Quantitative Proteomic Analyses Identify STO/BBX24 -Related Proteins Induced by UV-B
Abstract
:1. Introduction
2. Results
2.1. General Characteristics of STO-Dependent UV-B-Induecd Proteins
2.2. Changes of Candidate Proteins in Each Treatment Group
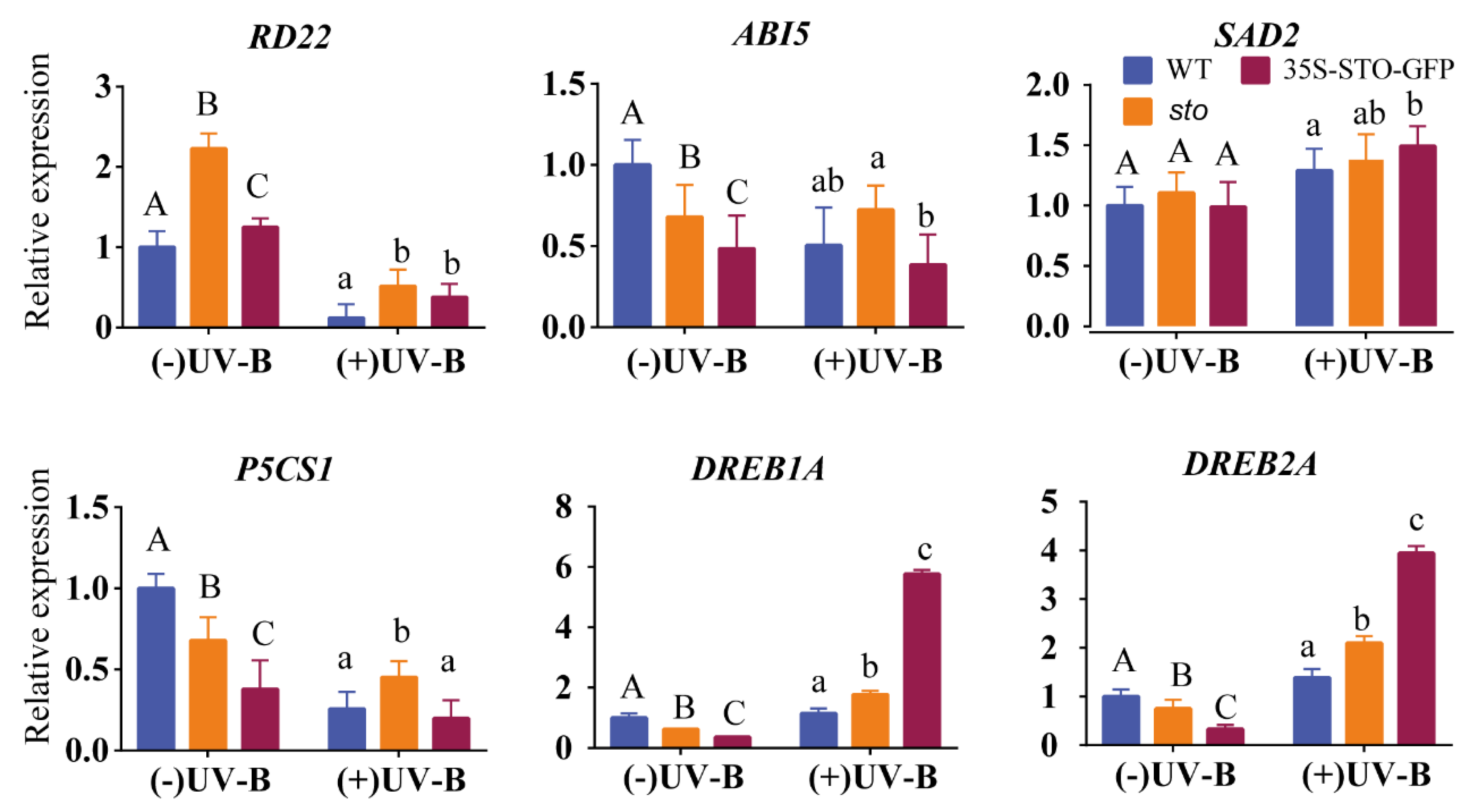
2.3. Verification of Differential Protein
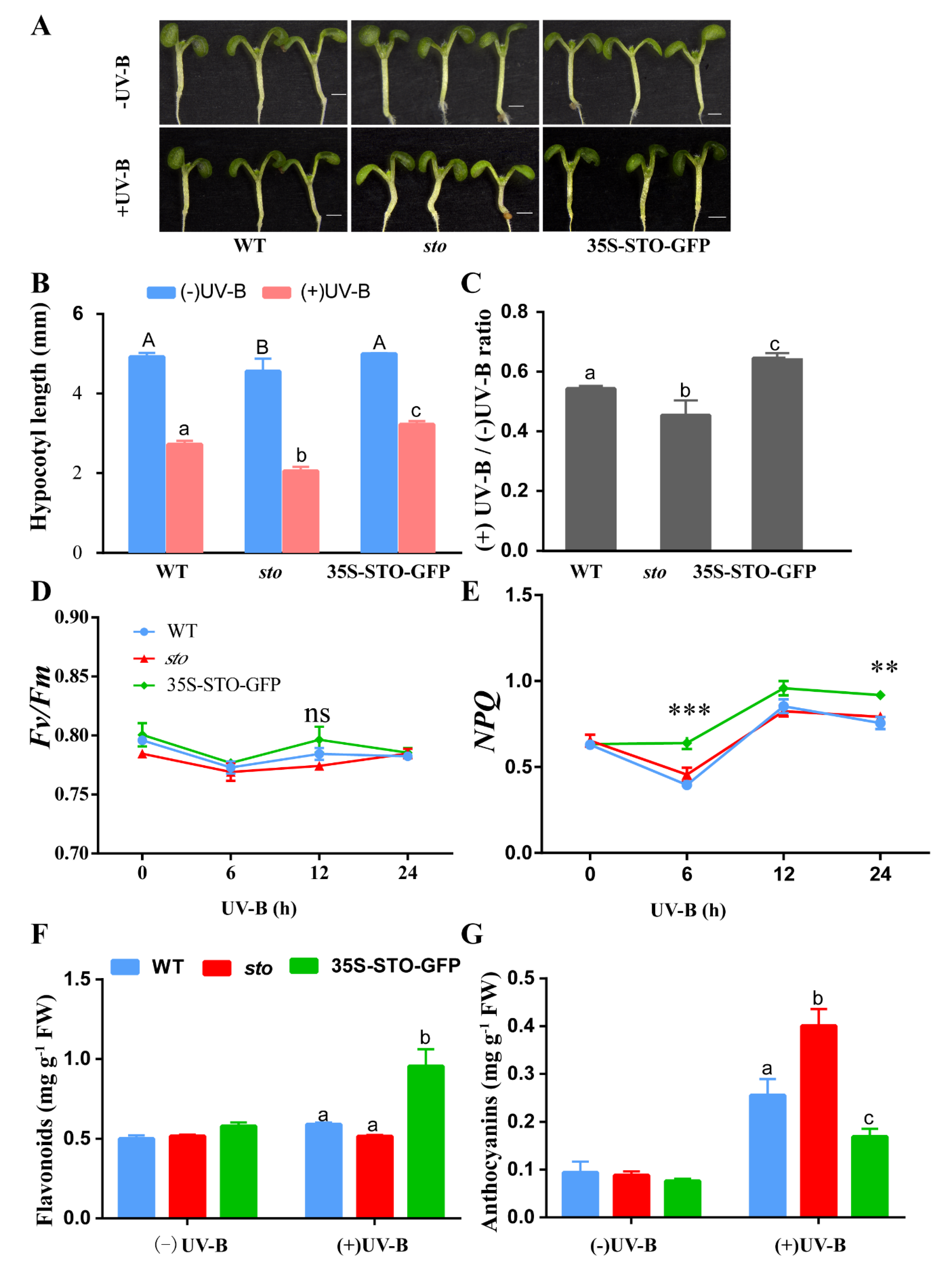
2.4. STO Regulates Hypocotyl Elongation and Protects the Photosynthetic System after UV-B Treatment
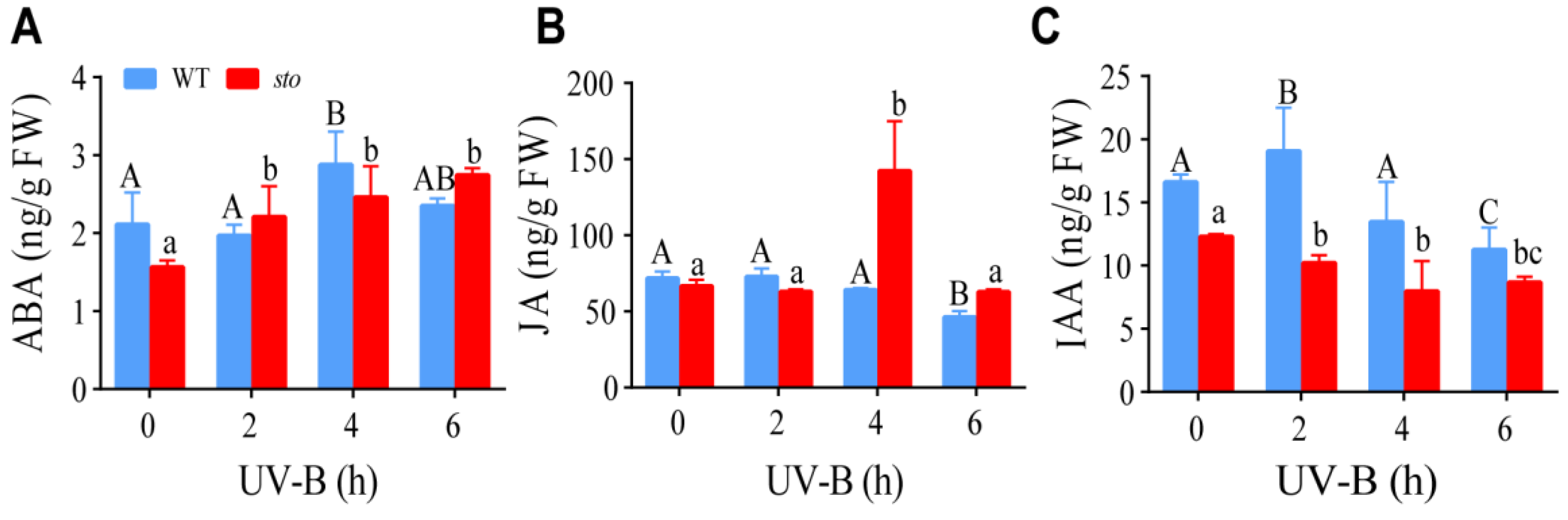
2.5. STO Affects the Content of ABA, JA, and IAA upon UV-B Exposure
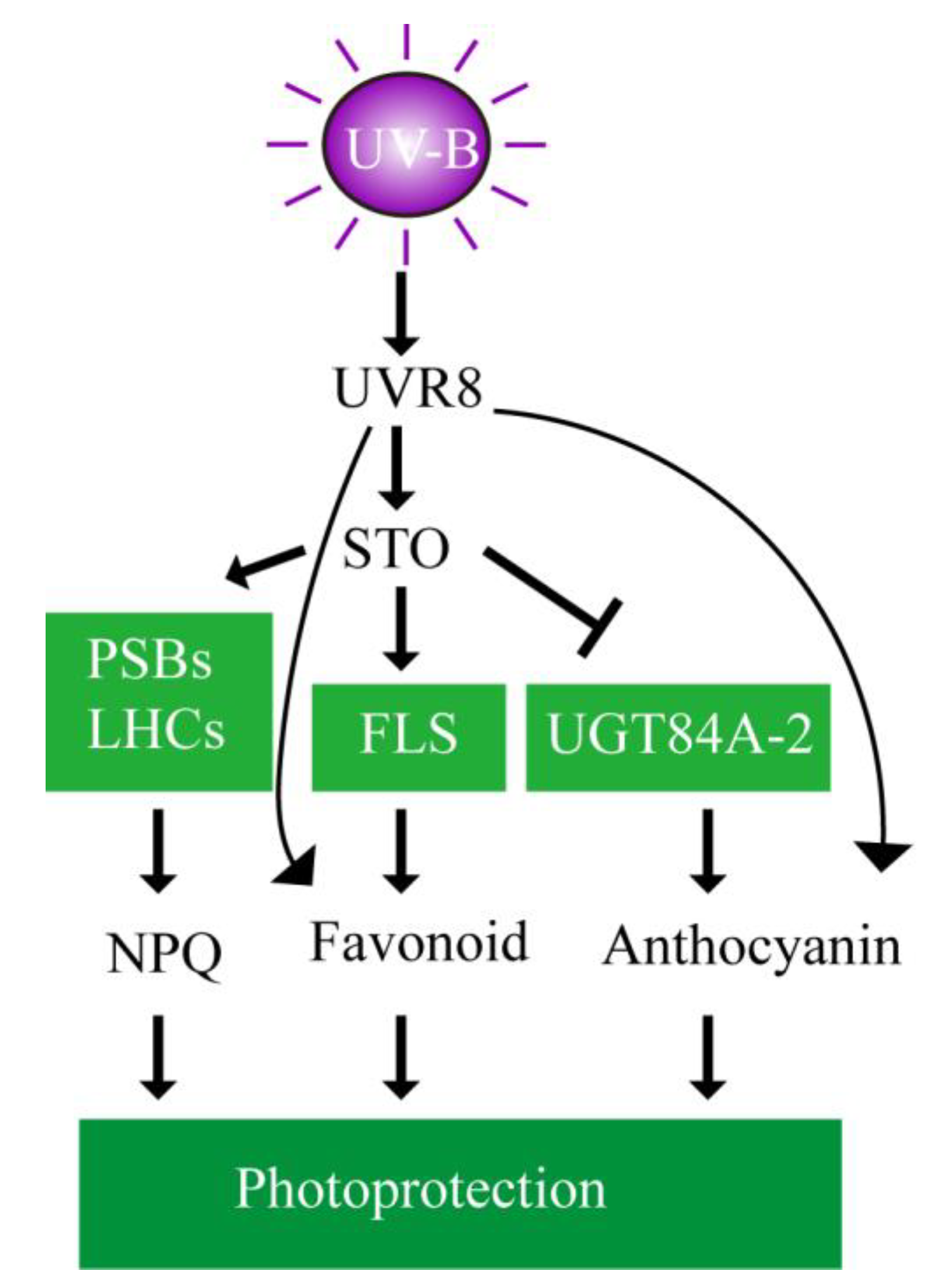
3. Discussion
3.1. The Accumulation of Flavonoids and Anthocyanins under UV-B Radiation is Independent of STO
3.2. STO Promotes Photosynthesis-Antenna Protein Accumulation
3.3. STO Had Crosstalk with ABA upon UV-B Radiation
4. Materials and Methods
4.1. Plant Materials and UV-B Treatment
4.2. Protein Extraction and Digestion
4.3. iTRAQ Labeling, Strong Cation Exchange (SCX) Fractionation, and LC-MS/MS Analysis
4.4. Protein Characterization
4.5. Function Annotation
4.6. Quantitative Real-Time PCR
4.7. Parallel Reaction Monitoring (PRM) Analysis
4.8. Flavonoid and Anthocyanin Measurement
4.9. Photochemical Activity Measurement
4.10. Measurements for IAA, ABA and JA
4.11. Data Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| STO | Salt Tolerance |
| HY5 | Elongated Hypocotyl 5 |
| iTRAQ | Isobaric Tags for Relative and Absolute Quantification |
| PRM | Parallel Reaction Monitoring |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| UVR8 | UV Resistance Locus 8 |
| COP1 | Constitutively Photomorphogenic 1 |
| RUPs | Repressor of UV-B Photomorphogenesis |
| PIF4 | Phytochrome Interacting Factor 4 |
| PAL | Phenylalanine Ammonia Lyase |
| CHS | Chalcone Synthase |
| DFR | Dihydroflavonol Reductase |
| FLS | Flavonol Synthase |
References
- Oravecz, A.; Baumann, A.; Máté, Z.; Brzezinska, A.; Molinier, J.; Oakeley, E.J.; Ádám, É.; Schäfer, E.; Nagy, F.; Ulm, R. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 2006, 18, 1975–1990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Favory, J.J.; Stec, A.; Gruber, H.; Rizzini, L.; Oravecz, A.; Funk, M.; Albert, A.; Cloix, C.; Jenkins, G.I.; Oakeley, E.J. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009, 28, 591–601. [Google Scholar] [CrossRef]
- Li, F.; Sun, J.; Wang, D.; Bai, S.; Clarke, A.K.; Holm, M. The B-box family gene STO (BBX24) in Arabidopsis thaliana regulates flowering time in different pathways. PLoS ONE 2014, 9, e87544. [Google Scholar] [CrossRef] [PubMed]
- Ulm, R.; Jenkins, G.I. Q & A: How do plants sense and respond to UV-B radiation? BMC Biol. 2015, 13, 45–50. [Google Scholar]
- Biever, J.J.; Gardner, G. The relationship between multiple UV-B perception mechanisms and DNA repair pathways in plants. Environ. Exp. Bot. 2016, 124, 89–99. [Google Scholar] [CrossRef]
- Jenkins, G.I. Structure and function of the UV-B photoreceptor UVR8. Curr. Opin. Struct. Biol. 2014, 29, 52–57. [Google Scholar] [CrossRef]
