TmSpz4 Plays an Important Role in Regulating the Production of Antimicrobial Peptides in Response to Escherichia coli and Candida albicans Infections
Abstract
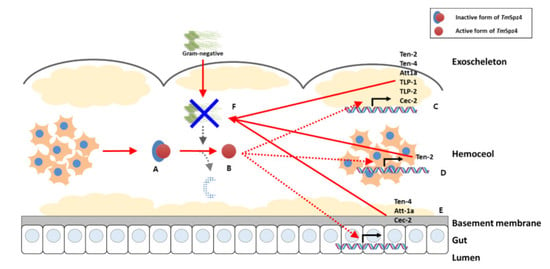
:1. Introduction
2. Results
2.1. Sequence Identification and Phylogenetic Analysis Of TmSpz4
2.2. Developmental and Tissue-Specific Expression Patterns of TmSpz4
2.3. Temporal Induction Pattern of TmSpz4
2.4. Effect of TmSpz4 RNAi on T. Molitor Survivability
2.5. Effects of TmSpz4 Gene Silencing on the Expression of AMPs
3. Discussion
4. Materials and Methods
4.1. Insect Culture
4.2. Preparation of Microorganisms
4.3. Identification and Cloning of Full-Length cDNA Sequence of TmSpz4
4.4. Domain and Phylogenetic Analysis
4.5. TmSpz4 Expression and Temporal Induction Pattern Analysis
4.6. Effect of TmSpz4 Gene Silencing in Response to Microorganisms
4.7. Effect of TmSpz4 RNAi on AMP Expression against Microbial Challenge
4.8. Effects of dsTmSpz4 on the Expression Patterns of NF-κB Genes
4.9. Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rosales, C.; Vonnie, S. Cellular and molecular mechanisms of insect immunity. In Insect Physiology and Ecology; InTeach Publication CCBY: London, UK, 2017; pp. 179–212. [Google Scholar]
- Jiang, Y.-Y. A review of researches on agglutinins of Lepidoptera insect. Entomol. J. E. China 2006, 15, 25–29. [Google Scholar]
- Janeway, C.A. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989, 54, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Imler, J.-L.; Bulet, P. Antimicrobial peptides in Drosophila: Structures, activities and gene regulation. In Mechanisms of Epithelial Defense; Karger Publishers: Basel, Switzerland, 2005; Volume 86, pp. 1–21. [Google Scholar]
- Wang, X.; Zhang, Y.; Zhang, R.; Zhang, J. The diversity of pattern recognition receptors (PRRs) involved with insect defence against pathogens. Curr. Opin. Insect Sci. 2019, 33, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.V.; Bokla, L.; Nüsslein-Volhard, C. Establishment of dorsal-ventral polarity in the Drosophila embryo: The induction of polarity by the Toll gene product. Cell 1985, 42, 791–798. [Google Scholar] [CrossRef]
- Levitin, A.; Whiteway, M. Drosophila innate immunity and response to fungal infections. Cell. Microbiol. 2008, 10, 1021–1026. [Google Scholar] [CrossRef]
- Ferrandon, D.; Imler, J.-L.; Hetru, C.; Hoffmann, J.A. The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 2007, 7, 862. [Google Scholar] [CrossRef]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef] [Green Version]
- El Chamy, L.; Leclerc, V.; Caldelari, I.; Reichhart, J.-M. Sensing of’danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways’ upstream’of Toll. Nat. Immunol. 2008, 9, 1165. [Google Scholar] [CrossRef] [Green Version]
- Roh, K.-B.; Kim, C.-H.; Lee, H.; Kwon, H.-M.; Park, J.-W.; Ryu, J.-H.; Kurokawa, K.; Ha, N.-C.; Lee, W.-J.; Lemaitre, B. Proteolytic cascade for the activation of the insect toll pathway induced by the fungal cell wall component. J. Biol. Chem. 2009, 284, 19474–19481. [Google Scholar] [CrossRef] [Green Version]
- Gobert, V.; Gottar, M.; Matskevich, A.A.; Rutschmann, S.; Royet, J.; Belvin, M.; Hoffmann, J.A.; Ferrandon, D. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science 2003, 302, 2126–2130. [Google Scholar] [CrossRef] [Green Version]
