Histone Deacetylation Inhibitors as Modulators of Regulatory T Cells
Abstract
:1. Introduction
2. Role of Tregs in Disease
2.1. Autoimmune Diseases
2.2. Transplant Rejection
2.3. Sepsis
3. HAT and HDAC Activities in Treg Differentiation
3.1. Foxp3
3.2. Cytotoxic T Lymphocyte-Associated Protein 4 (CTLA4 or CD152)
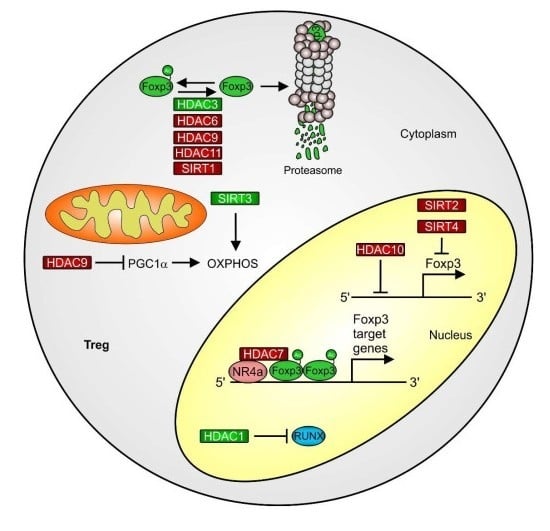
3.3. HDACs and HDACi as a Starting Point for Altering Treg Function
4. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Ø | diameter |
| Ac | acetylation |
| ALI | acute lung injury |
| ALS | amyolotrophic lateral sclerosis |
| AP1 | activator protein 1 |
| APC | antigen presenting cell |
| AS | ankylosis spondylitis |
| C | colitis |
| CAT | cardiac allograft transplantation |
| CD | cluster of differentiation |
| CF | cystic fibrosis |
| ChIP | chromatin immunoprecipitation |
| CIA | collagen-induced arthritis |
| CLP | cecal ligation and puncture |
| CNS | conserved non-coding region |
| CREB | cAMP-responsive element binding protein |
| CTLA4 | cytotoxic T lymphocyte associated protein 4 |
| ETS | E26-AMV virus oncogene cellular homologue |
| f | female |
| Foxp3 | Forkhead-box-protein P3 |
| g | gauge |
| H | histone |
| HAT | histone acetyltransferase |
| HDAC | histone deacetylase |
| HIF | hypoxia-inducible factor |
| HMGB1 | high-mobility-group-protein B1 |
| HP1 | heterochromatin protein-1 |
| i | in vitro |
| IBD | inflammatory bowel disease |
| ICOS | inducible T-cell costimulatory |
| Ikzf2 | ICAROS family zinc finger 2 |
| IPEX | immune dysregulation polyendocrinopathy and enteropathy |
| K | lysine |
| LPS | lipopolysaccharide |
| m | male |
| Me | methylation |
| MHC | major histocompatibility complex |
| MLN | mesenteric lymph nodes |
| MS | multiple sclerosis |
| NFAT | nuclear factor of activated T cells |
| NRa4 | nuclear receptor 4a |
| Nt5e | ecto-5’-nucleotidase |
| OXPHOS | oxidative phosphorylation |
| p | periphery |
| PC | peritoneal cavity |
| PGC1α | PPARgamma coactivator alpha |
| PIAS | protein inhibitor of activated STAT |
| PSO | psoriasis |
| PTEN | phosphatase and tensin homologue |
| RA | rheumatoid arthritis |
| Pde3b | phosphodiesterase 3b |
| RAR | retinoid acid receptor |
| RUNX | Runt-related transcription factor 1 |
| RXR | retinoid X receptor |
| SAHA | suberoylanilide hydroxamic acid |
| Satb1 | special AT-rich sequence binding protein |
| SIRT | silent information regulator |
| Smad | small mothers against decapentaplegic |
| SP | single positive |
| SSc | systemic sclerosis |
| STAT | signal transducer and activator of transcription |
| sumo | small ubiquitin-like modifier |
| t | thymus |
| T1D | type 1 diabetes |
| TCR | T cell receptor |
| TIP60 | HIV-Tat-interactive protein |
| TGF-β | transforming growth factor-beta |
| TLR | toll-like receptor |
| Treg | regulatory T cell |
| TSDR | Treg-cell specific region |
References
- Sakaguchi, S.; Sakaguchi, N.; Asano, M.; Itoh, M.; Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995, 155, 1151–1164. [Google Scholar] [PubMed]
- Abbas, A.K.; Benoist, C.; Bluestone, J.A.; Campbell, D.J.; Ghosh, S.; Hori, S.; Jiang, S.; Kuchroo, V.K.; Mathis, D.; Roncarolo, M.G.; et al. Regulatory T cells: Recommendations to simplify the nomenclature. Nat. Immunol. 2013, 14, 307–308. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zou, M.; Pezoldt, J.; Zhou, X.; Huehn, J. Thymus-derived Foxp3+ regulatory T cells upregulate RORγt expression under inflammatory conditions. J. Mol. Med. 2018, 96, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, S.; Ono, M.; Setoguchi, R.; Yagi, H.; Hori, S.; Fehervari, Z.; Shimizu, J.; Takahashi, T.; Nomura, T. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol. Rev. 2006, 212, 8–27. [Google Scholar] [CrossRef] [PubMed]
- Palmer, E.; Naeher, D. Affinity threshold for thymic selection through a T-cell receptor-co-receptor zipper. Nat. Rev. Immunol. 2009, 9, 207–213. [Google Scholar] [CrossRef]
- Koehli, S.; Naeher, D.; Galati-Fournier, V.; Zehn, D.; Palmer, E. Optimal T-cell receptor affinity for inducing autoimmunity. Proc. Natl. Acad. Sci. USA 2014, 111, 17248–17253. [Google Scholar] [CrossRef] [Green Version]
- Zehn, D.; Bevan, M.J. T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity 2006, 25, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Nishizuka, Y.; Sakakura, T. Thymus and reproduction: Sex-linked dysgenesia of the gonad after neonatal thymectomy in mice. Science 1969, 166, 753–755. [Google Scholar] [CrossRef]
- Asano, M.; Toda, M.; Sakaguchi, N.; Sakaguchi, S. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J. Exp. Med. 1996, 184, 387–396. [Google Scholar] [CrossRef]
- Fontenot, J.D.; Dooley, J.L.; Farr, A.G.; Rudensky, A.Y. Developmental regulation of Foxp3 expression during ontogeny. J. Exp. Med. 2005, 202, 901–906. [Google Scholar] [CrossRef] [Green Version]
- Surh, C.D.; Sprent, J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature 1994, 372, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Konkel, J.E.; Jin, W.; Abbatiello, B.; Grainger, J.R.; Chen, W. Thymocyte apoptosis drives the intrathymic generation of regulatory T cells. Proc. Natl. Acad. Sci. USA 2014, 111, E465–E473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marie, J.C.; Letterio, J.J.; Gavin, M.; Rudensky, A.Y. TGF-beta1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J. Exp. Med. 2005, 201, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Ramoji, A.; Ryabchykov, O.; Galler, K.; Tannert, A.; Markwart, R.; Requardt, R.P.; Rubio, I.; Bauer, M.; Bocklitz, T.; Popp, J.; et al. Raman Spectroscopy Follows Time-Dependent Changes in T Lymphocytes Isolated from Spleen of Endotoxemic Mice. Immunohorizons 2019, 3, 45–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apostolou, I.; Sarukhan, A.; Klein, L.; von Boehmer, H. Origin of regulatory T cells with known specificity for antigen. Nat. Immunol. 2002, 3, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.S.; Boesteanu, A.; Reed, A.J.; Petrone, A.L.; Holenbeck, A.E.; Lerman, M.A.; Naji, A.; Caton, A.J. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat. Immunol. 2001, 2, 301–306. [Google Scholar] [CrossRef]
- Zheng, Y.; Josefowicz, S.; Chaudhry, A.; Peng, X.P.; Forbush, K.; Rudensky, A.Y. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 2010, 463, 808–812. [Google Scholar] [CrossRef] [Green Version]