- Dotto, M.; Gómez, M.S.; Soto, M.S.; Casati, P. UV-B radiation delays flowering time through changes in the PRC2 complex activity and miR156 levels in Arabidopsis thaliana. Plant Cell Environ. 2018, 41, 1394–1406. [Google Scholar] [CrossRef]
- Rizzini, L.; Favory, J.J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schäfer, E.; Nagy, F.; Jenkins, G.I. Perception of UV-B by the Arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.; Hu, Q.; Yan, Z.; Chen, W.; Yan, C.; Huang, X.; Zhang, J.; Yang, P.; Deng, H.; Wang, J. Structural basis of ultraviolet-B perception by UVR8. Nature 2012, 484, 214–219. [Google Scholar] [PubMed]
- Fernández, M.B.; Tossi, V.; Lamattina, L.; Cassia, R. A comprehensive phylogeny reveals functional conservation of the UV-B photoreceptor UVR8 from green algae to higher plants. Front. Plant Sci. 2016, 7, 1698–1703. [Google Scholar] [CrossRef] [Green Version]
- Liang, T.; Yang, Y.; Liu, H. Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol. 2019, 221, 1247–1252. [Google Scholar] [CrossRef] [Green Version]
- Binkert, M.; Kozma-Bognár, L.; Terecskei, K.; De Veylder, L.; Nagy, F.; Ulm, R. UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 2014, 26, 4200–4213. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Liang, T.; Zhang, L.; Shao, K.; Gu, X.; Shang, R.; Shi, N.; Li, X.; Zhang, P.; Liu, H. UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat. Plants 2018, 4, 98–107. [Google Scholar] [CrossRef]
- Liang, T.; Mei, S.; Shi, C.; Yang, Y.; Peng, Y.; Ma, L.; Wang, F.; Li, X.; Huang, X.; Yin, Y. UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev. Cell 2018, 44, 512–523.e5. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Wang, Y.; Li, Q.F.; Björn, L.O.; He, J.X.; Li, S.S. Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 2012, 22, 1046–1057. [Google Scholar] [CrossRef] [Green Version]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [CrossRef]
- Hayes, S.; Velanis, C.N.; Jenkins, G.I.; Franklin, K.A. UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc. Natl. Acad. Sci. USA 2014, 111, 11894–11899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangappa, S.N.; Crocco, C.D.; Henrik, J.; Sourav, D.; Chamari, H.; Magnus, H.; Botto, J.F. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Job, N.; Yadukrishnan, P.; Bursch, K.; Datta, S.; Johansson, H. Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 2018, 176, 2963–2976. [Google Scholar] [CrossRef] [PubMed]
- Lyu, G.; Li, D.; Li, S.; Hu, H. STO and GA negatively regulate UV-B-induced Arabidopsis root growth inhibition. Plant Signal. Behav. 2019. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. Biorxiv 2018, 289660. [Google Scholar] [CrossRef]
