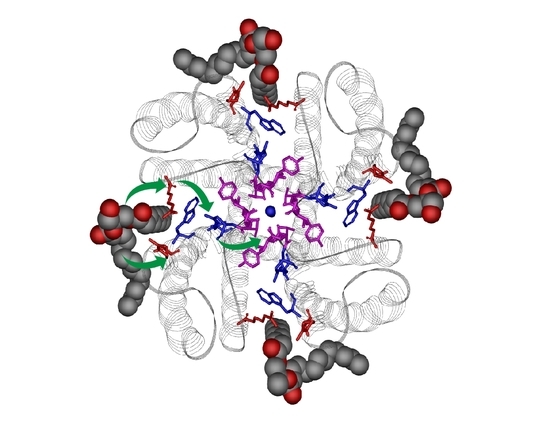
Modulation of Function, Structure and Clustering of K+ Channels by Lipids: Lessons Learnt from KcsA
Abstract
:1. Introduction
2. Lipids as Modulators of KcsA Function
3. Lipids as Structural Effectors of KcsA
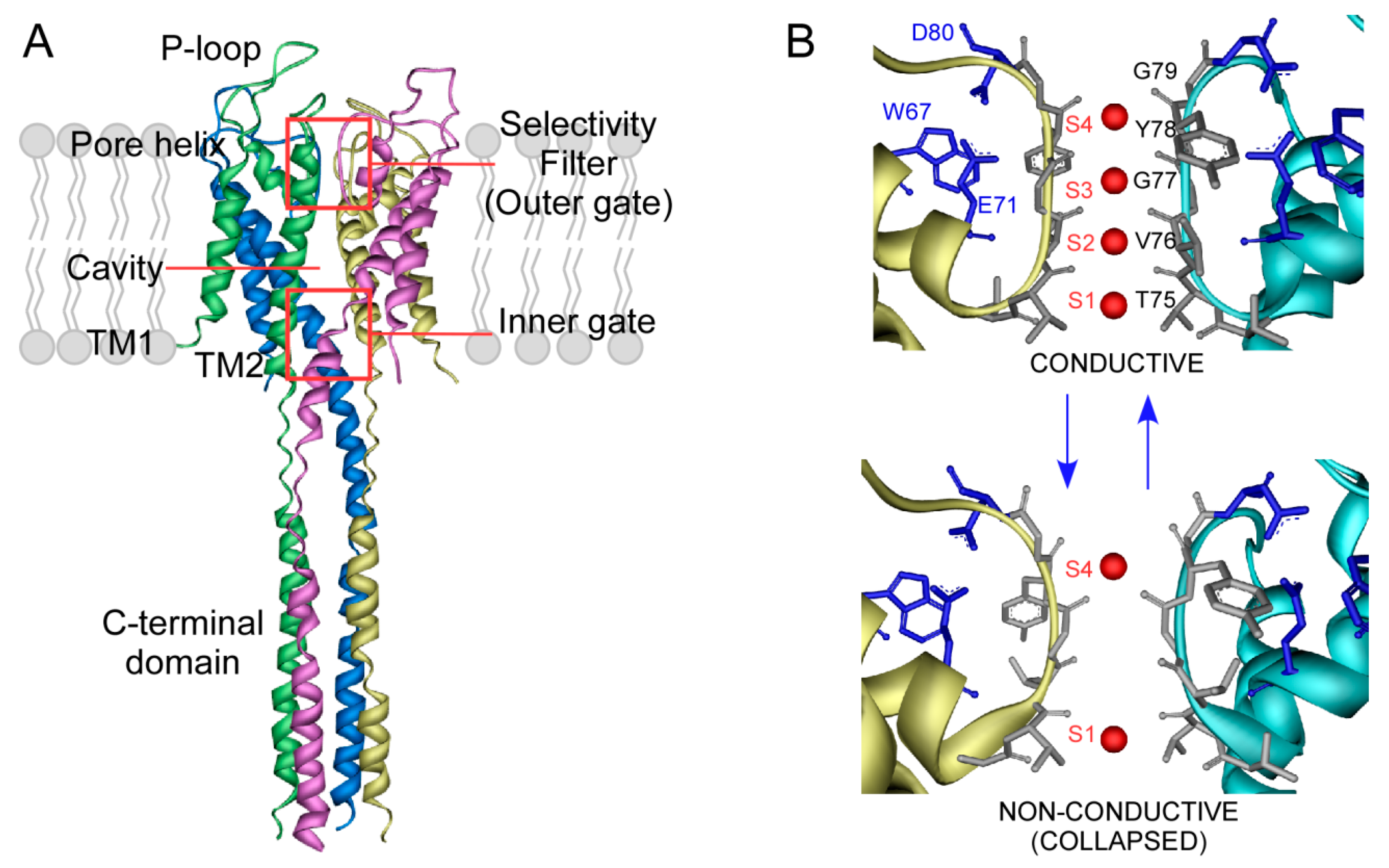
4. Ion Binding to the KcsA Selectivity Filter and Modulation by Lipids
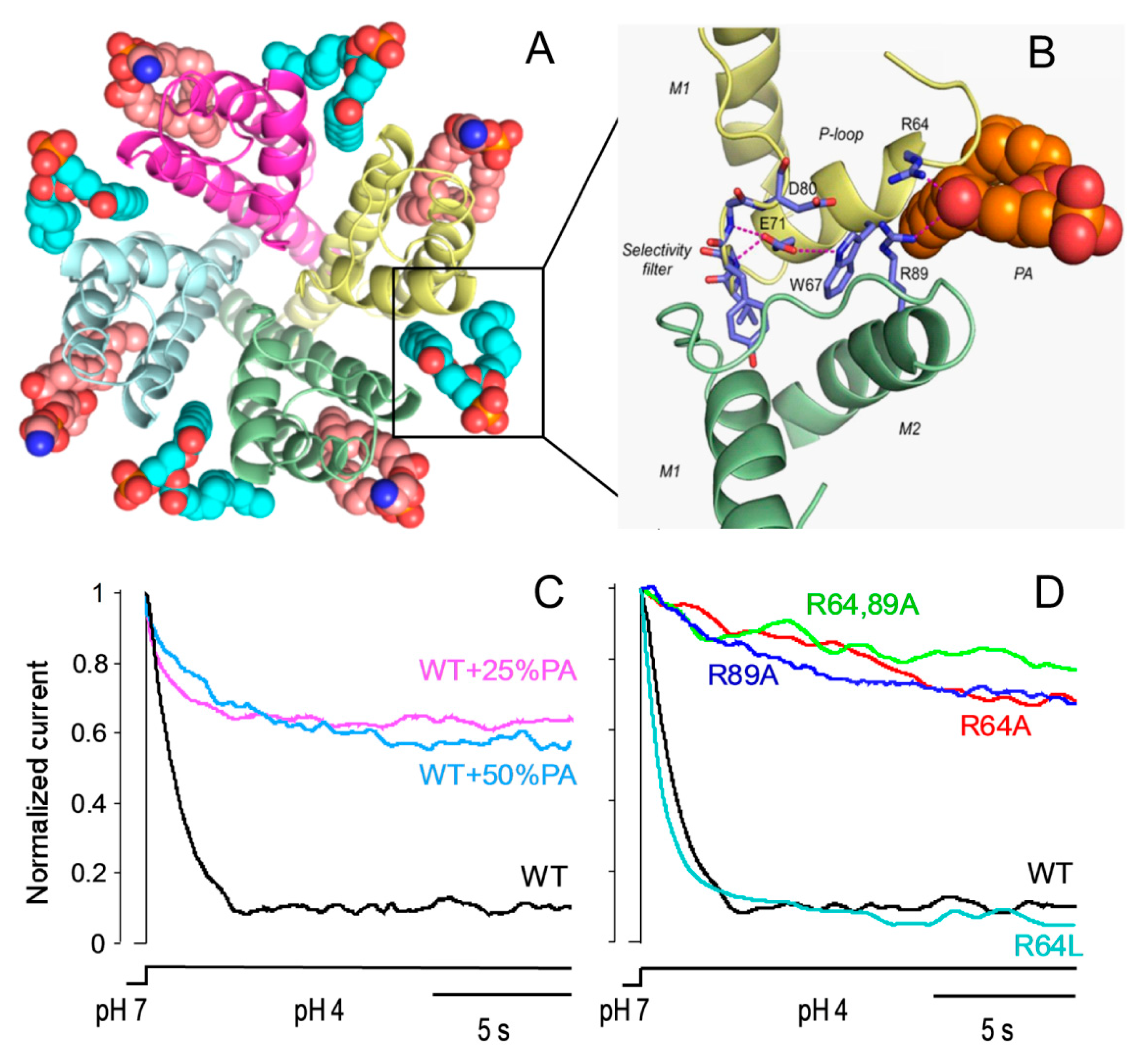
5. Lipid Modulation of Protein Assembly Processes in KcsA
5.1. Influence of Lipids on Folding and Tetramerization of KcsA
5.2. Effects of Lipids on the Formation of KcsA Clusters and Their Gating Properties
6. Concluding Remark
Author Contributions
Funding
Conflicts of Interest
References
- Lehmann-Horn, F.; Jurkat-Rott, K. Channelopathies; Lehmann-Horn, F., Jurkat-Rott, K., Eds.; Elsevier: Ulm, Germany, 2000; Volume 1, ISBN 978-0-444-50489-0. [Google Scholar]
- Kumar, P.; Kumar, D.; Jha, S.K.; Jha, N.K.; Ambasta, R.K. Ion Channels in Neurological Disorders. In Proceedings of the Advances in Protein Chemistry and Structural Biology; Academic Press Inc.: Cambridge, MA, USA, 2016; Volume 103, pp. 97–136. [Google Scholar]
- Demirbilek, H.; Galcheva, S.; Vuralli, D.; Al-Khawaga, S.; Hussain, K. Ion transporters, channelopathies, and glucose disorders. Int. J. Mol. Sci. 2019, 20, 2590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Morais-Cabral, J.H.; Kaufman, A.; Mackinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 2001, 414, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Morais-Cabral, J.H.; Zhou, Y.; MacKinnon, R. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature 2001, 414, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; MacKinnon, R. A mutant KcsA K(+) channel with altered conduction properties and selectivity filter ion distribution. J. Mol. Biol. 2004, 338, 839–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordero-Morales, J.F.; Cuello, L.G.; Zhao, Y.; Jogini, V.; Cortes, D.M.; Roux, B.; Perozo, E. Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 2006, 13, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Morales, J.F.; Jogini, V.; Lewis, A.; Vasquez, V.; Cortes, D.M.; Roux, B.; Perozo, E. Molecular driving forces determining potassium channel slow inactivation. Nat. Struct. Mol. Biol. 2007, 14, 1062–1069. [Google Scholar] [CrossRef]
- Cordero-Morales, J.F.; Jogini, V.; Chakrapani, S.; Perozo, E. A multipoint hydrogen-bond network underlying KcsA C-type inactivation. Biophys. J. 2011, 100, 2387–2393. [Google Scholar] [CrossRef] [Green Version]
- Cuello, L.G.; Cortes, D.M.; Perozo, E. The gating cycle of a K+ channel at atomic resolution. Elife 2017, 6, e28032. [Google Scholar] [CrossRef]
- Allen, T.W.; Kuyucak, S.; Chung, S.H. Molecular dynamics study of the KcsA potassium channel. Biophys. J. 1999, 77, 2502–2516. [Google Scholar] [CrossRef] [Green Version]
- Bernèche, S.; Roux, B. Molecular dynamics of the KcsA K(+) channel in a bilayer membrane. Biophys. J. 2000, 78, 2900–2917. [Google Scholar] [CrossRef] [Green Version]