- Bhairavabhotla, R.; Kim, Y.C.; Glass, D.D.; Escobar, T.M.; Patel, M.C.; Zahr, R.; Nguyen, C.K.; Kilaru, G.K.; Muljo, S.A.; Shevach, E.M. Transcriptome profiling of human FoxP3+ regulatory T cells. Hum. Immunol. 2016, 77, 201–213. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Josefowicz, S.Z.; Kas, A.; Chu, T.-T.; Gavin, M.A.; Rudensky, A.Y. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 2007, 445, 936–940. [Google Scholar] [CrossRef]
- Anandagoda, N.; Willis, J.C.; Hertweck, A.; Roberts, L.B.; Jackson, I.; Gökmen, M.R.; Jenner, R.G.; Howard, J.K.; Lord, G.M. microRNA-142-mediated repression of phosphodiesterase 3B critically regulates peripheral immune tolerance. J. Clin. Investig. 2019, 129, 1257–1271. [Google Scholar] [CrossRef] [PubMed]
- Getnet, D.; Grosso, J.F.; Goldberg, M.V.; Harris, T.J.; Yen, H.-R.; Bruno, T.C.; Durham, N.M.; Hipkiss, E.L.; Pyle, K.J.; Wada, S.; et al. A role for the transcription factor Helios in human CD4+CD25+ regulatory T cells. Mol. Immunol. 2010, 47, 1595–1600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfoertner, S.; Jeron, A.; Probst-Kepper, M.; Guzman, C.A.; Hansen, W.; Westendorf, A.M.; Toepfer, T.; Schrader, A.J.; Franzke, A.; Buer, J.; et al. Signatures of human regulatory T cells: An encounter with old friends and new players. Genome Biol. 2006, 7, R54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, M.; Yaguchi, H.; Ohkura, N.; Kitabayashi, I.; Nagamura, Y.; Nomura, T.; Miyachi, Y.; Tsukada, T.; Sakaguchi, S. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 2007, 446, 685–689. [Google Scholar] [CrossRef] [PubMed]
- D’Hennezel, E.; Ben-Shoshan, M.; Ochs, H.D.; Torgerson, T.R.; Russell, L.J.; Lejtenyi, C.; Noya, F.J.; Jabado, N.; Mazer, B.; Piccirillo, C.A. FOXP3 Forkhead Domain Mutation and Regulatory T Cells in the IPEX Syndrome. N. Engl. J. Med. 2009, 361, 1710–1713. [Google Scholar] [CrossRef]
- Lahl, K.; Loddenkemper, C.; Drouin, C.; Freyer, J.; Arnason, J.; Eberl, G.; Hamann, A.; Wagner, H.; Huehn, J.; Sparwasser, T. Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 2007, 204, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.M.; Rasmussen, J.P.; Rudensky, A.Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 2007, 8, 191–197. [Google Scholar] [CrossRef]
- Hoffmann, P.; Eder, R.; Kunz-Schughart, L.A.; Andreesen, R.; Edinger, M. Large-scale in vitro expansion of polyclonal human CD4+CD25high regulatory T cells. Blood 2004, 104, 895–903. [Google Scholar] [CrossRef]
- Koenecke, C.; Czeloth, N.; Bubke, A.; Schmitz, S.; Kissenpfennig, A.; Malissen, B.; Huehn, J.; Ganser, A.; Förster, R.; Prinz, I. Alloantigen-specific de novo-induced Foxp3+ Treg revert in vivo and do not protect from experimental GVHD. Eur. J. Immunol. 2009, 39, 3091–3096. [Google Scholar] [CrossRef]
- Roquilly, A.; McWilliam, H.E.G.; Jacqueline, C.; Tian, Z.; Cinotti, R.; Rimbert, M.; Wakim, L.; Caminschi, I.; Lahoud, M.H.; Belz, G.T.; et al. Local Modulation of Antigen-Presenting Cell Development after Resolution of Pneumonia Induces Long-Term Susceptibility to Secondary Infections. Immunity 2017, 47, 135–147. [Google Scholar] [CrossRef] [Green Version]
- Gupta, D.L.; Bhoi, S.; Mohan, T.; Galwnkar, S.; Rao, D.N. Coexistence of Th1/Th2 and Th17/Treg imbalances in patients with post traumatic sepsis. Cytokine 2016, 88, 214–221. [Google Scholar] [CrossRef]
- Carvelli, J.; Piperoglou, C.; Bourenne, J.; Farnarier, C.; Banzet, N.; Demerlé, C.; Gainnier, M.; Vély, F. Imbalance of Circulating Innate Lymphoid Cell Subpopulations in Patients With Septic Shock. Front. Immunol. 2019, 10, 2179. [Google Scholar] [CrossRef] [PubMed]
- Venet, F.; Chung, C.-S.; Kherouf, H.; Geeraert, A.; Malcus, C.; Poitevin, F.; Bohé, J.; Lepape, A.; Ayala, A.; Monneret, G. Increased circulating regulatory T cells (CD4+CD25+CD127−) contribute to lymphocyte anergy in septic shock patients. Intensive Care Med. 2009, 35, 678–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nascimento, D.C.; Melo, P.H.; Piñeros, A.R.; Ferreira, R.G.; Colón, D.F.; Donate, P.B.; Castanheira, F.V.; Gozzi, A.; Czaikoski, P.G.; Niedbala, W.; et al. IL-33 contributes to sepsis-induced long-term immunosuppression by expanding the regulatory T cell population. Nat. Commun. 2017, 8, 14919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizui, M.; Tsokos, G.C. Targeting Regulatory T Cells to Treat Patients With Systemic Lupus Erythematosus. Front. Immunol. 2018, 9, 786. [Google Scholar] [CrossRef]
- Visperas, A.; Vignali, D.A.A. Are Regulatory T Cells Defective in Type 1 Diabetes and Can We Fix Them? J. Immunol. 2016, 197, 3762–3770. [Google Scholar] [CrossRef] [Green Version]
- Sabat, R.; Wolk, K.; Loyal, L.; Döcke, W.D.; Ghoreschi, K. T cell pathology in skin inflammation. Semin. Immunopathol. 2019, 41, 359–377. [Google Scholar] [CrossRef] [Green Version]
- Esensten, J.H.; Muller, Y.D.; Bluestone, J.A.; Tang, Q. Regulatory T-cell therapy for autoimmune and autoinflammatory diseases: The next frontier. J. Allergy Clin. Immunol. 2018, 142, 1710–1718. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, L.M.R.; Muller, Y.D.; Bluestone, J.A.; Tang, Q. Next-generation regulatory T cell therapy. Nat. Rev. Drug Discov. 2019, 18, 749–769. [Google Scholar] [CrossRef] [Green Version]
- Marek-Trzonkowska, N.; Myśliwiec, M.; Dobyszuk, A.; Grabowska, M.; Derkowska, I.; Juścińska, J.; Owczuk, R.; Szadkowska, A.; Witkowski, P.; Młynarski, W.; et al. Therapy of type 1 diabetes with CD4+CD25highCD127-regulatory T cells prolongs survival of pancreatic islets—Results of one year follow-up. Clin. Immunol. 2014, 153, 23–30. [Google Scholar] [CrossRef]
- Bluestone, J.A.; Buckner, J.H.; Fitch, M.; Gitelman, S.E.; Gupta, S.; Hellerstein, M.K.; Herold, K.C.; Lares, A.; Lee, M.R.; Li, K.; et al. Type 1 diabetes immunotherapy using polyclonal regulatory T cells. Sci. Transl. Med. 2015, 7, 315ra189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strauss, L.; Czystowska, M.; Szajnik, M.; Mandapathil, M.; Whiteside, T.L. Differential responses of human regulatory T cells (Treg) and effector T cells to rapamycin. PLoS ONE 2009, 4, e5994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, D.; Bonilla, E.; Mirza, N.; Niland, B.; Perl, A. Rapamycin reduces disease activity and normalizes T cell activation-induced calcium fluxing in patients with systemic lupus erythematosus. Arthritis Rheum. 2006, 54, 2983–2988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Yang, K.; Burns, S.; Shrestha, S.; Chi, H. The S1P1-mTOR axis directs the reciprocal differentiation of TH1 and Treg cells. Nat. Immunol. 2010, 11, 1047–1056. [Google Scholar] [CrossRef]