- Imtiaz, M.; Yang, Y.; Liu, R.; Xu, Y.; Khan, M.A.; Wei, Q.; Gao, J.; Hong, B. Identification and functional characterization of the BBX24 promoter and gene from chrysanthemum in Arabidopsis. Plant Mol. Biol. 2015, 89, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ouyang, X.; Yang, P.; Lau, O.S.; Chen, L.; Wei, N.; Deng, X.W. Conversion from CUL4-based COP1–SPA E3 apparatus to UVR8–COP1–SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc. Natl. Acad. Sci. USA 2013, 110, 16669–16674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruber, H.; Heijde, M.; Heller, W.; Albert, A.; Seidlitz, H.K.; Ulm, R. Negative feedback regulation of UV-B–induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 20132–20137. [Google Scholar] [CrossRef] [Green Version]
- Treutter, D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol. 2005, 7, 581–591. [Google Scholar] [CrossRef]
- Choudhary, K.K.; Agrawal, S.B. Cultivar specificity of tropical mung bean (Vigna radiata L.) to elevated ultraviolet-B: Changes in antioxidative defense system, nitrogen metabolism and accumulation of jasmonic and salicylic acids. Environ. Exp. Bot. 2014, 99, 122–132. [Google Scholar] [CrossRef]
- Liu, X.; Li, Q.; Yue, M.; Zhang, X.; Zhang, R.; Zhang, B.; Wang, M. Nitric oxide is involved in integration of UV-B absorbing compounds among parts of clonal plants under a heterogeneous UV-B environment. Physiol. Plant. 2015, 155, 180–191. [Google Scholar] [CrossRef]
- Hideg, É.; Jansen, M.A.; Strid, Å. UV-B exposure, ROS, and stress: Inseparable companions or loosely linked associates? Trends Plant Sci. 2013, 18, 107–115. [Google Scholar] [CrossRef] [Green Version]
- Bais, A.F.; Lucas, R.M.; Bornman, J.F.; Williamson, C.E.; Sulzberger, B.; Austin, A.T.; Wilson, S.R.; Andrady, A.L.; Bernhard, G.; McKenzie, R.L. Environmental effects of ozone depletion, UV radiation and interactions with climate change: UNEP Environmental Effects Assessment Panel, update 2017. Photochem. Photobiol. Sci. 2018, 17, 127–179. [Google Scholar] [CrossRef]
- Morales, L.O.; Brosché, M.; Vainonen, J.; Jenkins, G.I.; Wargent, J.J.; Sipari, N.; Strid, Å.; Lindfors, A.V.; Tegelberg, R.; Aphalo, P.J. Multiple roles for UV RESISTANCE LOCUS8 in regulating gene expression and metabolite accumulation in Arabidopsis under solar ultraviolet radiation. Plant Physiol. 2013, 161, 744–759. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Abrahan, C.; Colquhoun, T.A.; Liu, C. A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in Arabidopsis. Plant Cell 2017, 29, 1157–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Jiang, S.; Zhang, Z.; Fang, H.; Xu, H.; Wang, Y.; Chen, X. Malus sieversii: The origin, flavonoid synthesis mechanism, and breeding of red-skinned and red-fleshed apples. Hortic. Res. 2018, 5, 70–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maier, A.; Schrader, A.; Kokkelink, L.; Falke, C.; Welter, B.; Iniesto, E.; Rubio, V.; Uhrig, J.F.; Hülskamp, M.; Hoecker, U. Light and the E3 ubiquitin ligase COP 1/SPA control the protein stability of the MYB transcription factors PAP 1 and PAP 2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013, 74, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Wang, H.; Li, D.; Yu, B.; Hui, Q.; Yan, S.; Huang, Z.; Cui, X.; Cao, B. Identification of candidate HY5-dependent and-independent regulators of anthocyanin biosynthesis in Tomato. Plant Cell Physiol. 2018, 60, 643–656. [Google Scholar] [CrossRef]