- Öster, C.; Hendriks, K.; Kopec, W.; Chevelkov, V.; Shi, C.; Michl, D.; Lange, S.; Sun, H.; de Groot, B.L.; Lange, A. The conduction pathway of potassium channels is water free under physiological conditions. Sci. Adv. 2019, 5, eaaw6756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuello, L.G.; Jogini, V.; Cortes, D.M.; Perozo, E. Structural mechanism of C-type inactivation in K(+) channels. Nature 2010, 466, 203–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Ostmeyer, J.; Boulanger, E.; Rui, H.; Perozo, E.; Roux, B. Chemical substitutions in the selectivity filter of potassium channels do not rule out constricted-like conformations for C-type inactivation. Proc. Natl. Acad. Sci. USA 2017, 114, 11145–11150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Ostmeyer, J.; Cuello, L.G.; Perozo, E.; Roux, B. Rapid constriction of the selectivity filter underlies C-type inactivation in the KcsA potassium channel. J. Gen. Physiol. 2018, 150, 1408–1420. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Focke, P.J.; Matulef, K.; Bian, X.; Moënne-Loccoz, P.; Valiyaveetil, F.I.; Lockless, S.W. Ion-binding properties of a K+ channel selectivity filter in different conformations. Proc. Natl. Acad. Sci. USA 2015, 112, 15096–15100. [Google Scholar] [CrossRef] [Green Version]
- Devaraneni, P.K.; Komarov, A.G.; Costantino, C.A.; Devereaux, J.J.; Matulef, K.; Valiyaveetil, F.I. Semisynthetic K+ channels show that the constricted conformation of the selectivity filter is not the C-type inactivated state. Proc. Natl. Acad. Sci. USA 2013, 110, 15698–15703. [Google Scholar] [CrossRef] [Green Version]
- Giudici, A.M.; Renart, M.L.; Díaz-García, C.; Morales, A.; Poveda, J.A.; González-Ros, J.M. Accessibility of Cations to the Selectivity Filter of KcsA in the Inactivated State: An Equilibrium Binding Study. Int. J. Mol. Sci. 2019, 20, 689. [Google Scholar] [CrossRef] [Green Version]
- Lockless, S.W.; Zhou, M.; MacKinnon, R. Structural and thermodynamic properties of selective ion binding in a K+ channel. PLoS Biol. 2007, 5, e121. [Google Scholar] [CrossRef]
- Zhou, Y.; MacKinnon, R. The occupancy of ions in the K+ selectivity filter: Charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates. J. Mol. Biol. 2003, 333, 965–975. [Google Scholar] [CrossRef]
- Montoya, E.; Lourdes Renart, M.; Marcela Giudici, A.; Poveda, J.A.; Fernández, A.M.; Morales, A.; González-Ros, J.M. Differential binding of monovalent cations to KcsA: Deciphering the mechanisms of potassium channel selectivity. Biochim. Biophys. Acta—Biomembr. 2017, 1859, 779–788. [Google Scholar] [CrossRef]
- Matulef, K.; Annen, A.W.; Nix, J.C.; Valiyaveetil, F.I. Individual Ion Binding Sites in the K(+) Channel Play Distinct Roles in C-type Inactivation and in Recovery from Inactivation. Structure 2016, 24, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Poveda, J.A.; Marcela Giudici, A.; Lourdes Renart, M.; Morales, A.; González-Ros, J.M. Towards understanding the molecular basis of ion channel modulation by lipids: Mechanistic models and current paradigms. Biochim. Biophys. Acta—Biomembr. 2017, 1859, 1507–1516. [Google Scholar] [CrossRef] [PubMed]
- Andersen, O.S.; Koeppe, R.E. Bilayer thickness and membrane protein function: An energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 107–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta—Biomembr. 2004, 1666, 62–87. [Google Scholar] [CrossRef] [Green Version]
- Marsh, D. Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta—Biomembr. 2008, 1778, 1545–1575. [Google Scholar] [CrossRef] [Green Version]