- Lai, Z.-W.; Hanczko, R.; Bonilla, E.; Caza, T.N.; Clair, B.; Bartos, A.; Miklossy, G.; Jimah, J.; Doherty, E.; Tily, H.; et al. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: A randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2012, 64, 2937–2946. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Choi, S.-C.; Xu, Z.; Perry, D.J.; Seay, H.; Croker, B.P.; Sobel, E.S.; Brusko, T.M.; Morel, L. Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl. Med. 2015, 7, 274ra18. [Google Scholar] [CrossRef] [Green Version]
- Koga, T.; Hedrich, C.M.; Mizui, M.; Yoshida, N.; Otomo, K.; Lieberman, L.A.; Rauen, T.; Crispín, J.C.; Tsokos, G.C. CaMK4-dependent activation of AKT/mTOR and CREM-α underlies autoimmunity-associated Th17 imbalance. J. Clin. Investig. 2014, 124, 2234–2245. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Zhang, X.; Wei, Y.; Sun, X.; Chen, Y.; Deng, J.; Jin, Y.; Gan, Y.; Hu, X.; Jia, R.; et al. Low-dose interleukin-2 treatment selectively modulates CD4+ T cell subsets in patients with systemic lupus erythematosus. Nat. Med. 2016, 22, 991–993. [Google Scholar] [CrossRef]
- Humrich, J.Y.; Spee-Mayer, C.; von Siegert, E.; Alexander, T.; Hiepe, F.; Radbruch, A.; Burmester, G.R.; Riemekasten, G. Rapid induction of clinical remission by low-dose interleukin-2 in a patient with refractory SLE. Ann. Rheum. Dis. 2015, 74, 791–792. [Google Scholar] [CrossRef]
- Spee-Mayer, C.; von Siegert, E.; Abdirama, D.; Rose, A.; Klaus, A.; Alexander, T.; Enghard, P.; Sawitzki, B.; Hiepe, F.; Radbruch, A.; et al. Low-dose interleukin-2 selectively corrects regulatory T cell defects in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2016, 75, 1407–1415. [Google Scholar] [CrossRef]
- Hartemann, A.; Bensimon, G.; Payan, C.A.; Jacqueminet, S.; Bourron, O.; Nicolas, N.; Fonfrede, M.; Rosenzwajg, M.; Bernard, C.; Klatzmann, D. Low-dose interleukin 2 in patients with type 1 diabetes: A phase 1/2 randomised, double-blind, placebo-controlled trial. Lancet Diabetes Endocrinol. 2013, 1, 295–305. [Google Scholar] [CrossRef]
- Raffin, C.; Vo, L.T.; Bluestone, J.A. Treg cell-based therapies: Challenges and perspectives. Nat. Rev. Immunol. 2020, 20, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Vincenti, F. Transplant trials with Tregs: Perils and promises. J. Clin. Investig. 2017, 127, 2505–2512. [Google Scholar] [CrossRef] [PubMed]
- Atif, M.; Conti, F.; Gorochov, G.; Oo, Y.H.; Miyara, M. Regulatory T cells in solid organ transplantation. Clin. Transl. Immunol. 2020, 9, e01099. [Google Scholar] [CrossRef] [PubMed]
- Todo, S.; Yamashita, K.; Goto, R.; Zaitsu, M.; Nagatsu, A.; Oura, T.; Watanabe, M.; Aoyagi, T.; Suzuki, T.; Shimamura, T.; et al. A pilot study of operational tolerance with a regulatory T-cell-based cell therapy in living donor liver transplantation. Hepatology 2016, 64, 632–643. [Google Scholar] [CrossRef] [Green Version]
- Chandran, S.; Tang, Q.; Sarwal, M.; Laszik, Z.G.; Putnam, A.L.; Lee, K.; Leung, J.; Nguyen, V.; Sigdel, T.; Tavares, E.C.; et al. Polyclonal Regulatory T Cell Therapy for Control of Inflammation in Kidney Transplants. Am. J. Transplant. 2017, 17, 2945–2954. [Google Scholar] [CrossRef]
- Mathew, J.M.; H-Voss, J.; LeFever, A.; Konieczna, I.; Stratton, C.; He, J.; Huang, X.; Gallon, L.; Skaro, A.; Ansari, M.J.; et al. A Phase I Clinical Trial with Ex Vivo Expanded Recipient Regulatory T cells in Living Donor Kidney Transplants. Sci. Rep. 2018, 8, 7428. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.L.; Trenado, A.; Vasey, D.; Klatzmann, D.; Salomon, B.L. CD4+CD25+ immunoregulatory T Cells: New therapeutics for graft-versus-host disease. J. Exp. Med. 2002, 196, 401–406. [Google Scholar] [CrossRef]
- Jones, S.C.; Murphy, G.F.; Korngold, R. Post-hematopoietic cell transplantation control of graft-versus-host disease by donor CD425 T cells to allow an effective graft-versus-leukemia response. Biol. Blood Marrow Transplant. 2003, 9, 243–256. [Google Scholar] [CrossRef] [Green Version]
- Taylor, P.A.; Lees, C.J.; Blazar, B.R. The infusion of ex vivo activated and expanded CD4+CD25+ immune regulatory cells inhibits graft-versus-host disease lethality. Blood 2002, 99, 3493–3499. [Google Scholar] [CrossRef]
- Mancusi, A.; Piccinelli, S.; Velardi, A.; Pierini, A. CD4+FOXP3+ Regulatory T Cell Therapies in HLA Haploidentical Hematopoietic Transplantation. Front. Immunol. 2019, 10, 2901. [Google Scholar] [CrossRef] [PubMed]
- Boomer, J.S.; To, K.; Chang, K.C.; Takasu, O.; Osborne, D.F.; Walton, A.H.; Bricker, T.L.; Jarman, S.D.; Kreisel, D.; Krupnick, A.S.; et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA 2011, 306, 2594–2605. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Fang, H.; Du, M.; Li, C.; Tang, R.; Liu, H.; Gao, Z.; Ji, Z.; Ke, B.; Chen, X.-L. The Modulation of Regulatory T Cells via HMGB1/PTEN/β-Catenin Axis in LPS Induced Acute Lung Injury. Front. Immunol. 2019, 10, 1612. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, S.; Vardon-Bounes, F.; Merlet-Dupuy, V.; Conil, J.M.; Buléon, M.; Fourcade, O.; Tack, I.; Minville, V. Sepsis modeling in mice: Ligation length is a major severity factor in cecal ligation and puncture. Intensive Care Med. Exp. 2016, 4, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, T.; Yin, H.; Chen, J.; Huang, L.; Jiang, J.; He, T.; Huang, H.; Hu, X. Survival advantage depends on cecal volume rather than cecal length in a mouse model of cecal ligation and puncture. J. Surg. Res. 2016, 203, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Knethen, A.; von Schäfer, A.; Kuchler, L.; Knape, T.; Christen, U.; Hintermann, E.; Fißlthaler, B.; Schröder, K.; Brandes, R.P.; Genz, B.; et al. Tolerizing CTL by Sustained Hepatic PD-L1 Expression Provides a New Therapy Approach in Mouse Sepsis. Theranostics 2019, 9, 2003–2016. [Google Scholar] [CrossRef]
- Gao, M.; Ou, H.; Jiang, Y.; Wang, K.; Peng, Y.; Zhang, H.; Yang, M.; Xiao, X. Tanshinone IIA attenuates sepsis-induced immunosuppression and improves survival rate in a mice peritonitis model. Biomed. Pharmacother. 2019, 112, 108609. [Google Scholar] [CrossRef]
- Gao, Y.L.; Chai, Y.F.; Qi, A.L.; Yao, Y.; Liu, Y.C.; Dong, N.; Wang, L.J.; Yao, Y.M. Neuropilin-1highCD4⁺CD25⁺ Regulatory T Cells Exhibit Primary Negative Immunoregulation in Sepsis. Mediat. Inflamm. 2016, 2016, 7132158. [Google Scholar] [CrossRef] [Green Version]
- Jeremias, I.C.; Victorino, V.J.; Barbeiro, H.V.; Kubo, S.A.; Prado, C.M.; Lima, T.M.; Soriano, F.G. The Role of Acetylcholine in the Inflammatory Response in Animals Surviving Sepsis Induced by Cecal Ligation and Puncture. Mol. Neurobiol. 2016, 53, 6635–6643. [Google Scholar] [CrossRef]
