In Vitro Biophysical and Biological Characterization of Lipid Nanoparticles Co-Encapsulating Oncosuppressors miR-199b-5p and miR-204-5p as Potentiators of Target Therapy in Metastatic Melanoma
Abstract
:1. Introduction
2. Results
2.1. Preparation of LNPs Encapsulating miRNAs
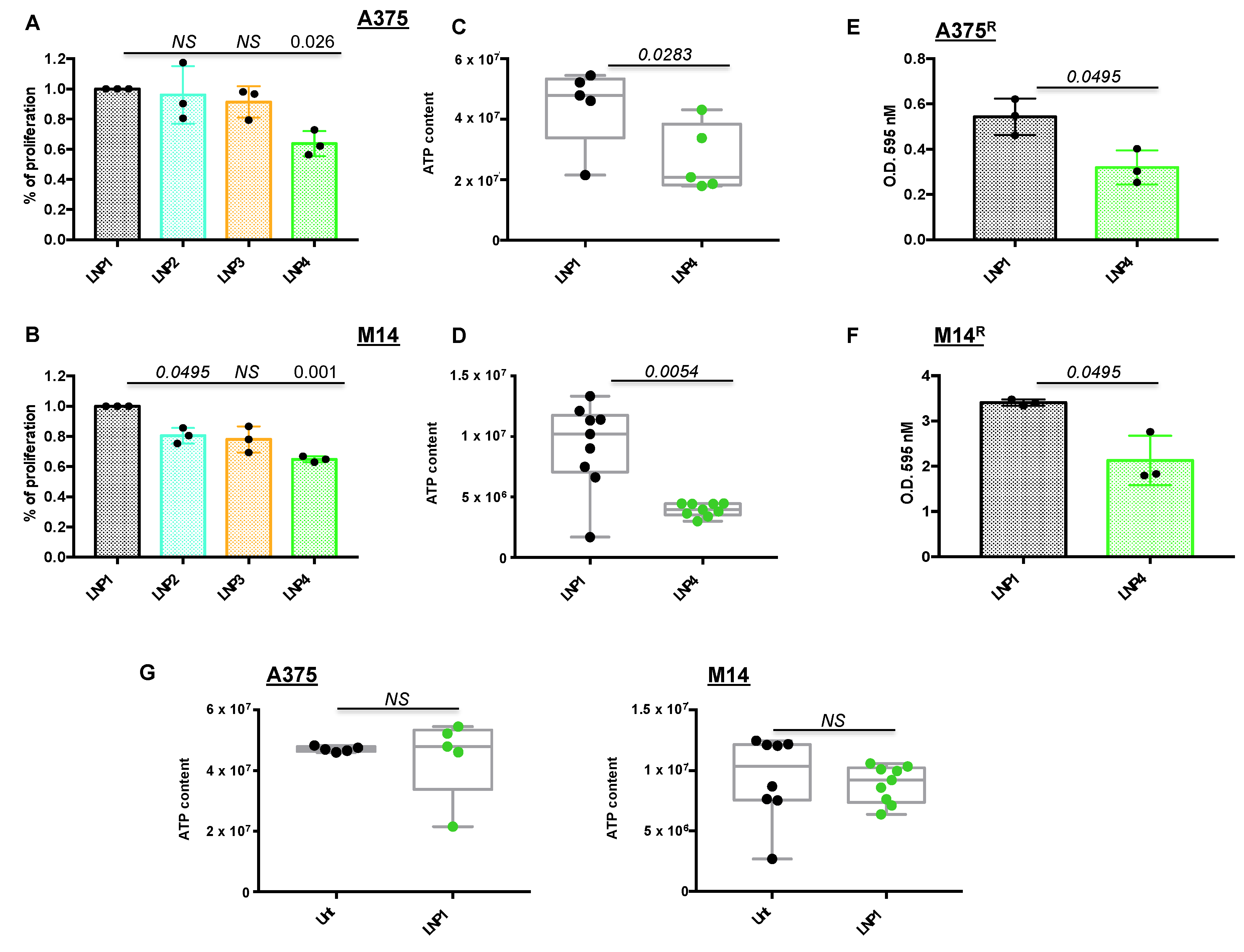
2.2. LNPs Encapsulating miRNAs Impair Proliferation of Melanoma Cell Lines Sensitive or Rendered Resistant to MAPKi
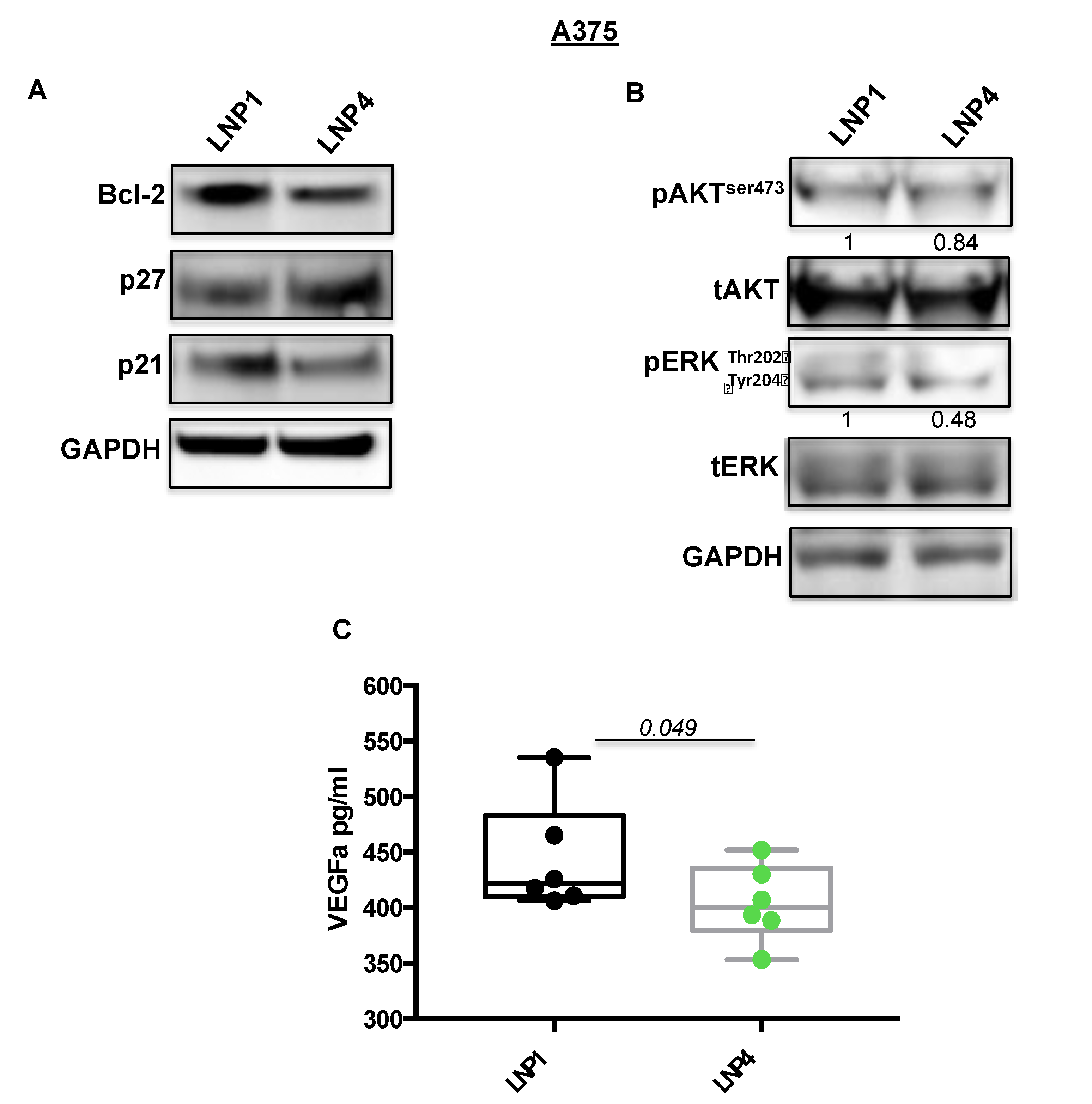
2.3. LNPs Carrying Therapeutic miRNAs Effectively Overexpress miR-204-5p and miR-199b-5p in Melanoma Cells and in Turn Inhibit Their Target Oncogenes Bcl-2 and VEGF-A
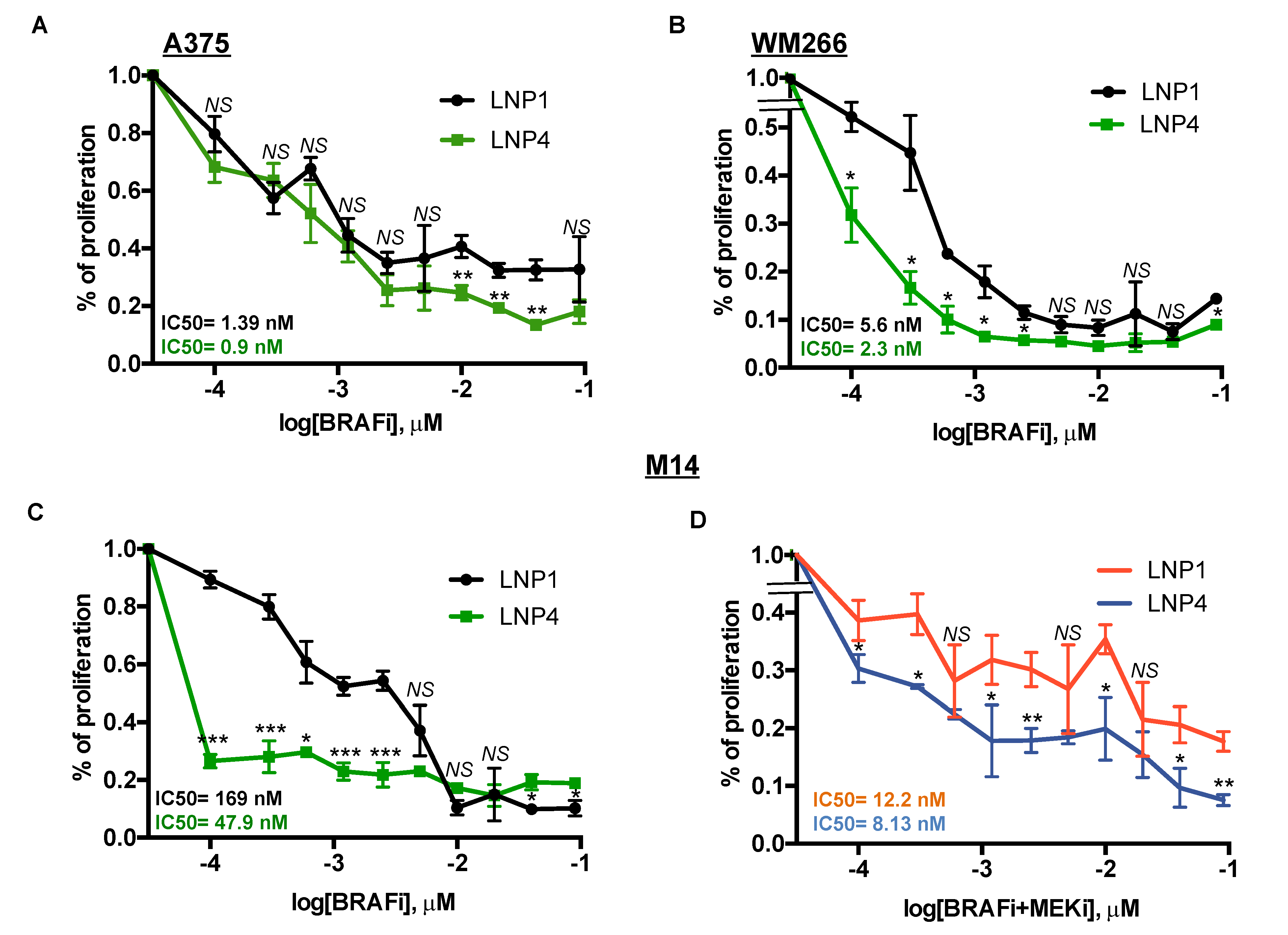
2.4. Therapeutic LNPs Potentiate MAPKi Action on BRAF-Mutant Melanoma Cell Growth
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of LNPs Encapsulating miRNAs
4.3. LNPs-miRNAs Characterization, Size and Polydispersity Index
4.4. Zeta Potential of LNPs
4.5. Lipid Dosage in LNPs
4.6. miRNA Encapsulation
4.7. Cell Lines and Treatments
4.8. Antibodies and Reagents
4.9. VEGF-A Detection
4.10. Western Blot Analysis
4.11. RNA Extraction and Real-Time
4.12. Cell Viability
4.13. Statistical Analyses
5. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MAPKi | BRAF/MEK inhibitors |
| LNP | Lipid nanoparticles |
| FBS | Fetal bovine serum |
References
- Domingues, B.; Lopes, J.M.; Soares, P.; Pòpulo, H. Melanoma treatment in review. ImmunoTargets Ther. 2018, 7, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Schilling, B.; Martens, A.; Geukes Foppen, M.H.; Gebhardt, C.; Hassel, J.C.; Rozeman, E.A.; Gesierich, A.; Gutzmer, R.; Kahler, K.C.; Livingstone, E.; et al. First-Line therapy-Stratified survival in BRAF-Mutant melanoma: A retrospective multicenter analysis. Cancer Immunol. Immunother. 2019, 68, 765–772. [Google Scholar] [CrossRef]
- Pelster, M.S.; Amaria, R.N. Combined targeted therapy and immunotherapy in melanoma: A review of the impact on the tumor microenvironment and outcomes of early clinical trials. Ther. Adv. Med Oncol. 2019, 11. [Google Scholar] [CrossRef] [Green Version]