- Poveda, J.A.; Giudici, A.M.; Renart, M.L.; Molina, M.L.; Montoya, E.; Fernández-Carvajal, A.; Fernández-Ballester, G.; Encinar, J.A.; González-Ros, J.M. Lipid modulation of ion channels through specific binding sites. Biochim. Biophys. Acta—Biomembr. 2014, 1838, 1560–1567. [Google Scholar] [CrossRef] [Green Version]
- Heginbotham, L.; Kolmakova-Partensky, L.; Miller, C. Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 1998, 111, 741–749. [Google Scholar] [CrossRef] [Green Version]
- Rusinova, R.; Kim, D.M.; Nimigean, C.M.; Andersen, O.S. Regulation of ion channel function by the host lipid bilayer examined by a stopped-flow spectrofluorometric assay. Biophys. J. 2014, 106, 1070–1078. [Google Scholar] [CrossRef] [Green Version]
- Raja, M.; Spelbrink, R.E.J.; de Kruijff, B.; Killian, J.A. Phosphatidic acid plays a special role in stabilizing and folding of the tetrameric potassium channel KcsA. FEBS Lett. 2007, 581, 5715–5722. [Google Scholar] [CrossRef] [Green Version]
- Triano, I.; Barrera, F.N.; Renart, M.L.; Molina, M.L.; Fernández-Ballester, G.; Poveda, J.A.; Fernández, A.M.; Encinar, J.A.; Ferrer-Montiel, A.V.; Otzen, D.; et al. Occupancy of nonannular lipid binding sites on KcsA greatly increases the stability of the tetrameric protein. Biochemistry 2010, 49, 5397–5404. [Google Scholar] [CrossRef]
- Barrera, F.N.; Renart, M.L.; Poveda, J.A.; De Kruijff, B.; Killian, J.A.; González-Ros, J.M. Protein self-assembly and lipid binding in the folding of the potassium channel KcsA. Biochemistry 2008, 47, 2123–2133. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Oiki, S. Amphipathic antenna of an inward rectifier K+ channel responds to changes in the inner membrane leaflet. Proc. Natl. Acad. Sci. USA 2013, 110, 749–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrantes, F.J. Structural basis for lipid modulation of nicotinic acetylcholine receptor function. Brain Res. Rev. 2004, 47, 71–95. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.G. Biological membranes: The importance of molecular detail. Trends Biochem. Sci. 2011, 36, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Yeagle, P.L. Non-covalent binding of membrane lipids to membrane proteins. Biochim. Biophys. Acta—Biomembr. 2014, 1838, 1548–1559. [Google Scholar] [CrossRef] [Green Version]
- Valiyaveetil, F.I.; Zhou, Y.; MacKinnon, R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry 2002, 41, 10771–10777. [Google Scholar] [CrossRef]
- Marius, P.; Zagnoni, M.; Sandison, M.E.; Malcolm East, J.; Morgan, H.; Lee, A.G. Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophys. J. 2008, 94, 1689–1698. [Google Scholar] [CrossRef] [Green Version]
- Deol, S.S.; Domene, C.; Bond, P.J.; Sansom, M.S.P. Anionic phospholipid interactions with the potassium channel KcsA: Simulation studies. Biophys. J. 2006, 90, 822–830. [Google Scholar] [CrossRef] [Green Version]
- Weingarth, M.; Prokofyev, A.; Van Der Cruijsen, E.A.W.; Nand, D.; Bonvin, A.M.J.J.; Pongs, O.; Baldus, M. Structural determinants of specific lipid binding to potassium channels. J. Am. Chem. Soc. 2013, 135, 3983–3988. [Google Scholar] [CrossRef] [Green Version]
- Poveda, J.A.; Giudici, A.M.; Renart, M.L.; Millet, O.; Morales, A.; González-Ros, J.M.; Oakes, V.; Furini, S.; Domene, C. Modulation of the potassium channel KcsA by anionic phospholipids: Role of arginines at the non-annular lipid binding sites. Biochim. Biophys. Acta—Biomembr. 2019, 1861, 183029–183043. [Google Scholar] [CrossRef]