- Luan, Y.; Yin, C.; Qin, Q.; Dong, N.; Zhu, X.; Sheng, Z.; Zhang, Q.; Yao, Y. Effect of Regulatory T Cells on Promoting Apoptosis of T Lymphocyte and Its Regulatory Mechanism in Sepsis. J. Interferon Cytokine Res. 2015, 35, 969–980. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Wu, Z.; Ni, H.; Ke, L.; Tong, Z.; Li, W.; Li, N.; Li, J. Codonopsis pilosula polysaccharide attenuates cecal ligation and puncture sepsis via circuiting regulatory T cells in mice. Shock 2014, 41, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Andrade, M.M.C.; Ariga, S.S.K.; Barbeiro, D.F.; Barbeiro, H.V.; Pimentel, R.N.; Petroni, R.C.; Soriano, F.G. Endotoxin tolerance modulates TREG and TH17 lymphocytes protecting septic mice. Oncotarget 2019, 10, 3451–3461. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.Q.; Yao, Y.; Chen, W.; Bian, J.; Zhao, L.; Chen, L.; Hong, G.; Lu, Z.; Zhao, G. Partial Depletion of Regulatory T Cells Enhances Host Inflammatory Response Against Acute Pseudomonas aeruginosa Infection After Sepsis. Inflammation 2018, 41, 1780–1790. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.J.; Kim, S.J.; Lee, S.M. Overexpression of HO-1 Contributes to Sepsis-Induced Immunosuppression by Modulating the Th1/Th2 Balance and Regulatory T-Cell Function. J. Infect. Dis. 2017, 215, 1608–1618. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kong, B.B.; Yang, W.P.; Zhao, X.; Zhang, R. Immunomodulatory intervention with Gamma interferon in mice with sepsis. Life Sci. 2017, 185, 85–94. [Google Scholar] [CrossRef]
- Restagno, D.; Venet, F.; Paquet, C.; Freyburger, L.; Allaouchiche, B.; Monneret, G.; Bonnet, J.M.; Louzier, V. Mice Survival and Plasmatic Cytokine Secretion in a “Two Hit” Model of Sepsis Depend on Intratracheal Pseudomonas Aeruginosa Bacterial Load. PLoS ONE 2016, 11, e0162109. [Google Scholar] [CrossRef]
- Sharma, A.; Yang, W.L.; Matsuo, S.; Wang, P. Differential alterations of tissue T-cell subsets after sepsis. Immunol. Lett. 2015, 168, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Molinaro, R.; Pecli, C.; Guilherme, R.F.; Alves-Filho, J.C.; Cunha, F.Q.; Canetti, C.; Kunkel, S.L.; Bozza, M.T.; Benjamim, C.F. CCR4 Controls the Suppressive Effects of Regulatory T Cells on Early and Late Events during Severe Sepsis. PLoS ONE 2015, 10, e0133227. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.W.; Yang, W.; Gao, L.; Kang, J.R.; Qin, J.J.; Liu, Y.P.; Lu, J.Y. Adoptive transfer of bone marrow-derived dendritic cells decreases inhibitory and regulatory T-cell differentiation and improves survival in murine polymicrobial sepsis. Immunology 2015, 145, 50–59. [Google Scholar] [CrossRef]
- Hasan, Z.; Palani, K.; Zhang, S.; Lepsenyi, M.; Hwaiz, R.; Rahman, M.; Syk, I.; Jeppsson, B.; Thorlacius, H. Rho kinase regulates induction of T-cell immune dysfunction in abdominal sepsis. Infect. Immun. 2013, 81, 2499–2506. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Luo, L.; Wang, Y.; Rahman, M.; Lepsenyi, M.; Syk, I.; Jeppsson, B.; Thorlacius, H. Simvastatin protects against T cell immune dysfunction in abdominal sepsis. Shock 2012, 38, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, J.; Sheng, Y.; Zou, Y.; Bo, L.; Wang, F.; Lou, J.; Fan, X.; Bao, R.; Wu, Y.; et al. Baicalin improves survival in a murine model of polymicrobial sepsis via suppressing inflammatory response and lymphocyte apoptosis. PLoS ONE 2012, 7, e35523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiraki, S.; Ono, S.; Tsujimoto, H.; Kinoshita, M.; Takahata, R.; Miyazaki, H.; Saitoh, D.; Hase, K. Neutralization of interleukin-10 or transforming growth factor-β decreases the percentages of CD4+ CD25+ Foxp3+ regulatory T cells in septic mice, thereby leading to an improved survival. Surgery 2012, 151, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yamada, M.; Tamura, Y.; Chang, K.; Mao, J.; Zou, L.; Feng, Y.; Kida, K.; Scherrer-Crosbie, M.; Chao, W.; et al. Farnesyltransferase inhibitor FTI-277 reduces mortality of septic mice along with improved bacterial clearance. J. Pharmacol. Exp. Ther. 2011, 339, 832–841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Bäumel, M.; Männel, D.N.; Howard, O.M.Z.; Oppenheim, J.J. Interaction of TNF with TNF receptor type 2 promotes expansion and function of mouse CD4+CD25+ T regulatory cells. J. Immunol. 2007, 179, 154–161. [Google Scholar] [CrossRef] [Green Version]
- Wisnoski, N.; Chung, C.S.; Chen, Y.; Huang, X.; Ayala, A. The contribution of CD4+ CD25+ T-regulatory-cells to immune suppression in sepsis. Shock 2007, 27, 251–257. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.N.; Alexander-Miller, M.; Yoza, B.K.; Vachharajani, V.; McCall, C.E. Sirtuin1 Targeting Reverses Innate and Adaptive Immune Tolerance in Septic Mice. J. Immunol. Res. 2018, 2018, 2402593. [Google Scholar] [CrossRef]
- Zhang, S.; Luo, L.; Wang, Y.; Gomez, M.F.; Thorlacius, H. Nuclear factor of activated T cells regulates neutrophil recruitment, systemic inflammation, and T-cell dysfunction in abdominal sepsis. Infect. Immun. 2014, 82, 3275–3288. [Google Scholar] [CrossRef] [Green Version]
- Shih, J.M.; Shih, Y.M.; Pai, M.H.; Hou, Y.C.; Yeh, C.L.; Yeh, S.L. Fish Oil-Based Fat Emulsion Reduces Acute Kidney Injury and Inflammatory Response in Antibiotic-Treated Polymicrobial Septic Mice. Nutrients 2016, 8, 165. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.S.; Kim, S.J.; Lee, S.M. Genipin attenuates sepsis-induced immunosuppression through inhibition of T lymphocyte apoptosis. Int. Immunopharmacol. 2015, 27, 15–23. [Google Scholar] [CrossRef]
- Mohr, A.; Polz, J.; Martin, E.M.; Griessl, S.; Kammler, A.; Pötschke, C.; Lechner, A.; Bröker, B.M.; Mostböck, S.; Männel, D.N. Sepsis leads to a reduced antigen-specific primary antibody response. Eur. J. Immunol. 2012, 42, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Yang, W.L.; Aziz, M.; Ma, G.; Wang, P. Therapeutic effect of human ghrelin and growth hormone: Attenuation of immunosuppression in septic aged rats. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2584–2593. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.H.; Wu, H.P.; Wu, K.H.; Tsai, Y.G.; Peng, C.T.; Lin, K.C.; Chao, W.R.; Lee, M.S.; Fu, Y.C. An increase in CD3+CD4+CD25+ regulatory T cells after administration of umbilical cord-derived mesenchymal stem cells during sepsis. PLoS ONE 2014, 9, e110338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, W.; Du, H.; Fu, Q.; Cui, N.; Du, C. The influence of Th1/Th2 and CD4+ regulatory t cells of mesenteric lymph nodes on systemic lipopolysaccharide. PJP 2014, 2, 125–129. [Google Scholar] [CrossRef]