- DeLotto, Y.; Smith, C.; DeLotto, R. Multiple isoforms of the Drosophila Spätzle protein are encoded by alternatively spliced maternal mRNAs in the precellular blastoderm embryo. Mol. Gen. Genet. MGG 2001, 264, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Arnot, C.J.; Gay, N.J.; Gangloff, M. Molecular mechanism that induces activation of Spätzle, the ligand for the Drosophila Toll receptor. J. Biol. Chem. 2010, 285, 19502–19509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, J.S.; Mizuguchi, K.; Gay, N.J. A family of proteins related to Spätzle, the toll receptor ligand, are encoded in the Drosophila genome. Proteins Struct. Funct. Bioinform. 2001, 45, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.; Li, C.-F.; He, Z.; Lu, Y.; Liu, X.-S.; Wang, Y.-F.; Ip, Y.T.; Strand, M.R.; Yu, X.-Q. Toll family members bind multiple Spätzle proteins and activate antimicrobial peptide gene expression in Drosophila. J. Biol. Chem. 2019, 294, 10172–10181. [Google Scholar] [CrossRef]
- Chowdhury, M.; He, Z.; Lu, Y.; Liu, X.; Wang, Y.; Yu, X.-Q. Multiple Toll-Spätzle Pathways in Drosophila melanogaster Immunity. bioRxiv 2018, 420679. [Google Scholar]
- Nonaka, S.; Kawamura, K.; Hori, A.; Salim, E.; Fukushima, K.; Nakanishi, Y.; Kuraishi, T. Characterization of Spz5 as a novel ligand for Drosophila Toll-1 receptor. Biochem. Biophys. Res. Commun. 2018, 506, 510–515. [Google Scholar] [CrossRef]
- Christophides, G.K.; Zdobnov, E.; Barillas-Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.T.; Collins, F.H.; Danielli, A.; Dimopoulos, G. Immunity-related genes and gene families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.W.; Bian, G.; Raikhel, A.S. A toll receptor and a cytokine, Toll5A and Spz1C, are involved in toll antifungal immune signaling in the mosquito Aedes aegypti. J. Biol. Chem. 2006, 281, 39388–39395. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Cheng, T.; Rayaprolu, S.; Zou, Z.; Xia, Q.; Xiang, Z.; Jiang, H. Proteolytic activation of pro-spätzle is required for the induced transcription of antimicrobial peptide genes in lepidopteran insects. Dev. Comp. Immunol. 2007, 31, 1002–1012. [Google Scholar] [CrossRef] [Green Version]
- An, C.; Jiang, H.; Kanost, M.R. Proteolytic activation and function of the cytokine Spätzle in the innate immune response of a lepidopteran insect, Manduca sexta. FEBS J. 2010, 277, 148–162. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Liu, J.; Li, F.; Wang, X.; Li, X.; Shen, Z.; Wu, J. Immune-Related Gene Spatzle4 and Its Differential Immune Responses against Microbes in the Silkworm, Bombyx Mori. Am. J. Clin. Exp. Med. 2015, 4, 344–349. [Google Scholar] [CrossRef] [Green Version]
- Yuan, K.; Yuan, F.-H.; Weng, S.-P.; He, J.-G.; Chen, Y.-H. Identification and functional characterization of a novel Spätzle gene in Litopenaeus vannamei. Dev. Comp. Immunol. 2017, 68, 46–57. [Google Scholar] [CrossRef]