- East, J.M.; Melville, D.; Lee, A.G. Exchange rates and numbers of annular lipids for the calcium and magnesium ion dependent adenosinetriphosphatase. Biochemistry 1985, 24, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Marsh, D.; Horváth, L.I. Structure, dynamics and composition of the lipid-protein interface. Perspectives from spin-labelling. Biochim. Biophys. Acta—Rev. Biomembr. 1998, 1376, 267–296. [Google Scholar] [CrossRef]
- Carney, J.; East, J.M.; Mall, S.; Marius, P.; Powl, A.M.; Wright, J.N.; Lee, A.G. Fluorescence Quenching Methods to Study Lipid-Protein Interactions. Curr. Protoc. Protein Sci. 2006, 45, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Alvis, S.J.; Williamson, I.M.; East, J.M.; Lee, A.G. Interactions of Anionic Phospholipids and Phosphatidylethanolamine with the Potassium Channel KcsA. Biophys. J. 2003, 85, 3828–3838. [Google Scholar] [CrossRef] [Green Version]
- Williamson, I.M.; Alvis, S.J.; Malcolm East, J.; Lee, A.G. Interactions of phospholipids with the potassium channel KcsA. Biophys. J. 2002, 83, 2026–2038. [Google Scholar] [CrossRef] [Green Version]
- Sehgal, P.; Otzen, D.E. Thermodynamics of unfolding of an integral membrane protein in mixed micelles. Protein Sci. 2006, 15, 890–899. [Google Scholar] [CrossRef] [Green Version]
- Sturtevant, J.M. Biochemical Applications of Differential Scanning Calorimetry. Annu. Rev. Phys. Chem. 1987, 38, 463–488. [Google Scholar] [CrossRef]
- Cooper, A.; Nutley, M.A.; Wadood, A. Differential scanning microcalorimetry. In Protein-Ligand Interactions: Hydrodynamics and Calorimetry; Harding, S.E., Chowdhry, B.Z., Eds.; Oxford University Press: Oxford, UK, 2000; pp. 287–318. [Google Scholar]
- Kooijman, E.E.; Tieleman, D.P.; Testerink, C.; Munnik, T.; Rijkers, D.T.S.; Burger, K.N.J.; de Kruijff, B. An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J. Biol. Chem. 2007, 282, 11356–11364. [Google Scholar] [CrossRef] [Green Version]
- Callahan, K.M.; Mondou, B.; Sasseville, L.; Schwartz, J.L.; D’Avanzo, N. The influence of membrane bilayer thickness on KcsA channel activity. Channels 2019, 13, 424–439. [Google Scholar] [CrossRef]
- Liu, S.; Bian, X.; Lockless, S.W. Preferential binding of K+ ions in the selectivity filter at equilibrium explains high selectivity of K+ channels. J. Gen. Physiol. 2012, 140, 671–679. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Lockless, S.W. Equilibrium selectivity alone does not create K + -selective ion conduction in K + channels. Nat. Commun. 2013, 4, 2746–2753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, K.A.; Tzitzilonis, C.; Kwiatkowski, W.; Choe, S.; Riek, R. Conformational dynamics of the KcsA potassium channel governs gating properties. Nat. Struct. Mol. Biol. 2007, 14, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Chill, J.H. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Sci. 2006, 15, 684–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, S.; Osawa, M.; Takeuchi, K.; Shimada, I. Structural basis underlying the dual gate properties of KcsA. Proc. Natl. Acad. Sci. USA 2010, 107, 6216–6221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renart, M.L.; Barrera, F.N.; Molina, M.L.; Encinar, J.A.; Poveda, J.A.; Fernández, A.M.; Gómez, J.; González-Ros, J.M. Effects of conducting and blocking ions on the structure and stability of the potassium channel KcsA. J. Biol. Chem. 2006, 281, 29905–29915. [Google Scholar] [CrossRef] [Green Version]