- Chen, H.H.; Chang, C.L.; Lin, K.C.; Sung, P.H.; Chai, H.T.; Zhen, Y.Y.; Chen, Y.C.; Wu, Y.C.; Leu, S.; Tsai, T.H.; et al. Melatonin augments apoptotic adipose-derived mesenchymal stem cell treatment against sepsis-induced acute lung injury. Am. J. Transl. Res. 2014, 6, 439–458. [Google Scholar]
- Chen, H.H.; Lin, K.C.; Wallace, C.G.; Chen, Y.T.; Yang, C.C.; Leu, S.; Chen, Y.C.; Sun, C.K.; Tsai, T.H.; Chen, Y.L.; et al. Additional benefit of combined therapy with melatonin and apoptotic adipose-derived mesenchymal stem cell against sepsis-induced kidney injury. J. Pineal Res. 2014, 57, 16–32. [Google Scholar] [CrossRef]
- Chang, C.L.; Leu, S.; Sung, H.C.; Zhen, Y.Y.; Cho, C.L.; Chen, A.; Tsai, T.H.; Chung, S.Y.; Chai, H.T.; Sun, C.K.; et al. Impact of apoptotic adipose-derived mesenchymal stem cells on attenuating organ damage and reducing mortality in rat sepsis syndrome induced by cecal puncture and ligation. J. Transl. Med. 2012, 10, 244. [Google Scholar] [CrossRef] [Green Version]
- Dhamne, C.; Chung, Y.; Alousi, A.M.; Cooper, L.J.N.; Tran, D.Q. Peripheral and thymic foxp3+ regulatory T cells in search of origin, distinction, and function. Front. Immunol. 2013, 4, 253. [Google Scholar] [CrossRef] [Green Version]
- Deng, G.; Song, X.; Fujimoto, S.; Piccirillo, C.A.; Nagai, Y.; Greene, M.I. Foxp3 Post-translational Modifications and Treg Suppressive Activity. Front. Immunol. 2019, 10, 2486. [Google Scholar] [CrossRef] [Green Version]
- Brunkow, M.E.; Jeffery, E.W.; Hjerrild, K.A.; Paeper, B.; Clark, L.B.; Yasayko, S.A.; Wilkinson, J.E.; Galas, D.; Ziegler, S.F.; Ramsdell, F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001, 27, 68–73. [Google Scholar] [CrossRef]
- Torgerson, T.R.; Ochs, H.D. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome: A model of immune dysregulation. Curr. Opin. Allergy Clin. Immunol. 2002, 2, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, Y.; Ohkura, N.; Kidani, Y.; Vandenbon, A.; Hirota, K.; Kawakami, R.; Yasuda, K.; Motooka, D.; Nakamura, S.; Kondo, M.; et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 2017, 18, 173–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, J.D.; Yasui, D.H.; Niida, H.; Joh, T.; Loh, D.Y.; Kohwi-Shigematsu, T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000, 14, 521–535. [Google Scholar] [PubMed]
- Chen, C.; Rowell, E.A.; Thomas, R.M.; Hancock, W.W.; Wells, A.D. Transcriptional regulation by Foxp3 is associated with direct promoter occupancy and modulation of histone acetylation. J. Biol. Chem. 2006, 281, 36828–36834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, C.; Tone, Y.; Tsuda, M.; Peter, C.; Waldmann, H.; Tone, M. TGF-β-mediated Foxp3 gene expression is cooperatively regulated by Stat5, Creb, and AP-1 through CNS2. J. Immunol. 2014, 192, 475–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polansky, J.K.; Kretschmer, K.; Freyer, J.; Floess, S.; Garbe, A.; Baron, U.; Olek, S.; Hamann, A.; Boehmer, H.; von Huehn, J. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 2008, 38, 1654–1663. [Google Scholar] [CrossRef]
- Li, X.; Liang, Y.; LeBlanc, M.; Benner, C.; Zheng, Y. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell 2014, 158, 734–748. [Google Scholar] [CrossRef] [Green Version]
- Hori, S. c-Rel: A pioneer in directing regulatory T-cell lineage commitment? Eur. J. Immunol. 2010, 40, 664–667. [Google Scholar] [CrossRef]
- Huehn, J.; Beyer, M. Epigenetic and transcriptional control of Foxp3+ regulatory T cells. Semin. Immunol. 2015, 27, 10–18. [Google Scholar] [CrossRef]
- Liu, B.; Tahk, S.; Yee, K.M.; Fan, G.; Shuai, K. The ligase PIAS1 restricts natural regulatory T cell differentiation by epigenetic repression. Science 2010, 330, 521–525. [Google Scholar] [CrossRef] [Green Version]
- Mantel, P.-Y.; Ouaked, N.; Rückert, B.; Karagiannidis, C.; Welz, R.; Blaser, K.; Schmidt-Weber, C.B. Molecular mechanisms underlying FOXP3 induction in human T cells. J. Immunol. 2006, 176, 3593–3602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouly, E.; Chemin, K.; Nguyen, H.V.; Chopin, M.; Mesnard, L.; Leite-de-Moraes, M.; Burlen-defranoux, O.; Bandeira, A.; Bories, J.-C. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J. Exp. Med. 2010, 207, 2113–2125. [Google Scholar] [CrossRef] [Green Version]
- Polansky, J.K.; Schreiber, L.; Thelemann, C.; Ludwig, L.; Krüger, M.; Baumgrass, R.; Cording, S.; Floess, S.; Hamann, A.; Huehn, J. Methylation matters: Binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J. Mol. Med. 2010, 88, 1029–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerdiles, Y.M.; Stone, E.L.; Beisner, D.R.; Beisner, D.L.; McGargill, M.A.; Ch’en, I.L.; Stockmann, C.; Katayama, C.D.; Hedrick, S.M. Foxo transcription factors control regulatory T cell development and function. Immunity 2010, 33, 890–904. [Google Scholar] [CrossRef] [Green Version]
- Sekiya, T.; Kashiwagi, I.; Yoshida, R.; Fukaya, T.; Morita, R.; Kimura, A.; Ichinose, H.; Metzger, D.; Chambon, P.; Yoshimura, A. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 2013, 14, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Yokota-Nakatsuma, A.; Ohoka, Y.; Kagechika, H.; Kato, C.; Song, S.-Y.; Iwata, M. Retinoid X receptor agonists modulate Foxp3⁺ regulatory T cell and Th17 cell differentiation with differential dependence on retinoic acid receptor activation. J. Immunol. 2013, 191, 3725–3733. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Kitani, A.; Stuelten, C.; McGrady, G.; Fuss, I.; Strober, W. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity 2010, 33, 313–325. [Google Scholar] [CrossRef] [Green Version]
- Bruno, L.; Mazzarella, L.; Hoogenkamp, M.; Hertweck, A.; Cobb, B.S.; Sauer, S.; Hadjur, S.; Leleu, M.; Naoe, Y.; Telfer, J.C.; et al. Runx proteins regulate Foxp3 expression. J. Exp. Med. 2009, 206, 2329–2337. [Google Scholar] [CrossRef]