- Ren, H.; Han, J.; Yang, P.; Mao, W.; Liu, X.; Qiu, L.; Qian, C.; Liu, Y.; Chen, Z.; Ouyang, X.; et al. Two E3 ligases antagonistically regulate the UV-B response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 4722–4731. [Google Scholar] [CrossRef] [Green Version]
- Stracke, R.; FAVORY, J.J.; Gruber, H.; Bartelniewoehner, L.; Bartels, S.; Binkert, M.; Funk, M.; Weisshaar, B.; Ulm, R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2010, 33, 88–103. [Google Scholar]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef] [Green Version]
- Ramsay, N.A.; Glover, B.J. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Tanaka, Y.; Ohmiya, A. Seeing is believing: Engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotechnol. 2008, 19, 190–197. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [Green Version]
- Shin, D.H.; Choi, M.G.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.-I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Cai, J.; Yang, Y.; Liu, Z. Overexpression of microRNA828 reduces anthocyanin accumulation in Arabidopsis. Plant Cell Tissue Organ Cult. (PCTOC) 2013, 115, 159–167. [Google Scholar] [CrossRef]
- Qin, X.; Pi, X.; Wang, W.; Han, G.; Zhu, L.; Liu, M.; Cheng, L.; Shen, J.-R.; Kuang, T.; Sui, S.-F. Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits. Nat. Plants 2019, 5, 263–272. [Google Scholar] [CrossRef]
- Yu, G.; Li, W.; Yuan, Z.; Cui, H.; Lv, C.; Gao, Z.; Han, B.; Gong, Y.; Chen, G. The effects of enhanced UV-B radiation on photosynthetic and biochemical activities in super-high-yield hybrid rice Liangyoupeijiu at the reproductive stage. Photosynthetica 2013, 51, 33–44. [Google Scholar] [CrossRef]
- Shi, S.B.; Zhu, Z.W.Y.; Li, H.M.; Zhou, D.W.; Fa, H.; Zhao, X.Q.; Tang, Y.H. Photosynthesis of Saussurea superba and Gentiana straminea is not reduced after long-term enhancement of UV-B radiation. Environ. Exp. Bot. 2004, 51, 75–83. [Google Scholar] [CrossRef]
- Kataria, S.; Jajoo, A.; Guruprasad, K.N. Impact of increasing Ultraviolet-B (UV-B) radiation on photosynthetic processes. J. Photochem. Photobiol. B Biol. 2014, 137, 55–66. [Google Scholar] [CrossRef]
- Xu, C.; Sullivan, J.H. Reviewing the technical designs for experiments with ultraviolet-B radiation and impact on photosynthesis, DNA and secondary metabolism. J. Integr. Plant Biol. 2010, 52, 377–387. [Google Scholar] [CrossRef]
- Allorent, G.; Lefebvre-Legendre, L.; Chappuis, R.; Kuntz, M.; Truong, T.B.; Niyogi, K.K.; Ulm, R.; Goldschmidt-Clermont, M. UV-B photoreceptor-mediated protection of the photosynthetic machinery in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2016, 113, 14864–14869. [Google Scholar] [CrossRef] [Green Version]
- Külheim, C.; Ågren, J.; Jansson, S. Rapid regulation of light harvesting and plant fitness in the field. Science 2002, 297, 91–93. [Google Scholar] [CrossRef]