- Renart, M.L.; Triano, I.; Poveda, J.A.; Encinar, J.A.; Fernández, A.M.; Ferrer-Montiel, A.V.; Gómez, J.; González Ros, J.M. Ion binding to KcsA: Implications in ion selectivity and channel gating. Biochemistry 2010, 49, 9480–9487. [Google Scholar] [CrossRef]
- Renart, M.L.; Montoya, E.; Fernández, A.M.; Molina, M.L.; Poveda, J.A.; Encinar, J.A.; Ayala, J.L.; Ferrer-Montiel, A.V.; Gómez, J.; Morales, A.; et al. Contribution of ion binding affinity to ion selectivity and permeation in KcsA, a model potassium channel. Biochemistry 2012, 51, 3891–3900. [Google Scholar] [CrossRef]
- Renart, M.L.; Giudici, A.M.; Poveda, J.A.; Fedorov, A.; Berberan-Santos, M.N.; Prieto, M.; Díaz-García, C.; González-Ros, J.M.; Coutinho, A. Conformational plasticity in the KcsA potassium channel pore helix revealed by homo-FRET studies. Sci. Rep. 2019, 9, 6215–6228. [Google Scholar] [CrossRef]
- Renart, M.L.; Montoya, E.; Giudici, A.M.; Poveda, J.A.; Fernández, A.M.; Morales, A.; González-Ros, J.M. Selective exclusion and selective binding both contribute to ion selectivity in KcsA, a model potassium channel. J. Biol. Chem. 2017, 292, 15552–15560. [Google Scholar] [CrossRef] [Green Version]
- Kratochvil, H.T.; Maj, M.; Matulef, K.; Annen, A.W.; Ostmeyer, J.; Perozo, E.; Roux, B.; Valiyaveetil, F.I.; Zanni, M.T. Probing the Effects of Gating on the Ion Occupancy of the K+ Channel Selectivity Filter Using Two-Dimensional Infrared Spectroscopy. J. Am. Chem. Soc. 2017, 139, 8837–8845. [Google Scholar] [CrossRef] [Green Version]
- Encinar, J.A.; Molina, M.L.; Poveda, J.A.; Barrera, F.N.; Renart, M.L.; Fernández, A.M.; González-Ros, J.M. The influence of a membrane environment on the structure and stability of a prokaryotic potassium channel, KcsA. FEBS Lett. 2005, 579, 5199–5204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chipot, C.; Dehez, F.; Schnell, J.R.; Zitzmann, N.; Pebay-Peyroula, E.; Catoire, L.J.; Miroux, B.; Kunji, E.R.S.; Veglia, G.; Cross, T.A.; et al. Perturbations of Native Membrane Protein Structure in Alkyl Phosphocholine Detergents: A Critical Assessment of NMR and Biophysical Studies. Chem. Rev. 2018, 118, 3559–3607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wylie, B.J.; Bhate, M.P.; McDermott, A.E. Transmembrane allosteric coupling of the gates in a potassium channel. Proc. Natl. Acad. Sci. USA 2014, 111, 185–190. [Google Scholar] [CrossRef] [Green Version]
- Bhate, M.P.; Wylie, B.J.; Tian, L.; McDermott, A.E. Conformational dynamics in the selectivity filter of KcsA in response to potassium ion concentration. J. Mol. Biol. 2010, 401, 155–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ader, C.; Schneider, R.; Hornig, S.; Velisetty, P.; Vardanyan, V.; Giller, K.; Ohmert, I.; Becker, S.; Pongs, O.; Baldus, M. Coupling of activation and inactivation gate in a K-channel: Potassium and ligand sensitivity. EMBO J. 2009, 28, 2825–2834. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Bhate, M.P.; McDermott, A.E. Transmembrane allosteric energetics characterization for strong coupling between proton and potassium ion binding in the KcsA channel. Proc. Natl. Acad. Sci. USA 2017, 114, 8788–8793. [Google Scholar] [CrossRef] [Green Version]