- Zorn, E.; Nelson, E.A.; Mohseni, M.; Porcheray, F.; Kim, H.; Litsa, D.; Bellucci, R.; Raderschall, E.; Canning, C.; Soiffer, R.J.; et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood 2006, 108, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- Takimoto, T.; Wakabayashi, Y.; Sekiya, T.; Inoue, N.; Morita, R.; Ichiyama, K.; Takahashi, R.; Asakawa, M.; Muto, G.; Mori, T.; et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 2010, 185, 842–855. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.O.; Nurieva, R.; Martinez, G.J.; Kang, H.S.; Chung, Y.; Pappu, B.P.; Shah, B.; Chang, S.H.; Schluns, K.S.; Watowich, S.S.; et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 2008, 29, 44–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Li, B.; Xiao, Y.; Chen, C.; Wang, Q.; Liu, Y.; Berezov, A.; Xu, C.; Gao, Y.; Li, Z.; et al. Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function. Cell Rep. 2012, 1, 665–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Nagai, Y.; Deng, G.; Ohtani, T.; Zhu, Z.; Zhou, Z.; Zhang, H.; Ji, M.Q.; Lough, J.W.; Samanta, A.; et al. Dynamic interactions between TIP60 and p300 regulate FOXP3 function through a structural switch defined by a single lysine on TIP60. Cell Rep. 2014, 7, 1471–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Loosdregt, J.; Vercoulen, Y.; Guichelaar, T.; Gent, Y.Y.J.; Beekman, J.M.; van Beekum, O.; Brenkman, A.B.; Hijnen, D.-J.; Mutis, T.; Kalkhoven, E.; et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood 2010, 115, 965–974. [Google Scholar] [CrossRef]
- Kwon, H.-S.; Lim, H.W.; Wu, J.; Schnölzer, M.; Verdin, E.; Ott, M. Three novel acetylation sites in the Foxp3 transcription factor regulate the suppressive activity of regulatory T cells. J. Immunol. 2012, 188, 2712–2721. [Google Scholar] [CrossRef] [Green Version]
- Beier, U.H.; Wang, L.; Bhatti, T.R.; Liu, Y.; Han, R.; Ge, G.; Hancock, W.W. Sirtuin-1 targeting promotes Foxp3+ T-regulatory cell function and prolongs allograft survival. Mol. Cell. Biol. 2011, 31, 1022–1029. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Quiros, J.; Mahuron, K.; Pai, C.C.; Ranzani, V.; Young, A.; Silveria, S.; Harwin, T.; Abnousian, A.; Pagani, M.; et al. Targeting EZH2 Reprograms Intratumoral Regulatory T Cells to Enhance Cancer Immunity. Cell Rep. 2018, 23, 3262–3274. [Google Scholar] [CrossRef]
- DuPage, M.; Chopra, G.; Quiros, J.; Rosenthal, W.L.; Morar, M.M.; Holohan, D.; Zhang, R.; Turka, L.; Marson, A.; Bluestone, J.A. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity 2015, 42, 227–238. [Google Scholar] [CrossRef] [Green Version]
- Kwon, H.K.; Chen, H.M.; Mathis, D.; Benoist, C. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat. Immunol. 2017, 18, 1238–1248. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Zou, J.; Wang, M.; Ding, X.; Chepelev, I.; Zhou, X.; Zhao, W.; Wei, G.; Cui, J.; Zhao, K.; et al. Critical role of histone demethylase Jmjd3 in the regulation of CD4+ T-cell differentiation. Nat. Commun. 2014, 5, 5780. [Google Scholar] [CrossRef] [Green Version]
- Mock, J.R.; Dial, C.F.; Tune, M.K.; Norton, D.L.; Martin, J.R.; Gomez, J.C.; Hagan, R.S.; Dang, H.; Doerschuk, C.M. Transcriptional analysis of Foxp3+ Tregs and functions of two identified molecules during resolution of ALI. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, R.; Zoeten, E.F.; de Ozkaynak, E.; Chen, C.; Wang, L.; Porrett, P.M.; Li, B.; Turka, L.A.; Olson, E.N.; Greene, M.I.; et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 2007, 13, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Samanta, A.; Li, B.; Song, X.; Bembas, K.; Zhang, G.; Katsumata, M.; Saouaf, S.J.; Wang, Q.; Hancock, W.W.; Shen, Y.; et al. TGF-beta and IL-6 signals modulate chromatin binding and promoter occupancy by acetylated FOXP3. Proc. Natl. Acad. Sci. USA 2008, 105, 14023–14027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akimova, T.; Ge, G.; Golovina, T.; Mikheeva, T.; Wang, L.; Riley, J.L.; Hancock, W.W. Histone/protein deacetylase inhibitors increase suppressive functions of human FOXP3+ Tregs. Clin. Immunol. 2010, 136, 348–363. [Google Scholar] [CrossRef] [Green Version]
- Wing, K.; Onishi, Y.; Prieto-Martin, P.; Yamaguchi, T.; Miyara, M.; Fehervari, Z.; Nomura, T.; Sakaguchi, S. CTLA-4 control over Foxp3+ regulatory T cell function. Science 2008, 322, 271–275. [Google Scholar] [CrossRef]
- Finn, P.W.; He, H.; Wang, Y.; Wang, Z.; Guan, G.; Listman, J.; Perkins, D.L. Synergistic induction of CTLA-4 expression by costimulation with TCR plus CD28 signals mediated by increased transcription and messenger ribonucleic acid stability. J. Immunol. 1997, 158, 4074–4081. [Google Scholar]
- Perkins, D.; Wang, Z.; Donovan, C.; He, H.; Mark, D.; Guan, G.; Wang, Y.; Walunas, T.; Bluestone, J.; Listman, J.; et al. Regulation of CTLA-4 expression during T cell activation. J. Immunol. 1996, 156, 4154–4159. [Google Scholar]
- Brunner-Weinzierl, M.C.; Rudd, C.E. CTLA-4 and PD-1 Control of T-Cell Motility and Migration: Implications for Tumor Immunotherapy. Front. Immunol. 2018, 9, 1972. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, T.; Tagami, T.; Yamazaki, S.; Uede, T.; Shimizu, J.; Sakaguchi, N.; Mak, T.W.; Sakaguchi, S. Immunologic self-tolerance maintained by CD25+CD4+ regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J. Exp. Med. 2000, 192, 303–310. [Google Scholar] [CrossRef]
- Read, S.; Malmström, V.; Powrie, F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25+CD4+ regulatory cells that control intestinal inflammation. J. Exp. Med. 2000, 192, 295–302. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, E.M.; Wang, C.J.; Ryan, G.A.; Clough, L.E.; Qureshi, O.S.; Goodall, M.; Abbas, A.K.; Sharpe, A.H.; Sansom, D.M.; Walker, L.S.K. Ctla-4 controls regulatory T cell peripheral homeostasis and is required for suppression of pancreatic islet autoimmunity. J. Immunol. 2009, 182, 274–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, T.; Kishi, A.; Osaki, M.; Morikawa, H.; Prieto-Martin, P.; Wing, K.; Saito, T.; Sakaguchi, S. Construction of self-recognizing regulatory T cells from conventional T cells by controlling CTLA-4 and IL-2 expression. Proc. Natl. Acad. Sci. USA 2013, 110, E2116–E2125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Han, S.; Kang, Y.; Guo, M.; Hong, S.; Liu, F.; Fu, S.; Wang, L.; Wang, Q.X. SAHA, an HDAC inhibitor, synergizes with tacrolimus to prevent murine cardiac allograft rejection. Cell. Mol. Immunol. 2012, 9, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Sakaguchi, S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017, 27, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koenen, H.J.P.M.; Smeets, R.L.; Vink, P.M.; van Rijssen, E.; Boots, A.M.H.; Joosten, I. Human CD25highFoxp3pos regulatory T cells differentiate into IL-17-producing cells. Blood 2008, 112, 2340–2352. [Google Scholar] [CrossRef] [Green Version]
- Bieliauskas, A.V.; Pflum, M.K.H. Isoform-selective histone deacetylase inhibitors. Chem. Soc. Rev. 2008, 37, 1402–1413. [Google Scholar] [CrossRef]
- Reichert, N.; Choukrallah, M.-A.; Matthias, P. Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell. Mol. Life Sci. 2012, 69, 2173–2187. [Google Scholar] [CrossRef] [Green Version]