- Shi, X.-Z.; Zhang, R.-R.; Jia, Y.-P.; Zhao, X.-F.; Yu, X.-Q.; Wang, J.-X. Identification and molecular characterization of a Spätzle-like protein from Chinese shrimp (Fenneropenaeus chinensis). Fish Shellfish. Immunol. 2009, 27, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.-H.; Kim, S.-J.; Kan, H.; Kwon, H.-M.; Roh, K.-B.; Jiang, R.; Yang, Y.; Park, J.-W.; Lee, H.-H.; Ha, N.-C. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced toll pathway in an insect. J. Biol. Chem. 2008, 283, 7599–7607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, A.N.; Gangloff, M.; Moncrieffe, M.C.; Hyvert, Y.; Imler, J.-L.; Gay, N.J. Role of the Spätzle pro-domain in the generation of an active Toll receptor ligand. J. Biol. Chem. 2007, 282, 13522–13531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhu, S. Evolutionary and functional epitopes of the Spatzle protein: New insights into activation of the Toll receptor. Cell. Mol. Life Sci. 2009, 66, 1595–1602. [Google Scholar] [CrossRef] [PubMed]
- Imler, J.-L.; Hoffmann, J.A. Signaling mechanisms in the antimicrobial host defense of Drosophila. Curr. Opin. Microbiol. 2000, 3, 16–22. [Google Scholar] [CrossRef]
- Yu, Y.; Park, J.-W.; Kwon, H.-M.; Hwang, H.-O.; Jang, I.-H.; Masuda, A.; Kurokawa, K.; Nakayama, H.; Lee, W.-J.; Dohmae, N. Diversity of innate immune recognition mechanism for bacterial polymeric meso-diaminopimelic acid-type peptidoglycan in insects. J. Biol. Chem. 2010, 285, 32937–32945. [Google Scholar] [CrossRef] [Green Version]
- Thummel, C.S. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev. Cell 2001, 1, 453–465. [Google Scholar] [CrossRef] [Green Version]
- Dimarcq, J.-L.; Imler, J.-L.; Lanot, R.; Ezekowitz, R.A.B.; Hoffmann, J.A.; Janeway, C.A.; Lagueux, M. Treatment of l (2) mbn Drosophila tumorous blood cells with the steroid hormone ecdysone amplifies the inducibility of antimicrobial peptide gene expression. Insect Biochem. Mol. Biol. 1997, 27, 877–886. [Google Scholar] [CrossRef]
- Borst, D.W.; Bollenbacher, W.E.; O’Connor, J.D.; King, D.S.; Fristrom, J.W. Ecdysone levels during metamorphosis ofDrosophila melanogaster. Dev. Biol. 1974, 39, 308–316. [Google Scholar] [CrossRef]
- Ling, E.; Shirai, K.; Kanekatsu, R.; Kiguchi, K. Hemocyte differentiation in the hematopoietic organs of the silkworm, Bombyx mori: Prohemocytes have the function of phagocytosis. Cell Tissue Res. 2005, 320, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Merchant, D.; Ertl, R.L.; Rennard, S.I.; Stanley, D.W.; Miller, J.S. Eicosanoids mediate insect hemocyte migration. J. Insect Physiol. 2008, 54, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Tsakas, S.; Marmaras, V. Insect immunity and its signalling: An overview. Invertebr. Surviv. J. 2010, 7, 228–238. [Google Scholar]
- Agaisse, H.; Petersen, U.-M.; Boutros, M.; Mathey-Prevot, B.; Perrimon, N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 2003, 5, 441–450. [Google Scholar] [CrossRef] [Green Version]
- Edosa, T.T.; Jo, Y.H.; Keshavarz, M.; Bae, Y.M.; Kim, D.H.; Lee, Y.S.; Han, Y.S. TmSpz6 Is Essential for Regulating the Immune Response to Escherichia Coli and Staphylococcus Aureus Infection in Tenebrio Molitor. Insects 2020, 11, 105. [Google Scholar] [CrossRef] [Green Version]
- Bulet, P.; Cociancich, S.; Dimarcq, J.-L.; Lambert, J.; Reichhart, J.-M.; Hoffmann, D.; Hetru, C.; Hoffmann, J. Insect immunity. Isolation from a coleopteran insect of a novel inducible antibacterial peptide and of new members of the insect defensin family. J. Biol. Chem. 1991, 266, 24520–24525. [Google Scholar]