- Funk, C.; Schroeder, W.P.; Napiwotzki, A.; Tjus, S.E.; Renger, G.; Andersson, B. The PSII-S protein of higher plants: A new type of pigment-binding protein. Biochemistry 1995, 34, 11133–11141. [Google Scholar] [CrossRef]
- Müller, P.; Li, X.P.; Niyogi, K.K. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001, 125, 1558–1566. [Google Scholar] [CrossRef] [Green Version]
- Bec̆ková, M.; Yu, J.; Krynická, V.; Kozlo, A.; Shao, S.; Koník, P.; Komenda, J.; Murray, J.W.; Nixon, P.J. Structure of Psb29/Thf1 and its association with the FtsH protease complex involved in photosystem II repair in cyanobacteria. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160394. [Google Scholar] [CrossRef]
- Alboresi, A.; Ballottari, M.; Hienerwadel, R.; Giacometti, G.M.; Morosinotto, T. Antenna complexes protect photosystem I from photoinhibition. BMC Plant Biol. 2009, 9, 71–80. [Google Scholar] [CrossRef] [Green Version]
- Hieber, A.D.; Bugos, R.C.; Yamamoto, H.Y. Plant lipocalins: Violaxanthin de-epoxidase and zeaxanthin epoxidase. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 2000, 1482, 84–91. [Google Scholar] [CrossRef]
- Goss, R.; Lepetit, B. Biodiversity of NPQ. J. Plant Physiol. 2015, 172, 13–32. [Google Scholar] [CrossRef]
- Schaller, S.; Wilhelm, C.; Strzałka, K.; Goss, R. Investigating the interaction between the violaxanthin cycle enzyme zeaxanthin epoxidase and the thylakoid membrane. J. Photochem. Photobiol. B Biol. 2012, 114, 119–125. [Google Scholar] [CrossRef]
- Thole, J.M.; Beisner, E.R.; Liu, J.; Venkova, S.V.; Strader, L.C. Abscisic acid regulates root elongation through the activities of auxin and ethylene in Arabidopsis thaliana. G3 Genes Genomes Genet. 2014, 4, 1259–1274. [Google Scholar] [CrossRef] [Green Version]
- Forgione, I.; Wołoszyńska, M.; Pacenza, M.; Chiappetta, A.; Greco, M.; Araniti, F.; Abenavoli, M.R.; Van Lijsebettens, M.; Bitonti, M.B.; Bruno, L. Hypomethylated drm1 drm2 cmt3 mutant phenotype of Arabidopsis thaliana is related to auxin pathway impairment. Plant Sci. 2019, 280, 383–396. [Google Scholar] [CrossRef]
- Ronemus, M.J.; Galbiati, M.; Ticknor, C.; Chen, J.; Dellaporta, S.L. Demethylation-induced developmental pleiotropy in Arabidopsis. Science 1996, 273, 654–657. [Google Scholar] [CrossRef]
- Greco, M.; Sáez, C.A.; Contreras, R.A.; Rodríguez-Rojas, F.; Bitonti, M.B.; Brown, M.T. Cadmium and/or copper excess induce interdependent metal accumulation, DNA methylation, induction of metal chelators and antioxidant defences in the seagrass Zostera marina. Chemosphere 2019, 224, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Zhang, L.; Zhao, K.; Niu, R.; Zhai, H.; Zhang, J. VaERD15, a transcription factor gene associated with cold-tolerance in Chinese Wild Vitis amurensis. Front. Plant Sci. 2017, 8, 297. [Google Scholar] [CrossRef] [Green Version]