- Asfaha, Y.; Schrenk, C.; Alves Avelar, L.A.; Hamacher, A.; Pflieger, M.; Kassack, M.U.; Kurz, T. Recent advances in class IIa histone deacetylases research. Bioorg. Med. Chem. 2019, 27, 115087. [Google Scholar] [CrossRef]
- Felice, C.; Lewis, A.; Armuzzi, A.; Lindsay, J.O.; Silver, A. Review article: Selective histone deacetylase isoforms as potential therapeutic targets in inflammatory bowel diseases. Aliment. Pharmacol. Ther. 2015, 41, 26–38. [Google Scholar] [CrossRef]
- Warren, J.L.; MacIver, N.J. Regulation of Adaptive Immune Cells by Sirtuins. Front. Endocrinol. (Lausanne) 2019, 10, 466. [Google Scholar] [CrossRef]
- Yanginlar, C.; Logie, C. HDAC11 is a regulator of diverse immune functions. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.; Gao, J.; Su, L. Foxp3 inhibits HDAC1 activity to modulate gene expression in human T cells. Virology 2011, 421, 12–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murken, D.; Aufhauser, D., Jr.; Concors, S.; Wang, Z.; Ge, G. Nuclear Co-Repressor Complex Inhibition Reverses Benefit of HDAC2 Deletion in Renal Ischemia. Am. J. Transplant. 2017, 17 (Suppl. 3), 555. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, W.; Wang, J.; Malovannaya, A.; Xi, Y.; Li, W.; Guerra, R.; Hawke, D.H.; Qin, J.; Chen, J. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol. Syst. Biol. 2015, 11, 775. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Han, R.; Beier, U.H.; Bhatti, T.R.; Akimova, T.; Greene, M.I.; Hiebert, S.W.; Hancock, W.W. FOXP3⁺ regulatory T cell development and function require histone/protein deacetylase 3. J. Clin. Investig. 2015, 125, 1111–1123. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Beier, U.H.; Akimova, T.; Dahiya, S.; Han, R.; Samanta, A.; Levine, M.H.; Hancock, W.W. Histone/protein deacetylase inhibitor therapy for enhancement of Foxp3+ T-regulatory cell function posttransplantation. Am. J. Transplant. 2018, 18, 1596–1603. [Google Scholar] [CrossRef]
- Xiao, H.; Jiao, J.; Wang, L.; O’Brien, S.; Newick, K.; Wang, L.C.S.; Falkensammer, E.; Liu, Y.; Han, R.; Kapoor, V.; et al. HDAC5 controls the functions of Foxp3+ T-regulatory and CD8+ T cells. Int. J. Cancer 2016, 138, 2477–2486. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Wang, L.; Dahiya, S.; Beier, U.H.; Han, R.; Samanta, A.; Bergman, J.; Sotomayor, E.M.; Seto, E.; Kozikowski, A.P.; et al. Histone/protein deacetylase 11 targeting promotes Foxp3+ Treg function. Sci. Rep. 2017, 7, 8626. [Google Scholar] [CrossRef] [Green Version]
- Kasler, H.G.; Lim, H.W.; Mottet, D.; Collins, A.M.; Lee, I.S.; Verdin, E. Nuclear export of histone deacetylase 7 during thymic selection is required for immune self-tolerance. EMBO J. 2012, 31, 4453–4465. [Google Scholar] [CrossRef] [Green Version]
- Beier, U.H.; Wang, L.; Han, R.; Akimova, T.; Liu, Y.; Hancock, W.W. Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci. Signal. 2012, 5, ra45. [Google Scholar] [CrossRef] [Green Version]
- De Zoeten, E.F.; Wang, L.; Sai, H.; Dillmann, W.H.; Hancock, W.W. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology 2010, 138, 583–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Zoeten, E.F.; Wang, L.; Butler, K.; Beier, U.H.; Akimova, T.; Sai, H.; Bradner, J.E.; Mazitschek, R.; Kozikowski, A.P.; Matthias, P.; et al. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3+ T-regulatory cells. Mol. Cell. Biol. 2011, 31, 2066–2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalin, J.H.; Butler, K.V.; Akimova, T.; Hancock, W.W.; Kozikowski, A.P. Second-generation histone deacetylase 6 inhibitors enhance the immunosuppressive effects of Foxp3+ T-regulatory cells. J. Med. Chem. 2012, 55, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Bodas, M.; Mazur, S.; Min, T.; Vij, N. Inhibition of histone-deacetylase activity rescues inflammatory cystic fibrosis lung disease by modulating innate and adaptive immune responses. Respir. Res. 2018, 19, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nijhuis, L.; Peeters, J.G.C.; Vastert, S.J.; van Loosdregt, J. Restoring T Cell Tolerance, Exploring the Potential of Histone Deacetylase Inhibitors for the Treatment of Juvenile Idiopathic Arthritis. Front. Immunol. 2019, 10, 151. [Google Scholar] [CrossRef]
- Regna, N.L.; Vieson, M.D.; Luo, X.M.; Chafin, C.B.; Puthiyaveetil, A.G.; Hammond, S.E.; Caudell, D.L.; Jarpe, M.B.; Reilly, C.M. Specific HDAC6 inhibition by ACY-738 reduces SLE pathogenesis in NZB/W mice. Clin. Immunol. 2016, 162, 58–73. [Google Scholar] [CrossRef] [Green Version]
- Dahiya, S.; Wang, L.; Beier, U.H.; Han, R.; Hancock, W.W. HDAC10 Targeting Regulates Foxp3 Promoter, Enhances T-regulatory (Treg) Function and Suppresses Autoimmune Colitis. J. Immunol. 2018, 200, 54.11. [Google Scholar]
- Géraldy, M.; Morgen, M.; Sehr, P.; Steimbach, R.R.; Moi, D.; Ridinger, J.; Oehme, I.; Witt, O.; Malz, M.; Nogueira, M.S.; et al. Selective Inhibition of Histone Deacetylase 10: Hydrogen Bonding to the Gatekeeper Residue is Implicated. J. Med. Chem. 2019, 62, 4426–4443. [Google Scholar] [CrossRef]
- Akimova, T.; Xiao, H.; Liu, Y.; Bhatti, T.R.; Jiao, J.; Eruslanov, E.; Singhal, S.; Wang, L.; Han, R.; Zacharia, K.; et al. Targeting sirtuin-1 alleviates experimental autoimmune colitis by induction of Foxp3+ T-regulatory cells. Mucosal Immunol. 2014, 7, 1209–1220. [Google Scholar] [CrossRef]
- Levine, M.H.; Wang, Z.; Xiao, H.; Jiao, J.; Wang, L.; Bhatti, T.R.; Hancock, W.W.; Beier, U.H. Targeting Sirtuin-1 prolongs murine renal allograft survival and function. Kidney Int. 2016, 89, 1016–1026. [Google Scholar] [CrossRef] [Green Version]
- Shu, L.; Xu, C.Q.; Yan, Z.Y.; Yan, Y.; Jiang, S.Z.; Wang, Y.R. Post-Stroke Microglia Induce Sirtuin2 Expression to Suppress the Anti-inflammatory Function of Infiltrating Regulatory T Cells. Inflammation 2019, 42, 1968–1979. [Google Scholar] [CrossRef] [PubMed]
- Beier, U.H.; Angelin, A.; Akimova, T.; Wang, L.; Liu, Y.; Xiao, H.; Koike, M.A.; Hancock, S.A.; Bhatti, T.R.; Han, R.; et al. Essential role of mitochondrial energy metabolism in Foxp3⁺ T-regulatory cell function and allograft survival. FASEB J. 2015, 29, 2315–2326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, W.; Chen, W.; Liu, W.; Xu, Z.; Zhang, L. Sirtuin4 suppresses the anti-neuroinflammatory activity of infiltrating regulatory T cells in the traumatically injured spinal cord. Immunology 2019, 158, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Dequiedt, F.; van Lint, J.; Lecomte, E.; van Duppen, V.; Seufferlein, T.; Vandenheede, J.R.; Wattiez, R.; Kettmann, R. Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J. Exp. Med. 2005, 201, 793–804. [Google Scholar] [CrossRef]