- Carlsson, A.; Nyström, T.; de Cock, H.; Bennich, H. Attacin-an insect immune protein-binds LPS and triggers the specific inhibition of bacterial outer-membrane protein synthesis. Microbiology 1998, 144, 2179–2188. [Google Scholar] [CrossRef] [Green Version]
- Bechinger, B.; Lohner, K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta BBA Biomembr. 2006, 1758, 1529–1539. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, L.; Xu, X.; Freed, S.; Gao, Y.; Yu, J.; Wang, S.; Ju, W.; Zhang, Y.; Jin, F. Cecropins from Plutella xylostella and Their Interaction with Metarhizium anisopliae. PLoS ONE 2015, 10, e0142451. [Google Scholar] [CrossRef]
- Carlsson, A.; Engström, P.; Palva, E.T.; Bennich, H. Attacin, an antibacterial protein from Hyalophora cecropia, inhibits synthesis of outer membrane proteins in Escherichia coli by interfering with omp gene transcription. Infect. Immun. 1991, 59, 3040–3045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, Y.H.; Park, S.; Park, K.B.; Noh, M.Y.; Cho, J.H.; Ko, H.J.; Kim, C.E.; Patnaik, B.B.; Kim, J.; Won, R. In silico identification, characterization and expression analysis of attacin gene family in response to bacterial and fungal pathogens in Tenebrio molitor. Entomol. Res. 2018, 48, 45–54. [Google Scholar] [CrossRef]
- Chae, J.-H.; Kurokawa, K.; So, Y.-I.; Hwang, H.O.; Kim, M.-S.; Park, J.-W.; Jo, Y.-H.; Lee, Y.S.; Lee, B.L. Purification and characterization of tenecin 4, a new anti-Gram-negative bacterial peptide, from the beetle Tenebrio molitor. Dev. Comp. Immunol. 2012, 36, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Maistrou, S.; Paris, V.; Jensen, A.B.; Rolff, J.; Meyling, N.V.; Zanchi, C. A constitutively expressed antifungal peptide protects Tenebrio molitor during a natural infection by the entomopathogenic fungus Beauveria bassiana. Dev. Comp. Immunol. 2018, 86, 26–33. [Google Scholar] [CrossRef]
- Yang, Y.T.; Lee, M.R.; Lee, S.J.; Kim, S.; Nai, Y.S.; Kim, J.S. Tenebrio molitor Gram-negative-binding protein 3 (TmGNBP3) is essential for inducing downstream antifungal Tenecin 1 gene expression against infection with Beauveria bassiana JEF-007. Insect Sci. 2018, 25, 969–977. [Google Scholar] [CrossRef]
- Thevissen, K.; Warnecke, D.C.; Francois, I.E.; Leipelt, M.; Heinz, E.; Ott, C.; Zahringer, U.; Thomma, B.P.; Ferket, K.K.; Cammue, B.P. Defensins from insects and plants interact with fungal glucosylceramides. J. Biol. Chem. 2004, 279, 3900–3905. [Google Scholar] [CrossRef] [Green Version]
- Ganesan, S.; Aggarwal, K.; Paquette, N.; Silverman, N. NF-κB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr. Top. Microbiol. Immunol. 2011, 349, 25–60. [Google Scholar] [CrossRef] [Green Version]
- Valanne, S.; Wang, J.-H.; Rämet, M. The Drosophila toll signaling pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef] [Green Version]
- Yaffe, H.; Buxdorf, K.; Shapira, I.; Ein-Gedi, S.; Moyal-Ben Zvi, M.; Fridman, E.; Moshelion, M.; Levy, M. LogSpin: A simple, economical and fast method for RNA isolation from infected or healthy plants and other eukaryotic tissues. BMC Res. Notes 2012, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]











| Primer Name | Sequence (5′-3′) |
|---|---|
| TmSpz4-qPCR-Fw | GGCGATGCTCTTCCAGGAC |