- Dunaeva, M.; Adamska, I. Identification of genes expressed in response to light stress in leaves of Arabidopsis thaliana using RNA differential display. Eur. J. Biochem. 2001, 268, 5521–5529. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Li, M.; Zhang, K.; Long, R.; Sun, Y.; Kang, J.; Zhang, T.; Cao, S. iTRAQ-based comparative proteomic analysis reveals tissue-specific and novel early-stage molecular mechanisms of salt stress response in Carex Rigescens. Environ. Exp. Bot. 2017, 143, 99–114. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Minoru, K.; Susumu, G.; Yoko, S.; Miho, F.; Mao, T. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011, 40, D109–D114. [Google Scholar]
- Peterson, A.C.; Russell, J.D.; Bailey, D.J.; Westphall, M.S.; Coon, J.J. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteom. 2012, 11, 1475–1488. [Google Scholar] [CrossRef] [Green Version]
- Kucera, B.; Leubner-Metzger, G.; Wellmann, E. Distinct ultraviolet-signaling pathways in bean leaves. DNA damage is associated with β-1, 3-glucanase gene induction, but not with flavonoid formation. Plant Physiol. 2003, 133, 1445–1452. [Google Scholar] [CrossRef] [Green Version]
- Weiss, D.; Halevy, A.H. Stamens and gibberellin in the regulation of corolla pigmentation and growth in Petunia hybrida. Planta 1989, 179, 89–96. [Google Scholar] [CrossRef]
- Meng, X.; Wang, X. Regulation of flower development and anthocyanin accumulation in Gerbera hybrida. J. Hortic. Sci. Biotechnol. 2004, 79, 131–137. [Google Scholar] [CrossRef]
- Sedej, T.T.; Gaberščik, A. The effects of enhanced UV-B radiation on physiological activity and growth of Norway spruce planted outdoors over 5 years. Trees Struct. Funct. 2008, 22, 423–435. [Google Scholar] [CrossRef]
- Zhou, L.; Xiao, L.; Xue, H. Dynamic cytology and transcriptional regulation of rice lamina joint development. Plant Physiol. 2017, 174, 1728–1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]


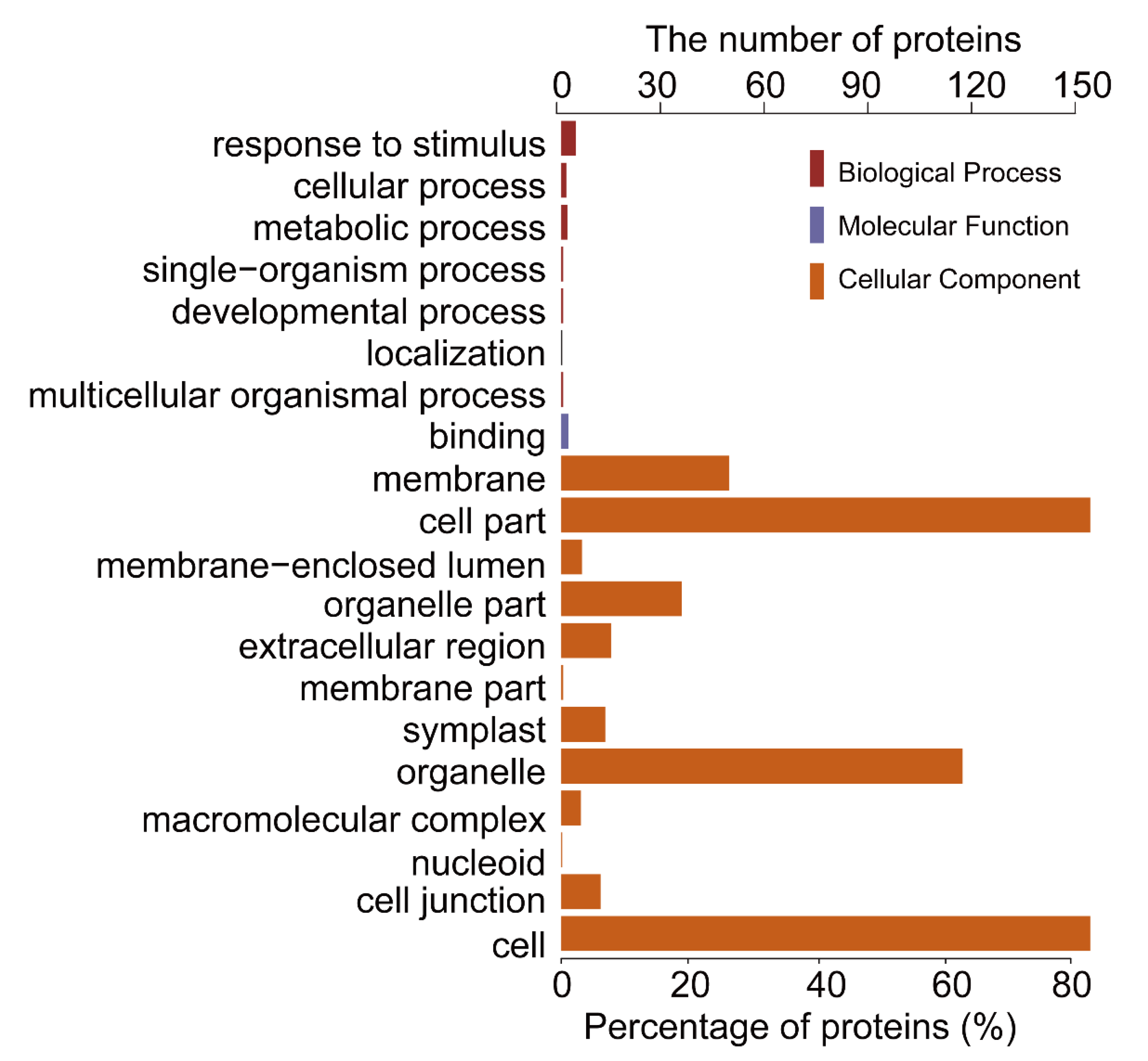
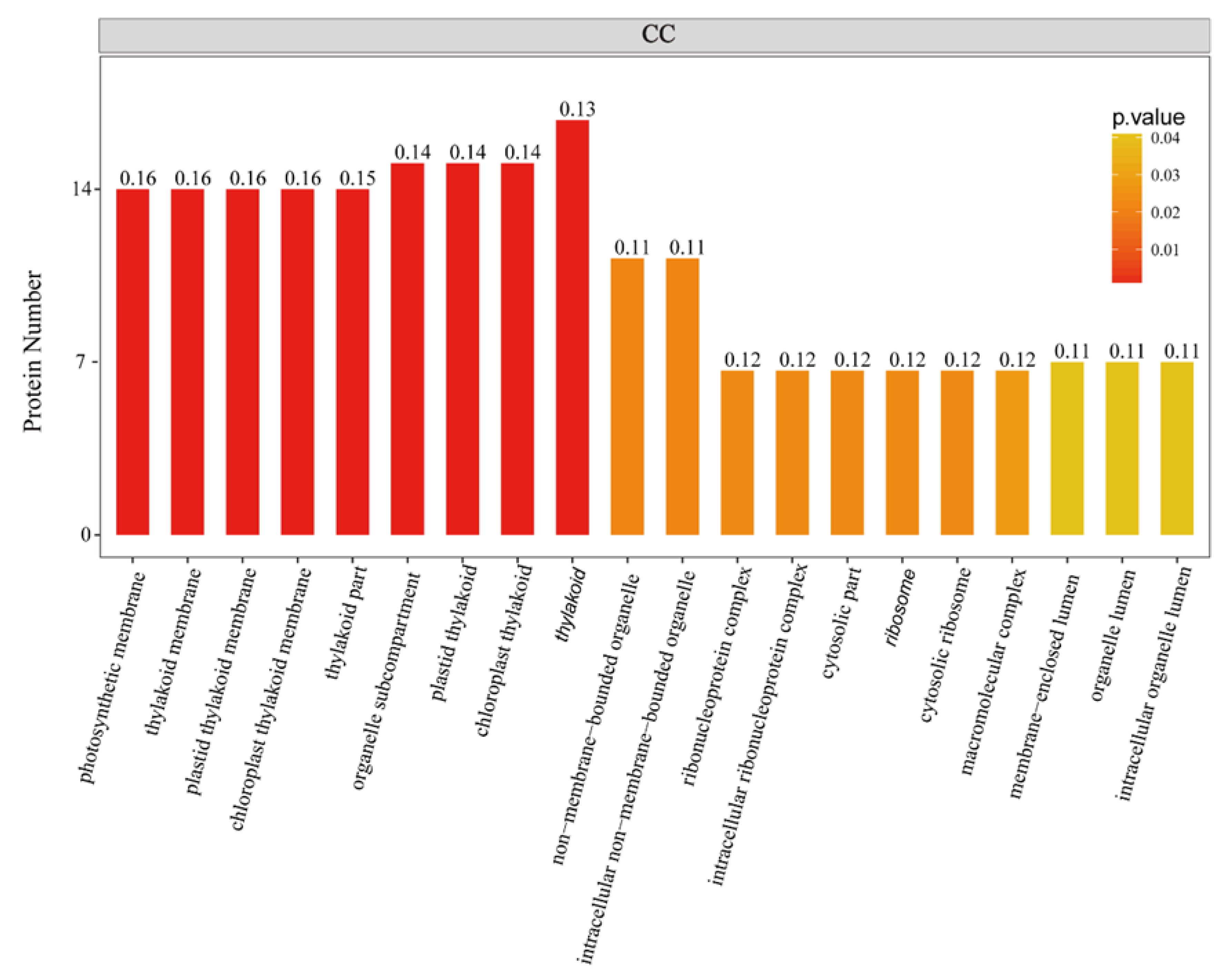
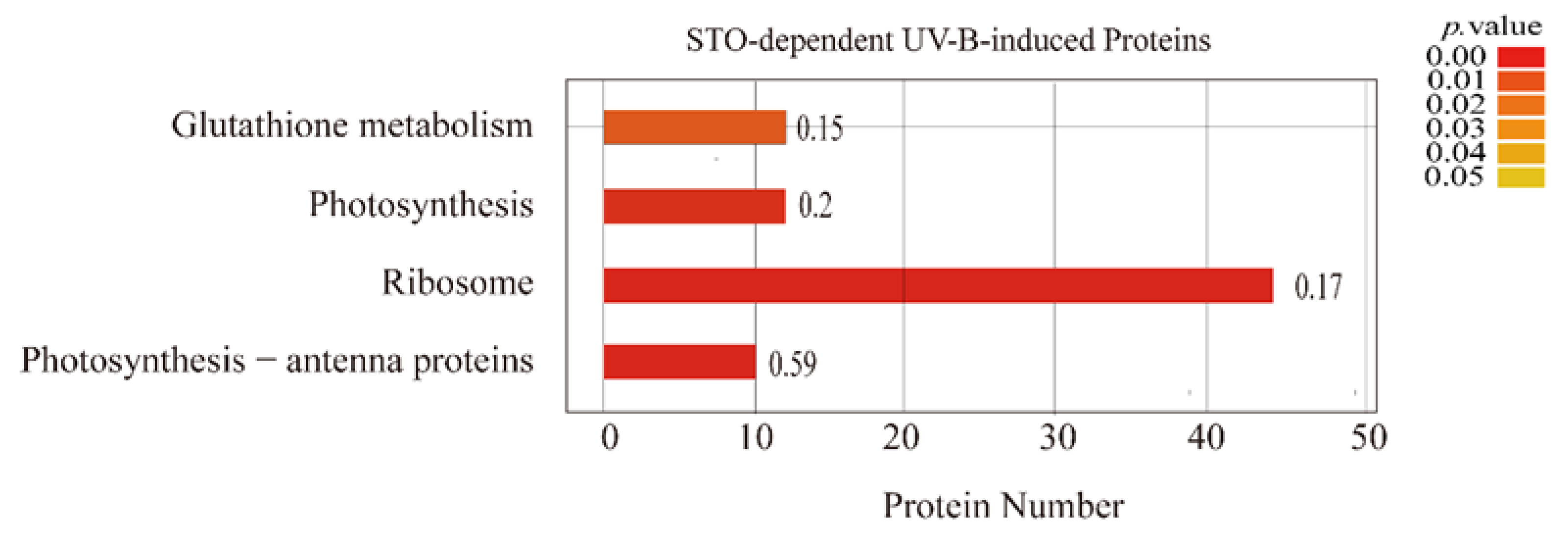


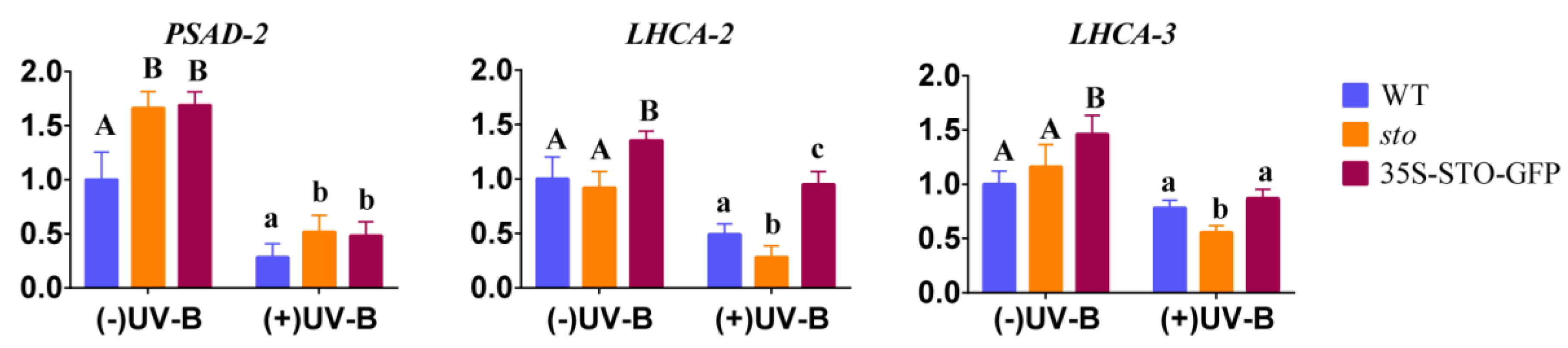
| Database | NO. | Total Spectra | Peptide Spectrum Match | Peptides | Unique Peptides | Protein Groups |
|---|---|---|---|---|---|---|
| TAIR | 1 | 312,550 | 94,209 | 41,480 | 36,822 | 7306 |
| TAIR | 2 | 307,230 | 94,057 | 40,076 | 35,436 | 7130 |
| TAIR | 3 | 306,008 | 93,046 | 38,283 | 33,967 | 6931 |
| TAIR | Total | 925,788 | 281,312 | 54,992 | 48,791 | 8212 |
| Comparison Group | Up-Regulated | Down-Regulated | In Total |
|---|---|---|---|
| (+)WT/(-)WT | 27 | 14 | 41 |
| (+)sto/(-)sto | 213 | 109 | 322 |
| (+)sto/(+)WT | 292 | 322 | 614 |
| (-)sto/(-)WT | 45 | 39 | 84 |
| Gene ID | Description | (+)WT/ (-)WT | (+)sto/ (-)sto | (-)sto/ (-)WT | (+)sto/ (+)WT |
|---|---|---|---|---|---|
| AT2G37040.1 | PAL1, phenylalanine ammonia-lyase 1 | 1.6 | 1.31 | 1.13(NS) | 0.92 |
| AT3G53260.1 | PAL2, phenylalanine ammonia-lyase 2 | 1.35 | 1.18 | 0.99(NS) | 0.87 |
| AT5G13930.1 | CHS, chalcone and stilbene synthase family protein | 1.86 | 1.39 | 1.23 | 0.92 |