- Manca, A.; Lissia, A.; Capone, M.; Ascierto, P.A.; Botti, G.; Caracò, C.; Stanganelli, I.; Colombino, M.; Sini, M.; Cossu, A.; et al. Activating PIK3CA mutations coexist with BRAF or NRAS mutations in a limited fraction of melanomas. J. Transl. Med. 2015, 13, 37. [Google Scholar] [CrossRef] [Green Version]
- Dummer, R.; Ascierto, P.A.; Gogas, H.J.; Arance, A.; Mandala, M.; Liszkay, G.; Garbe, C.; Schadendorf, D.; Krajsova, I.; Gutzmer, R.; et al. Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF-Mutant melanoma (COLUMBUS): A multicentre, open-Label, randomised phase 3 trial. Lancet Oncol. 2018, 19, 603–615. [Google Scholar] [CrossRef] [Green Version]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2014, 372, 30–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.; Piva, M.; Sun, L.; Hong, A.; Moriceau, G.; Kong, X.; Zhang, H.; Lomeli, S.; Qian, J.; Yu, C.C.; et al. Recurrent Tumor Cell-Intrinsic and -Extrinsic Alterations during MAPKi-Induced Melanoma Regression and Early Adaptation. Cancer Discov. 2017, 7, 1248–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madonna, G.; Ballesteros-Merino, C.; Feng, Z.; Bifulco, C.; Capone, M.; Giannarelli, D.; Mallardo, D.; Simeone, E.; Grimaldi, A.M.; Caracò, C.; et al. PD-L1 expression with immune-infiltrate evaluation and outcome prediction in melanoma patients treated with ipilimumab. Oncoimmunology 2018, 7, e1405206. [Google Scholar] [CrossRef] [Green Version]
- Pistillo, M.P.; Fontana, V.; Morabito, A.; Dozin, B.; Laurent, S.; Carosio, R.; Banelli, B.; Ferrero, F.; Spano, L.; Tanda, E.; et al. Soluble CTLA-4 as a favorable predictive biomarker in metastatic melanoma patients treated with ipilimumab: An Italian melanoma intergroup study. Cancer Immunol. Immunother. 2019, 68, 97–107. [Google Scholar] [CrossRef]
- Hodi, F.S.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.; Rutkowski, P.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-Year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. 2018, 19, 1480–1492. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fattore, L.; Mancini, R.; Ascierto, P.A.; Ciliberto, G. The potential of BRAF-Associated non-Coding RNA as a therapeutic target in melanoma. Expert Opin. Ther. Targets 2019, 23, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Moriceau, G.; Hugo, W.; Hong, A.; Shi, H.; Kong, X.; Yu, C.C.; Koya, R.C.; Samatar, A.A.; Khanlou, N.; Braun, J.; et