| TmSpz4-qPCR-Rv | CGCGTTCACTCCTTTCATTTGG |
| TmSpz4-T7-Fw | TAATACGACTCACTATAGGGTCCAGATGTACTGTCGCGATG |
| TmSpz4-T7-Rv | TAATACGACTCACTATAGGGTTTCCTTCTGTACCAGTCGGG |
| TmSpz4-cloning-Fw | ACCGACACAACCACCAAAAG |
| TmSpz4-cloning-Rv | ATCCGTGATCTCCGGAAAAA |
| TmSpz4-cloning-FullORF-Fw | GACGGTGCCCACGAACTAT |
| TmSpz4-cloning-FullORF-Rv | AAGAGCACGAGCCTTGACAT |
| TmL27a_qPCR_Fw | TCATCCTGAAGGCAAAGCTCCAGT |
| TmL27a_qPCR_Rv | AGGTTGGTTAGGCAGGCACCTTTA |
| dsEGFP_Fw | TAATACGACTCACTATAGGGTCGTAAACGGCCACAAGTTC |
| dsEGFP_Rv | TAATACGACTCACTATAGGGT TGCTCAGGTAGTGTTGTCG |
| TmTenecin-1_Fw | CAGCTGAAGAAATCGAACAAGG |
| TmTenecin-1_Rv | CAGACCCTCTTTCCGTTACAGT |
| TmTenecin-2_Fw | CAGCAAAACGGAGGATGGTC |
| TmTenecin-2_Rv | CGTTGAAATCGTGATCTTGTCC |
| TmTenecin-3_Fw | GATTTGCTTGATTCTGGTGGTC |
| TmTenecin-3_Rv | CTGATGGCCTCCTAAATGTCC |
| TmTenecin-4_Fw | GGACATTGAAGATCCAGGAAAG |
| TmTenecin-4_Rv | CGGTGTTCCTTATGTAGAGCTG |
| TmDefensin-1_Fw | AAATCGAACAAGGCCAACAC |
| TmDefencin-1_Rv | GCAAATGCAGACCCTCTTTC |
| TmDefencin-2_Fw | GGGATGCCTCATGAAGATGTAG |
| TmDefencin-2_Rv | CCAATGCAAACACATTCGTC |
| TmColoptericin-1_Fw | GGACAGAATGGTGGATGGTC |
| TmColoptericin-1_Rv | CTCCAACATTCCAGGTAGGC |
| TmColoptericin-2_Fw | GGACGGTTCTGATCTTCTTGAT |
| TmColoptericin-2_Rv | CAGCTGTTTGTTTGTTCTCGTC |
| TmAttacin-1a_Fw | GAAACGAAATGGAAGGTGGA |
| TmAttacin-1a_Rv | TGCTTCGGCAGACAATACAG |
| TmAttacin-1b_Fw | GAGCTGTGAATGCAGGACAA |
| TmAttacin-1b_Rv | CCCTCTGATGAAACCTCCAA |
| TmAttacin-2_Fw | AACTGGGATATTCGCACGTC |
| TmAttacin-2_Rv | CCCTCCGAAATGTCTGTTGT |
| TmCecropin-2_Fw | TACTAGCAGCGCCAAAACCT |
| TmCecropin-2_Rv | CTGGAACATTAGGCGGAGAA |
| TmThaumatin-likeprotein-1_Fw | CTCAAAGGACACGCAGGACT |
| TmThaumatin-like protein-1_Rv | ACTTTGAGCTTCTCGGGACA |
| TmThaumatin-like protein-2_Fw | CCGTCTGGCTAGGAGTTCTG |
| TmThaumatin-like protein-2_Rv | ACTCCTCCAGCTCCGTTACA |
| TmDorsal-X1_qPCR_Fw | AGCGTTGAGGTTTCGGTATG |
| TmDorsal-X1_qPCR_Rv | TCTTTGGTGACGCAAGACAC |
| TmDorsal-X2_qPCR_Fw | ACACCCCCGAAATCACAAAC |
| TmDorsal-X2_qPCR_Rv | TTTCAGAGCGCCAGGTTTTG |
| TmRelish_qPCR_Fw | AGCGTCAAGTTGGAGCAGAT |
| TmRelish_qPCR_Rv | GTCCGGACCTCAAGTGT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Edosa, T.T.; Jo, Y.H.; Keshavarz, M.; Bae, Y.M.; Kim, D.H.; Lee, Y.S.; Han, Y.S. TmSpz4 Plays an Important Role in Regulating the Production of Antimicrobial Peptides in Response to Escherichia coli and Candida albicans Infections. Int. J. Mol. Sci. 2020, 21, 1878. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051878
Edosa TT, Jo YH, Keshavarz M, Bae YM, Kim DH, Lee YS, Han YS. TmSpz4 Plays an Important Role in Regulating the Production of Antimicrobial Peptides in Response to Escherichia coli and Candida albicans Infections. International Journal of Molecular Sciences. 2020; 21(5):1878. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051878
Chicago/Turabian StyleEdosa, Tariku Tesfaye, Yong Hun Jo, Maryam Keshavarz, Young Min Bae, Dong Hyun Kim, Yong Seok Lee, and Yeon Soo Han. 2020. "TmSpz4 Plays an Important Role in Regulating the Production of Antimicrobial Peptides in Response to Escherichia coli and Candida albicans Infections" International Journal of Molecular Sciences 21, no. 5: 1878. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21051878