| AT5G08640.1 | Flavonol synthase 1 | 1.32 | 1.12 | 1.07(NS) | 0.9 |
| AT3G21560.1 | UDP-Glycosyltransferase 84A2 | 1.57 | 1.35 | 1.04(NS) | 0.89 |
| AT1G43620.1 | UDP-Glycosyltransferase superfamily protein | 1.22 | 1.05(NS) | 0.96(NS) | 0.82 |
| Gene ID | Description | (+)WT/ (-)WT | (+) sto/ (-) sto | (-)sto/ (-)WT | (+)sto/ (+)WT |
|---|---|---|---|---|---|
| AT2G05620.1 | PGR5, response to water deprivation, photosynthetic electron transport in PSI | 1.10(NS) | 0.90(NS) | 0.95(NS) | 0.78 |
| AT3G54890.4 | Chlorophyll binding component of the light harvesting complex associated with PSI | 1.08(NS) | 0.93(NS) | 0.90(NS) | 0.77 |
| AT1G31330.1 | PSAF, PSI subunit F | 1.11(NS) | 1.00(NS) | 0.91(NS) | 0.81 |
| ATCG01010.1 | NADH dehydrogenase activity | 1.12(NS) | 0.92(NS) | 0.95(NS) | 0.78 |
| AT5G58260.1 | Oxidoreductases, acting on NADH or NADPH, quinone or similar compound as acceptor | 1.07(NS) | 0.96(NS) | 0.92(NS) | 0.82 |
| AT3G01440.1 | Oxygen evolving enhancer 3 (PsbQ) family protein | 1.05(NS) | 0.97(NS) | 0.9 | 0.83 |
| ATCG00710.1 | A component of the photosystem II oxygen evolving core | 1.19 | 0.94(NS) | 1.04(NS) | 0.82 |
| AT2G40100.1 | LHCB4.3 (light harvesting complex PSc), chlorophyll binding | 1.03(NS) | 0.85 | 0.97(NS) | 0.8 |
| AT1G15820.1 | LHCB6, PSII antenna complex | 1.09(NS) | 0.96(NS) | 0.92(NS) | 0.81 |
| AT1G61520.2 | LHCA3, chlorophyll binding | 1.13(NS) | 0.93(NS) | 1.00(NS) | 0.83 |
| AT2G05070.1 | LHCB2.2, constitute the antenna system of the photosynthetic apparatus, chlorophyll binding | 1.08(NS) | 0.98(NS) | 0.91(NS) | 0.83 |
| AT1G19150.1 | LHCA6, chlorophyll binding PSI type II chlorophyll a/b-binding | 1.11(NS) | 0.94(NS) | 0.89(NS) | 0.76 |
| AT2G34430.1 | LHB1B1, chlorophyll binding PSII type I chlorophyll a/b-binding | 1.07(NS) | 0.91(NS) | 0.79 | 0.67 |
| AT2G34420.1 | LHB1B2, chlorophyll binding | 0.94(NS) | 0.88 | 0.89 | 0.83 |
| AT2G20890.1 | PSB29, controls the assembly of the PSII complex | 1.14(NS) | 0.99(NS) | 0.91(NS) | 0.79 |
| Gene ID | Description | (+)WT/ (-)WT | (+)sto/ (-)sto | (-)sto/ (-)WT | (+)sto/ (+)WT |
|---|---|---|---|---|---|
| AT2G41430.1 | ERD15, an attenuator of plant ABA, response to light intensity, water deprivation, cold tolerance, salt stress | 0.78 | 1.62 | 0.92(NS) | 1.91 |
| AT2G39800.1 | P5CS1, response to ABA, desiccation, oxidative stress, salt stress, water deprivation, root development | 1.00(NS) | 1.09(NS) | 0.75 | 0.82 |
| AT5G25610.1 | RD22, response to abscisic acid, response to desiccation, salt stress | 0.97(NS) | 0.91(NS) | 0.8 | 0.75 |
| AT5G59320.1 | LTP3, involved in lipid transport, response to ABA, water deprivation | 1.19(NS) | 0.99(NS) | 0.78(NS) | 0.69 |
| AT5G01990.1 | Auxin efflux carrier family protein | 0.94(NS) | 1.79 | 0.86(NS) | 1.64 |
| AT1G54990.1 | AXR4/RGR1, auxin-resistant root growth | 1.04(NS) | 1.4 | 1.04 | 1.41 |
| AT2G33830.1 | DRM2, response to stress and environmental factors | 0.76 | 0.86 | 1.11 | 1.26 |