al. Tunable-Combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell 2015, 27, 240–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hugo, W.; Shi, H.; Sun, L.; Piva, M.; Song, C.; Kong, X.; Moriceau, G.; Hong, A.; Dahlman, K.B.; Johnson, D.B.; et al. Non-Genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell 2015, 162, 1271–1285. [Google Scholar] [CrossRef] [Green Version]
- Fattore, L.; Sacconi, A.; Mancini, R.; Ciliberto, G. MicroRNA-Driven deregulation of cytokine expression helps development of drug resistance in metastatic melanoma. Cytokine Growth Factor Rev. 2017, 36, 39–48. [Google Scholar] [CrossRef]
- Pisanu, M.E.; Maugeri-Saccà, M.; Fattore, L.; Bruschini, S.; De Vitis, C.; Tabbì, E.; Bellei, B.; Migliano, E.; Kovacs, D.; Camera, E.; et al. Inhibition of Stearoyl-CoA desaturase 1 reverts BRAF and MEK inhibition-Induced selection of cancer stem cells in BRAF-Mutated melanoma. J. Exp. Clin. Cancer Res. CR 2018, 37, 318. [Google Scholar] [CrossRef]
- Fattore, L.; Marra, E.; Pisanu, M.E.; Noto, A.; de Vitis, C.; Belleudi, F.; Aurisicchio, L.; Mancini, R.; Torrisi, M.R.; Ascierto, P.A.; et al. Activation of an early feedback survival loop involving phospho-ErbB3 is a general response of melanoma cells to RAF/MEK inhibition and is abrogated by anti-ErbB3 antibodies. J. Transl. Med. 2013, 11, 180. [Google Scholar] [CrossRef] [Green Version]
- Ruggiero, C.F.; Malpicci, D.; Fattore, L.; Madonna, G.; Vanella, V.; Mallardo, D.; Liguoro, D.; Salvati, V.; Capone, M.; Bedogni, B.; et al. ErbB3 Phosphorylation as Central Event in Adaptive Resistance to Targeted Therapy in Metastatic Melanoma: Early Detection in CTCs during Therapy and Insights into Regulation by Autocrine Neuregulin. Cancers (Basel) 2019, 11, 425. [Google Scholar] [CrossRef] [Green Version]
- Fattore, L.; Malpicci, D.; Marra, E.; Belleudi, F.; Noto, A.; De Vitis, C.; Pisanu, M.E.; Coluccia, P.; Camerlingo, R.; Roscilli, G.; et al. Combination of antibodies directed against different ErbB3 surface epitopes prevents the establishment of resistance to BRAF/MEK inhibitors in melanoma. Oncotarget 2015, 6, 24823–24841. [Google Scholar] [CrossRef] [Green Version]