- Qasim, A.; Sher, I.; Hirschhorn, O.; Shaked, H.; Qasem, Z.; Ruthstein, S.; Chill, J.H. Investigation of a KcsA Cytoplasmic pH Gate in Lipoprotein Nanodiscs. Chembiochem 2019, 20, 813–821. [Google Scholar] [CrossRef]
- Dörr, J.M.; Koorengevel, M.C.; Schäfer, M.; Prokofyev, A.V.; Scheidelaar, S.; van der Cruijsen, E.A.W.; Dafforn, T.R.; Baldus, M.; Killian, J.A. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. USA 2014, 111, 18607–18612. [Google Scholar] [CrossRef] [Green Version]
- Shenkarev, Z.O.; Lyukmanova, E.N.; Solozhenkin, O.I.; Gagnidze, I.E.; Nekrasova, O.V.; Chupin, V.V.; Tagaev, A.A.; Yakimenko, Z.A.; Ovchinnikova, T.V.; Kirpichnikov, M.P.; et al. Lipid-protein nanodiscs: Possible application in high-resolution NMR investigations of membrane proteins and membrane-active peptides. Biochemistry. (Mosc.) 2009, 74, 756–765. [Google Scholar] [CrossRef]
- van Dalen, A.; Schrempf, H.; Killian, J.A.; de Kruijff, B. Efficient membrane assembly of the KcsA potassium channel in Escherichia coli requires the protonmotive force. EMBO Rep. 2000, 1, 340–346. [Google Scholar] [CrossRef] [Green Version]
- Van Dalen, A.; Hegger, S.; Killian, J.A.; De Kruijff, B. Influence of lipids on membrane assembly and stability of the potassium channel KcsA. FEBS Lett. 2002, 525, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Ando, M.; Akiyama, M.; Okuno, D.; Hirano, M.; Ide, T.; Sawada, S.; Sasaki, Y.; Akiyoshi, K. Liposome chaperon in cell-free membrane protein synthesis: One-step preparation of KcsA-integrated liposomes and electrophysiological analysis by the planar bilayer method. Biomater. Sci. 2016, 4, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Focke, P.J.; Hein, C.; Hoffmann, B.; Matulef, K.; Bernhard, F.; Dötsch, V.; Valiyaveetil, F.I. Combining in Vitro Folding with Cell Free Protein Synthesis for Membrane Protein Expression. Biochemistry 2016, 55, 4212–4219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaish, A.; Guo, S.; Murray, R.M.; Grandsard, P.J.; Chen, Q. On-chip membrane protein cell-free expression enables development of a direct binding assay: A curious case of potassium channel KcsA-Kv1.3. Anal. Biochem. 2018, 556, 70–77. [Google Scholar] [CrossRef]
- Barrera, F.N.; Renart, M.L.; Molina, M.L.; Poveda, J.A.; Encinar, J.A.; Fernández, A.M.; Neira, J.L.; González-Ros, J.M. Unfolding and refolding in vitro of a tetrameric, alpha-helical membrane protein: The prokaryotic potassium channel KcsA. Biochemistry 2005, 44, 14344–14352. [Google Scholar] [CrossRef]
- van den Brink-van der Laan, E.; Chupin, V.; Killian, J.A.; de Kruijff, B. Stability of KcsA tetramer depends on membrane lateral pressure. Biochemistry 2004, 43, 4240–4250. [Google Scholar] [CrossRef] [Green Version]
- Raja, M. The role of extramembranous cytoplasmic termini in assembly and stability of the tetrameric K(+)-channel KcsA. J. Membr. Biol. 2010, 235, 51–61. [Google Scholar] [CrossRef] [Green Version]
- Molina, M.L.; Barrera, F.N.; Fernandez, A.M.; Poveda, J.A.; Renart, M.L.; Encinar, J.A.; Riquelme, G.; Gonzalez-Ros, J.M. Clustering and coupled gating modulate the activity in KcsA, a potassium channel model. J. Biol. Chem. 2006, 281, 18837–18848. [Google Scholar] [CrossRef] [Green Version]
