juratli, M.A.; Nedosekin, D.A.; Foster, S.R.; Jamshidi-Parsian, A.; Siegel, E.R.; Makhoul, I.; et al. In vivo liquid biopsy using Cytophone platform for photoacoustic detection of circulating tumor cells in patients with melanoma. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Kim, Y.R.; Yoo, J.K.; Jeong, C.W.; Choi, J.W. Selective killing of circulating tumor cells prevents metastasis and extends survival. J. Hematol. Oncol. 2018, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Galanzha, E.I.; Weingold, R.; Nedosekin, D.A.; Sarimollaoglu, M.; Nolan, J.; Harrington, W.; Kuchyanov, A.S.; Parkhomenko, R.G.; Watanabe, F.; Nima, Z.; et al. Spaser as a biological probe. Nat. Commun. 2017, 8, 15528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menyailo, M.E.; Tretyakova, M.S.; Denisov, E.V. Heterogeneity of Circulating Tumor Cells in Breast Cancer: Identifying Metastatic Seeds. Int. J. Mol. Sci. 2020, 21, 1696. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051696
Menyailo ME, Tretyakova MS, Denisov EV. Heterogeneity of Circulating Tumor Cells in Breast Cancer: Identifying Metastatic Seeds. International Journal of Molecular Sciences. 2020; 21(5):1696. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051696
Chicago/Turabian StyleMenyailo, Maxim E., Maria S. Tretyakova, and Evgeny V. Denisov. 2020. "Heterogeneity of Circulating Tumor Cells in Breast Cancer: Identifying Metastatic Seeds" International Journal of Molecular Sciences 21, no. 5: 1696. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051696