- Fattore, L.; Costantini, S.; Malpicci, D.; Ruggiero, C.F.; Ascierto, P.A.; Croce, C.M.; Mancini, R.; Ciliberto, G. MicroRNAs in melanoma development and resistance to target therapy. Oncotarget 2017, 8, 22262–22278. [Google Scholar] [CrossRef] [Green Version]
- Fattore, L.; Mancini, R.; Acunzo, M.; Romano, G.; Lagana, A.; Pisanu, M.E.; Malpicci, D.; Madonna, G.; Mallardo, D.; Capone, M.; et al. miR-579-3p controls melanoma progression and resistance to target therapy. Proc. Natl. Acad. Sci. USA 2016, 113, E5005–E5013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fattore, L.; Ruggiero, C.F.; Pisanu, M.E.; Liguoro, D.; Cerri, A.; Costantini, S.; Capone, F.; Acunzo, M.; Romano, G.; Nigita, G.; et al. Reprogramming miRNAs global expression orchestrates development of drug resistance in BRAF-mutated melanoma. Cell Death Differ. 2019, 26, 1267–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Gao, D.Y.; Huang, L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv. Drug Deliv. Rev. 2015, 81, 128–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Martino, M.T.; Campani, V.; Misso, G.; Gallo Cantafio, M.E.; Gullà, A.; Foresta, U.; Guzzi, P.H.; Castellano, M.; Grimaldi, A.; Gigantino, V.; et al. In Vivo Activity of MiR-34a Mimics Delivered by Stable Nucleic Acid Lipid Particles (SNALPs) against Multiple Myeloma. PLoS ONE 2014, 9, e90005. [Google Scholar] [CrossRef]
- Semple, S.C.; Klimuk, S.K.; Harasym, T.O.; Dos Santos, N.; Ansell, S.M.; Wong, K.F.; Maurer, N.; Stark, H.; Cullis, P.R.; Hope, M.J.; et al. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: Formation of novel small multilamellar vesicle structures. Biochim. Biophys. Acta 2001, 1510, 152–166. [Google Scholar] [CrossRef] [Green Version]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-Based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef]
- Scognamiglio, I.; Di Martino, M.T.; Campani, V.; Virgilio, A.; Galeone, A.; Gulla, A.; Gallo Cantafio, M.E.; Misso, G.; Tagliaferri, P.; Tassone, P.; et al. Transferrin-Conjugated SNALPs encapsulating 2’-O-Methylated miR-34a for the treatment of multiple myeloma. Biomed. Res. Int. 2014. [Google Scholar] [CrossRef]