- Molina, M.L.; Encinar, J.A.; Barrera, F.N.; Fernandez-Ballester, G.; Riquelme, G.; Gonzalez-Ros, J.M. Influence of C-terminal protein domains and protein-lipid interactions on tetramerization and stability of the potassium channel KcsA. Biochemistry 2004, 43, 14924–14931. [Google Scholar] [CrossRef] [Green Version]
- Seeger, H.M.; Bortolotti, C.A.; Alessandrini, A.; Facci, P. Phase-transition-induced protein redistribution in lipid bilayers. J. Phys. Chem. B 2009, 113, 16654–16659. [Google Scholar] [CrossRef]
- Giudici, A.M.; Molina, M.L.; Ayala, J.L.; Montoya, E.; Renart, M.L.; Fernández, A.M.; Encinar, J.A.; Ferrer-Montiel, A.V.; Poveda, J.A.; González-Ros, J.M. Detergent-labile, supramolecular assemblies of KcsA: Relative abundance and interactions involved. Biochim. Biophys. Acta 2013, 1828, 193–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegermann, J.; Overbeck, J.; Schrempf, H. In vivo monitoring of the potassium channel KcsA in Streptomyces lividans hyphae using immuno-electron microscopy and energy-filtering transmission electron microscopy. Microbiology 2006, 152, 2831–2841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raja, M.; Vales, E. Effects of sodium chloride on membrane fusion and on the formation of aggregates of potassium channel KcsA in Escherichia coli membrane. Biophys. Chem. 2009, 142, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Ujwal, R.; Cascio, D.; Chaptal, V.; Ping, P.; Abramson, J. Crystal packing analysis of murine VDAC1 crystals in a lipidic environment reveals novel insights on oligomerization and orientation. Channels (Austin) 2009, 3, 167–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina, M.L.; Giudici, A.M.; Poveda, J.A.; Fernández-Ballester, G.; Montoya, E.; Renart, M.L.; Fernández, A.M.; Encinar, J.A.; Riquelme, G.; Morales, A.; et al. Competing Lipid-Protein and Protein-Protein Interactions Determine Clustering and Gating Patterns in the Potassium Channel from Streptomyces lividans (KcsA). J. Biol. Chem. 2015, 290, 25745–25755. [Google Scholar] [CrossRef] [Green Version]
- Schrempf, H.; Schmidt, O.; Kümmerlen, R.; Hinnah, S.; Müller, D.; Betzler, M.; Steinkamp, T.; Wagner, R. A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. EMBO J. 1995, 14, 5170–5178. [Google Scholar] [CrossRef] [Green Version]
- Cuello, L.G.; Romero, J.G.; Cortes, D.M.; Perozo, E. pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry 1998, 37, 3229–3236. [Google Scholar] [CrossRef]
- Meuser, D.; Splitt, H.; Wagner, R.; Schrempf, H. Exploring the open pore of the potassium channel from Streptomyces lividans. FEBS Lett. 1999, 462, 447–452. [Google Scholar] [CrossRef] [Green Version]
- Kazuteru, D.; Tatsunori, O.; Tan, S.H.; Nanda, V. Mutations stabilizing an open conformation within the external region of the permeation pathway of the potassium channel KcsA. Eur. Biophys. J. 2001, 30, 385–391. [Google Scholar]
- Navedo, M.F.; Cheng, E.P.; Yuan, C.; Votaw, S.; Molkentin, J.D.; Scott, J.D.; Santana, L.F. Increased coupled gating of L-type Ca2+ channels during hypertension and Timothy syndrome. Circ. Res. 2010, 106, 748–756. [Google Scholar] [CrossRef] [Green Version]