- Xu, K.; Yang, W.Y.; Nanayakkara, G.K.; Shao, Y.; Yang, F.; Hu, W.; Choi, E.T.; Wang, H.; Yang, X. GATA3, HDAC6, and BCL6 Regulate FOXP3+ Treg Plasticity and Determine Treg Conversion into Either Novel Antigen-Presenting Cell-Like Treg or Th1-Treg. Front. Immunol. 2018, 9, 45. [Google Scholar] [CrossRef] [Green Version]
- Ellmeier, W. Molecular control of CD4+ T cell lineage plasticity and integrity. Int. Immunopharmacol. 2015, 28, 813–817. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, M.; Wang, Y.; Dorfman, R.G.; Liu, H.; Yu, T.; Chen, X.; Tang, D.; Xu, L.; Yin, Y.; et al. Faecalibacterium prausnitzii Produces Butyrate to Maintain Th17/Treg Balance and to Ameliorate Colorectal Colitis by Inhibiting Histone Deacetylase 1. Inflamm. Bowel Dis. 2018, 24, 1926–1940. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Buechler, N.L.; Woodruff, A.G.; Long, D.L.; Zabalawi, M.; Yoza, B.K.; McCall, C.E.; Vachharajani, V. Sirtuins and Immuno-Metabolism of Sepsis. Int. J. Mol. Sci. 2018, 19, 2738. [Google Scholar] [CrossRef] [Green Version]
- Vachharajani, V.T.; Liu, T.; Wang, X.; Hoth, J.J.; Yoza, B.K.; McCall, C.E. Sirtuins Link Inflammation and Metabolism. J. Immunol. Res. 2016, 2016, 8167273. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.J.; Hu, X.F.; Yan, M.; Zhang, W.Y.; Mao, X.B.; Shu, Y.W. Human umbilical vein endothelial cells promote the inhibitory activation of CD4+CD25+Foxp3+ regulatory T cells via PD-L1. Atherosclerosis 2016, 244, 108–112. [Google Scholar] [CrossRef]


| Strain | Sex | Weight | Age | CLP | Tregs | Ref. | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| [g] | [weeks] | Ligation | Needle Ø | Perfo-Ration | Dura-tion | Organ | ||||
| Mice | BALB/c | m | - | 8 | immediately distal to the ileocecal valve | twice | ↑ (spleen) | [67] | ||
| m | 20 ± 2 | 6–8 | 1/3, 2/3, 3/3 | 23G | single | 24 h | ↑ (spleen) | [68] | ||
| m | 20–25 | 8 | 50% | 21G | once | 15 d | ↑ (spleen) | [69] | ||
| m | 18–22 | - | 50% | 21G | twice | 24 + 48 h | ↑ (spleen) | [70] | ||
| m | 20 ± 1 | 6–8 | below the ileocecal valve | 18G | once | 1/2/3/4 d | ↑ (blood) | [71] | ||
| C57BL/6 | m | 25 | 8 | caecum ligated at its base | 18G | twice | ↓ (blood) | [72] | ||
| f | - | 6–8 | 50% | 27G | twice | 3/7 d | ↑ (spleen) | [73] | ||
| m | 25–27 | - | - | 23G | - | 48 h | ↑ (spleen) | [74] | ||
| m | 25–35 | - | 50% | 18G | twice | 24 h | ↑ (spleen) | [75] | ||
| m | 20–25 | 7–9 | 30% of its length | 21G | twice | 5 d | ↑ (spleen) | [76] | ||
| m | 20–25 | 8–10 | 1.5 cm from the tip | 22G | twice | 20 h | ↑ (spleen) | [77] | ||
| m/f | 6–8 | below the ileocecal valve | 21G | nine | 24 h | ↑ (PC, MLN) | [78] | |||
| m | 22–25 | 6–8 | 22G | once | 3 d | ↑ (spleen) | [79] | |||
| m | 20–25 | - | 75% | 21G | twice | 24 h | ↑ (spleen) | [80] | ||
| m | 20–25 | - | 75% | 21G | twice | 24 h | ↑ (spleen) | [81] | ||
| m | 22–30 | 8–10 | below the ileocecal valve | 22G | twice | 24 h | ↑ (spleen) | [82] | ||
| m | 25 | 8 | at its base | 21G | once | 1/3 d | ↑ (MLN) | [83] | ||
| m | - | 7 | 1 cm from the apex | 18G | twice | 16 h | ↑ (spleen) | [84] | ||
| f | - | 8–12 | 30% | 27G | once | 24 + 48 h | ↑ (spleen) | [85] | ||
| m | - | 8–12 | 22G | twice | 24 h | ↑ (spleen) | [86] | |||
| m | - | 6–8 | 22G | twice | 30 h | ↑ (spleen) | [87] | |||
| FVB/N 9xNFAT luc | - | - | - | 75% | 21G | twice | 24 h | ↑ (spleen) | [88] | |
| ICR | m | 30–35 | 6–8 | 50% | 23G | twice | 24 + 72 h | ↑ (blood) | [89] | |
| m | 27–29 | - | at its distal site | 20G | twice | 26 h | ↑ (spleen) | [90] | ||
| NMRI | 20–30 | - | 30% | 27G | once | 1/2/3 d | ↑ (spleen) | [91] | ||
| Rats | Fischer | m | - | 104 | 70% of its length | 18G | twice | 20 h | ↑ (spleen) | [92] |
| Wistar | m | 250–300 | 50% | 18G | twice | 18 h | ↓ (blood) | [93] | ||
| Wistar Hannover | m | 200–250 | 8 | below the ileocecal valve | 18G | twice | 24 h | ↑ (MLN) | [94] | |
| Sprague Dawley | m | 350–400 | - | distal ligation | 18G | twice | 3 d | ↑ (blood) | [95] | |
| Sprague Dawley | m | 320–350 | - | distal ligation | 18G | twice | 3 d | ↑ (spleen) | [96] | |
| Sprague Dawley | m | 400–450 | - | distal ligation | 18G | twice | 72 h | ↑ (blood + spleen) | [97] | |
| Class | Isoform | Localization | Effect of HDAC Targeting | Specific HDACi | HDAC-Foxp3 Interaction | Models | Ref. |
|---|---|---|---|---|---|---|---|
| I | HDAC1 | nucleus | ↓ | no | inhibits HDAC1 | CAT, C | [152,153] |
| HDAC2 | nucleus | ↑ | in progress | associates with Foxp3 | CAT, C | [153,154] | |
| HDAC3 | nucleus/cytosol | ↓ | no | destabilizes Foxp3 | CAT, C | [155] | |
| HDAC8 | nucleus | ↓ | available | ? | CAT | [155,156] | |
| IIa | HDAC5 | nucleus/cytosol | ↓ | no | ? | CAT | [157] |
| HDAC7 | nucleus/cytosol | ↓ | no | forms a transcriptional complex with Foxp3 | thymic positive and negative T cell selection | [158,159] | |
| HDAC9 | nucleus/cytosol | ↑ | no | destabilizes Foxp3 | C | [132,160,161] | |
| IIb | HDAC6 | nucleus/cytosol | ↑ | available | destabilizes Foxp3 | CF, CIA, JIA, lupus prone mice | [160,162,163,164,165,166] |
| HDAC10 | nucleus/cytosol | ↑ | in progress | destabilizes Foxp3, re-presses Foxp3 transcription | CAT, C | [167,168,169] | |
| III | SIRT1 | nucleus | ↑ | available | destabilizes Foxp3 | CLP, heterotrophic cardiac and ortho-tropic renal allo-graft, C | [87,126,160,169,170] |
| SIRT2 | cytosol | ↑ | no | destabilizes Foxp3 | MCAO | [171] | |
| SIRT3 | mito | ↓ | no | - | CAT | [172] | |
| SIRT4 | mito | ↑ | no | inhibits Foxp3 expression | mouse spinal cord compression in-jury | [173] | |
| IV | HDAC11 | nucleus | ↑ | available | destabilizes Foxp3 | CAT | [158] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
von Knethen, A.; Heinicke, U.; Weigert, A.; Zacharowski, K.; Brüne, B. Histone Deacetylation Inhibitors as Modulators of Regulatory T Cells. Int. J. Mol. Sci. 2020, 21, 2356. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072356
von Knethen A, Heinicke U, Weigert A, Zacharowski K, Brüne B. Histone Deacetylation Inhibitors as Modulators of Regulatory T Cells. International Journal of Molecular Sciences. 2020; 21(7):2356. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072356
Chicago/Turabian Stylevon Knethen, Andreas, Ulrike Heinicke, Andreas Weigert, Kai Zacharowski, and Bernhard Brüne. 2020. "Histone Deacetylation Inhibitors as Modulators of Regulatory T Cells" International Journal of Molecular Sciences 21, no. 7: 2356. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072356