- Campani, V.; Salzano, G.; Lusa, S.; De Rosa, G. Lipid Nanovectors to Deliver RNA Oligonucleotides in Cancer. Nanomaterials (Basel Switz.) 2016, 6, 131. [Google Scholar] [CrossRef] [Green Version]
- Sacconi, A.; Biagioni, F.; Canu, V.; Mori, F.; Di Benedetto, A.; Lorenzon, L.; Ercolani, C.; Di Agostino, S.; Cambria, A.M.; Germoni, S.; et al. miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 2012, 3, e423. [Google Scholar] [CrossRef]
- Dai, L.; Lou, W.; Zhu, J.; Zhou, X.; Di, W. MiR-199a inhibits the angiogenic potential of endometrial stromal cells under hypoxia by targeting HIF-1alpha/VEGF pathway. Int. J. Clin. Exp. Pathol. 2015, 8, 4735–4744. [Google Scholar] [PubMed]
- Wang, C.; Zhang, Y.; Zhou, D.; Cao, G.; Wu, Y. miR-204 enhances p27 mRNA stability by targeting Brd4 in head and neck squamous cell carcinoma. Oncol. Lett. 2018, 16, 4179–4184. [Google Scholar] [CrossRef] [PubMed]
- Karimian, A.; Ahmadi, Y.; Yousefi, B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 2016, 42, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Fattore, L.; Ruggiero, C.F.; Liguoro, D.; Mancini, R.; Ciliberto, G. Single cell analysis to dissect molecular heterogeneity and disease evolution in metastatic melanoma. Cell Death Dis. 2019, 10, 827. [Google Scholar] [CrossRef] [PubMed]
- Vescarelli, E.; Gerini, G.; Megiorni, F.; Anastasiadou, E.; Pontecorvi, P.; Solito, L.; De Vitis, C.; Camero, S.; Marchetti, C.; Mancini, R.; et al. MiR-200c sensitizes Olaparib-Resistant ovarian cancer cells by targeting Neuropilin 1. J. Exp. Clin. Cancer Res. CR 2020, 39, 3. [Google Scholar] [CrossRef]
- Kozar, I.; Cesi, G.; Margue, C.; Philippidou, D.; Kreis, S. Impact of BRAF kinase inhibitors on the miRNomes and transcriptomes of melanoma cells. Biochim. Biophys. Acta. Gen. Subj. 2017, 1861, 2980–2992. [Google Scholar] [CrossRef]
- Caporali, S.; Amaro, A.; Levati, L.; Alvino, E.; Lacal, P.M.; Mastroeni, S.; Ruffini, F.; Bonmassar, L.; Antonini Cappellini, G.C.; Felli, N.; et al. miR-126-3p down-Regulation contributes to dabrafenib acquired resistance in melanoma by up-regulating ADAM9 and VEGF-A. J. Exp. Clin. Cancer Res. CR 2019, 38, 272. [Google Scholar] [CrossRef]