- Grage, S.L.; Keleshian, A.M.; Turdzeladze, T.; Battle, A.R.; Tay, W.C.; May, R.P.; Holt, S.A.; Contera, S.A.; Haertlein, M.; Moulin, M.; et al. Bilayer-mediated clustering and functional interaction of MscL channels. Biophys. J. 2011, 100, 1252–1260. [Google Scholar] [CrossRef] [PubMed]
- Vivas, O.; Moreno, C.M.; Santana, L.F.; Hille, B. Proximal clustering between BK and CaV1.3 channels promotes functional coupling and BK channel activation at low voltage. Elife 2017, 6, e28029. [Google Scholar] [CrossRef] [PubMed]
- Spira, F.; Mueller, N.S.; Beck, G.; Von Olshausen, P.; Beig, J.; Wedlich-Söldner, R. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat. Cell Biol. 2012, 14, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.S.; Wedlich-Söldner, R.; Spira, F. From mosaic to patchwork: Matching lipids and proteins in membrane organization. Mol. Membr. Biol. 2012, 29, 186–196. [Google Scholar] [CrossRef]
- Choi, K.-H. Cooperative gating between ion channels. Gen. Physiol. Biophys. 2014, 33, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Kim, G.E.; Kronengold, J.; Barcia, G.; Quraishi, I.H.; Martin, H.C.; Blair, E.; Taylor, J.C.; Dulac, O.; Colleaux, L.; Nabbout, R.; et al. Human slack potassium channel mutations increase positive cooperativity between individual channels. Cell Rep. 2014, 9, 1661–1672. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.E.; Moreno, C.M.; Yuan, C.; Opitz-Araya, X.; Binder, M.D.; Navedo, M.F.; Santana, L.F. Graded Ca2+/calmodulin-dependent coupling of voltage-gated CaV1.2 channels. Elife 2015, 4, e05608. [Google Scholar] [CrossRef]
- Moreno, C.M.; Dixon, R.E.; Tajada, S.; Yuan, C.; Opitz-Araya, X.; Binder, M.D.; Santana, L.F. Ca(2+) entry into neurons is facilitated by cooperative gating of clustered CaV1.3 channels. Elife 2016, 5, e15744. [Google Scholar] [CrossRef] [Green Version]
- Gianoli, F.; Risler, T.; Kozlov, A.S. Lipid bilayer mediates ion-channel cooperativity in a model of hair-cell mechanotransduction. Proc. Natl. Acad. Sci. USA 2017, 114, E11010–E11019. [Google Scholar] [CrossRef] [Green Version]
- Clatot, J.; Hoshi, M.; Wan, X.; Liu, H.; Jain, A.; Shinlapawittayatorn, K.; Marionneau, C.; Ficker, E.; Ha, T.; Deschênes, I. Voltage-gated sodium channels assemble and gate as dimers. Nat. Commun. 2017, 8, 2077–2091. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renart, M.L.; Giudici, A.M.; Díaz-García, C.; Molina, M.L.; Morales, A.; González-Ros, J.M.; Poveda, J.A. Modulation of Function, Structure and Clustering of K+ Channels by Lipids: Lessons Learnt from KcsA. Int. J. Mol. Sci. 2020, 21, 2554. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072554
Renart ML, Giudici AM, Díaz-García C, Molina ML, Morales A, González-Ros JM, Poveda JA. Modulation of Function, Structure and Clustering of K+ Channels by Lipids: Lessons Learnt from KcsA. International Journal of Molecular Sciences. 2020; 21(7):2554. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072554
Chicago/Turabian StyleRenart, María Lourdes, Ana Marcela Giudici, Clara Díaz-García, María Luisa Molina, Andrés Morales, José M. González-Ros, and José Antonio Poveda. 2020. "Modulation of Function, Structure and Clustering of K+ Channels by Lipids: Lessons Learnt from KcsA" International Journal of Molecular Sciences 21, no. 7: 2554. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072554