| AT3G45140.1 | LOX2, response to water deprivation, JA, oxidation-reduction process, lipid oxidation | 1.03(NS) | 1.02(NS) | 0.79 | 0.78 |
| AT1G62380.1 | ACO2, response to fatty acid, defense response, ethylene stimulus, ethylene biosynthetic process, oxidation-reduction process, cytokinin, salt stress | 0.97(NS) | 0.93(NS) | 1.4 | 1.34 |
| Accession Number | Gene Symbol | Signature Peptides | PRM | iTRAQ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | A | B | C | D | |||
| AT2G37040.1 | PAL1 | LAGISSGFFDLQPK | 2.89 | 1.95 | 1.60 | 1.14 | 1.60 | 1.3 | 1.13 | 0.92 |
| FLNAGIFGSTK | ||||||||||
| EELGTELLTGEK | ||||||||||
| AT3G21560.1 | UGT84A2 | YDFFDDGLPEDDEASR | 2.59 | 2.59 | 1.14 | 1.30 | 1.57 | 1.35 | 1.04 | 0.89 |
| IVEWCSQEK | ||||||||||
| AT1G15820.1 | LHCB6 | WVDFFNPDSQSVEWATPWSK | 1.16 | 1.07 | 1.00 | 0.79 | 1.09 | 0.96 | 0.92 | 0.81 |
| DGVYEPDFEK | ||||||||||
| SWIPAVK | ||||||||||
| AT1G03130.1 | PSAD-2 | AQVEEFYVITWNSPK | 1.31 | 0.66 | 1.14 | 0.52 | 1.10 | 0.94 | 0.91 | 0.78 |
| ITYQFYR | ||||||||||
| EGPNLLK | ||||||||||
| Name | Primer Name | Sequence |
|---|---|---|
| Actin | ACTIN2-F | GTTGGGATGAACCAGAAGGA |
| ACTIN2-R | GCTCTTCAGGAGCAATACGAAG | |
| PAL1 | PAL1-QF | GATTATGGATTCAAGGGAG |
| PAL1-QR | TTTGCGAGACGAGATTAG | |
| FLS | FLS-QF | ATACAGGGAGGTGAATGAA |
| FLS-QR | ACACGGCGGATAATAGTT | |
| CHS | CHS-QF | ACGTCACGTGTTGAGCGAGTATGG |
| CHS-QR | GAGGAACGCTGTGCAAGACGACTG | |
| LHCA3 | LHCA3-QF | TTCTTCACTTACCTCCTCTG |
| LHCA3-QR | GTCTGTTGGCTCCTTGCT | |
| LHCA2 | LHCA2-QF | TAGCCTCCCTGGTGACTT |
| LHCA2-QR | GGATTCCGATCTTCGTTAG | |
| PSAD-2 | PSAD-2-QF | GCCATAACAACCACTACTTC |
| PSAD-2-QR | ACTGGAGCTTCTTTCACG | |
| DREB2A | DREB2A-QF | TGAAAGGTAAAGGAGGAC |
| DREB2A-QR | CCAAAGGACCATACATAGC | |
| DREB1A | DREB1A-QF | GTGGGTTTGTGAGGTTAGAG |
| DREB1A-QR | CCTTAGCGCAAGTTGATT | |
| RD22 | RD22-QF | CCAAACACTCCCATTCCC |
| RD22-QR | ACACCTCCCTTTCCAACG | |
| ABI5 | ABI5-QF | GGTGAGAATCATCCGTTTA |
| ABI5-QR | TCCTCTGCGTTCCAAATA | |
| P5CS1 | P5CS1-QF | AGGGAAAGTTCCAGAAAG |
| P5CS1-QR | CATAACTAAGCGAGCCAC | |
| SAD2 | SAD2-QF | CTTATGACCGACAGAAACA |
| SAD2-QR | CAACAGTGAGACGCAGAT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, G.; Li, D.; Xiong, H.; Xiao, L.; Tong, J.; Ning, C.; Wang, P.; Li, S. Quantitative Proteomic Analyses Identify STO/BBX24 -Related Proteins Induced by UV-B. Int. J. Mol. Sci. 2020, 21, 2496. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072496
Lyu G, Li D, Xiong H, Xiao L, Tong J, Ning C, Wang P, Li S. Quantitative Proteomic Analyses Identify STO/BBX24 -Related Proteins Induced by UV-B. International Journal of Molecular Sciences. 2020; 21(7):2496. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072496
Chicago/Turabian StyleLyu, Guizhen, Dongbing Li, Hui Xiong, Langtao Xiao, Jianhua Tong, Chanjuan Ning, Ping Wang, and Shaoshan Li. 2020. "Quantitative Proteomic Analyses Identify STO/BBX24 -Related Proteins Induced by UV-B" International Journal of Molecular Sciences 21, no. 7: 2496. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072496