- Litzinger, D.C.; Buiting, A.M.J.; van Rooijen, N.; Huang, L. Effect of liposome size on the circulation time and intraorgan distribution of amphipathic poly(ethylene glycol)-Containing liposomes. Biochim. Biophys. Acta (BBA)-Biomembr. 1994, 1190, 99–107. [Google Scholar] [CrossRef]
- Stewart, J.C. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem. 1980, 104, 10–14. [Google Scholar] [CrossRef]
- Megiorni, F.; Camero, S.; Ceccarelli, S.; McDowell, H.P.; Mannarino, O.; Marampon, F.; Pizer, B.; Shukla, R.; Pizzuti, A.; Marchese, C.; et al. DNMT3B in vitro knocking-Down is able to reverse embryonal rhabdomyosarcoma cell phenotype through inhibition of proliferation and induction of myogenic differentiation. Oncotarget 2016, 7, 79342–79356. [Google Scholar] [CrossRef] [Green Version]
- Costanzo, P.; Santini, A.; Fattore, L.; Novellino, E.; Ritieni, A. Toxicity of aflatoxin B1 towards the vitamin D receptor (VDR). Food Chem. Toxicol. 2015, 76, 77–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salemi, R.; Falzone, L.; Madonna, G.; Polesel, J.; Cinà, D.; Mallardo, D.; Ascierto, P.A.; Libra, M.; Candido, S. MMP-9 as a Candidate Marker of Response to BRAF Inhibitors in Melanoma Patients with BRAF(V600E) Mutation Detected in Circulating-Free DNA. Front. Pharmacol. 2018, 9, 856. [Google Scholar] [CrossRef] [PubMed]
- D′Amici, S.; Ceccarelli, S.; Vescarelli, E.; Romano, F.; Frati, L.; Marchese, C.; Angeloni, A. TNFa modulates Fibroblast Growth Factor Receptor 2 gene expression through the pRB/E2F1 pathway: Identification of a non-Canonical E2F binding motif. PLoS ONE 2013, 8, e61491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruschini, S.; di Martino, S.; Pisanu, M.E.; Fattore, L.; De Vitis, C.; Laquintana, V.; Buglioni, S.; Tabbì, E.; Cerri, A.; Visca, P.; et al. CytoMatrix for a reliable and simple characterization of lung cancer stem cells from malignant pleural effusions. J. Cell. Physiol. 2020, 235, 1877–1887. [Google Scholar] [CrossRef] [Green Version]
- De Vitis, C.; Corleone, G.; Salvati, V.; Ascenzi, F.; Pallocca, M.; De Nicola, F.; Fanciulli, M.; di Martino, S.; Bruschini, S.; Napoli, C.; et al. B4GALT1 Is a New Candidate to Maintain the Stemness of Lung Cancer Stem Cells. J. Clin. Med. 2019, 8, 1928. [Google Scholar] [CrossRef] [Green Version]
- Ascierto, P.A.; Capone, M.; Grimaldi, A.M.; Mallardo, D.; Simeone, E.; Madonna, G.; Roder, H.; Meyer, K.; Asmellash, S.; Oliveira, C.; et al. Proteomic test for anti-PD-1 checkpoint blockade treatment of metastatic melanoma with and without BRAF mutations. J. Immunother. Cancer 2019, 7, 91. [Google Scholar] [CrossRef]

| Formulation | miRNA | Diameter (nm) ± SD | PI ± SD | ZP (mV ± SD) | Actual Loading (μg miRNA/mg Lipids) | EE% ± SD |
|---|---|---|---|---|---|---|
| LNP1 | - | 138 ± 9 | 0.19 ± 0.02 | −11 ± 1 | - | - |
| LNP2 | miR-204-5p | 157 ± 3 | 0.17 ± 0.05 | −28 ± 8 | 186 ± 0.005 | 93 ± 3 |
| LNP3 | miR-199b-5p | 160.4 ± 11 | 0.18 ± 0.08 | −18 ± 8 | 199 ± 0.001 | 99 ± 1 |
| LNP4 | miR-204-5p/199b-5p | 167 ± 5 | 0.15 ± 0.05 | −26 ± 9 | 190 ± 0.009 | 95 ± 5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fattore, L.; Campani, V.; Ruggiero, C.F.; Salvati, V.; Liguoro, D.; Scotti, L.; Botti, G.; Ascierto, P.A.; Mancini, R.; De Rosa, G.; et al. In Vitro Biophysical and Biological Characterization of Lipid Nanoparticles Co-Encapsulating Oncosuppressors miR-199b-5p and miR-204-5p as Potentiators of Target Therapy in Metastatic Melanoma. Int. J. Mol. Sci. 2020, 21, 1930. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21061930
Fattore L, Campani V, Ruggiero CF, Salvati V, Liguoro D, Scotti L, Botti G, Ascierto PA, Mancini R, De Rosa G, et al. In Vitro Biophysical and Biological Characterization of Lipid Nanoparticles Co-Encapsulating Oncosuppressors miR-199b-5p and miR-204-5p as Potentiators of Target Therapy in Metastatic Melanoma. International Journal of Molecular Sciences. 2020; 21(6):1930. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21061930
Chicago/Turabian StyleFattore, Luigi, Virginia Campani, Ciro Francesco Ruggiero, Valentina Salvati, Domenico Liguoro, Lorena Scotti, Gerardo Botti, Paolo Antonio Ascierto, Rita Mancini, Giuseppe De Rosa, and et al. 2020. "In Vitro Biophysical and Biological Characterization of Lipid Nanoparticles Co-Encapsulating Oncosuppressors miR-199b-5p and miR-204-5p as Potentiators of Target Therapy in Metastatic Melanoma" International Journal of Molecular Sciences 21, no. 6: 1930. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21061930





