Histone Deacetylase TaHDT701 Functions in TaHDA6-TaHOS15 Complex to Regulate Wheat Defense Responses to Blumeria graminis f.sp. tritici
Abstract
:1. Introduction
2. Results
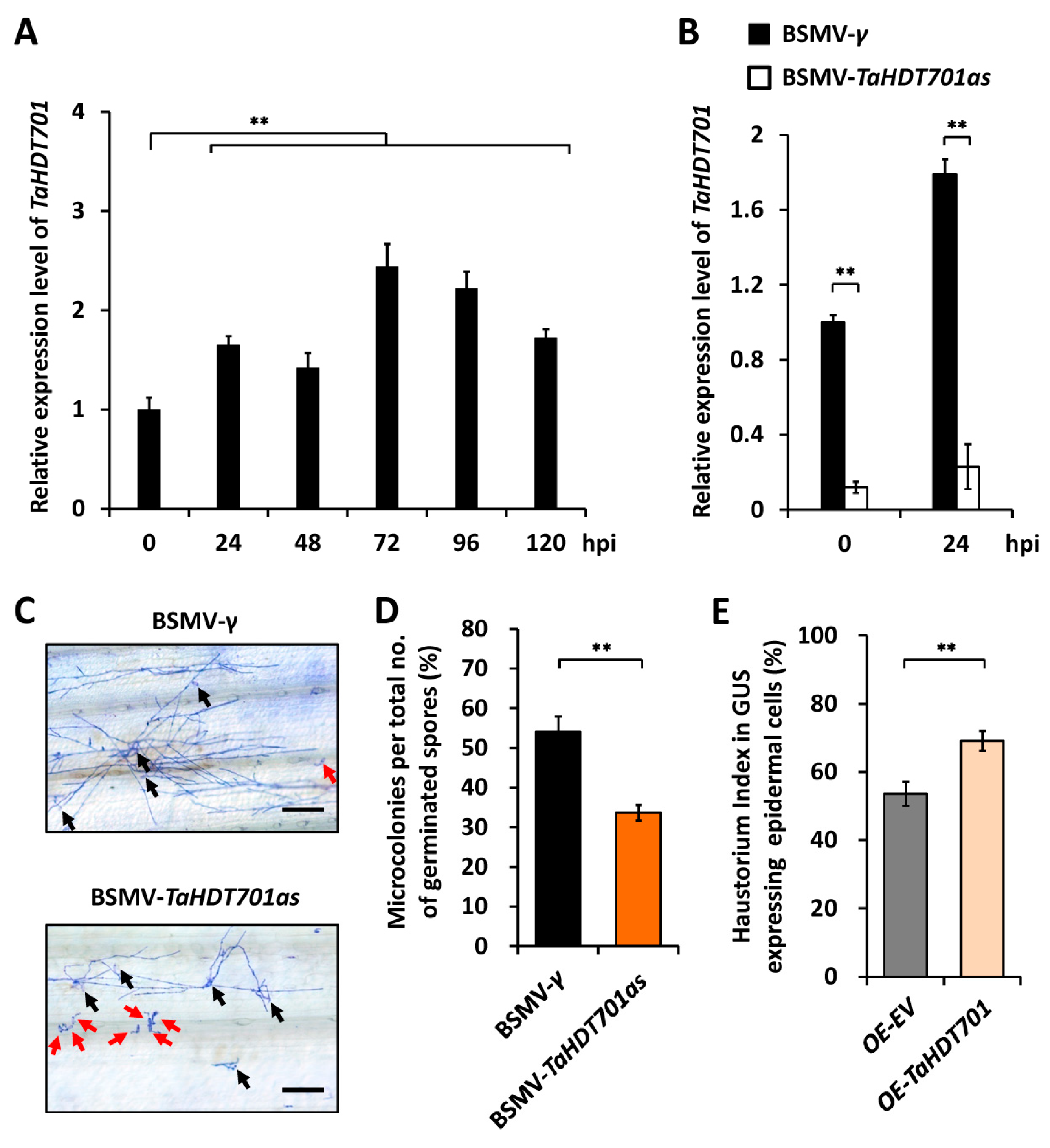
2.1. TaHDT701 Negatively Regulates The Wheat Defense Responses to Powdery Mildew
2.2. Wheat TaHDT701 Associates with TaHDA6 and TaHOS15 in The Histone Deacetylase Complex
2.3. Knock-Down of TaHDT701, TaHDA6, and TaHOS15 Compromised Wheat Susceptibility to Powdery Mildew
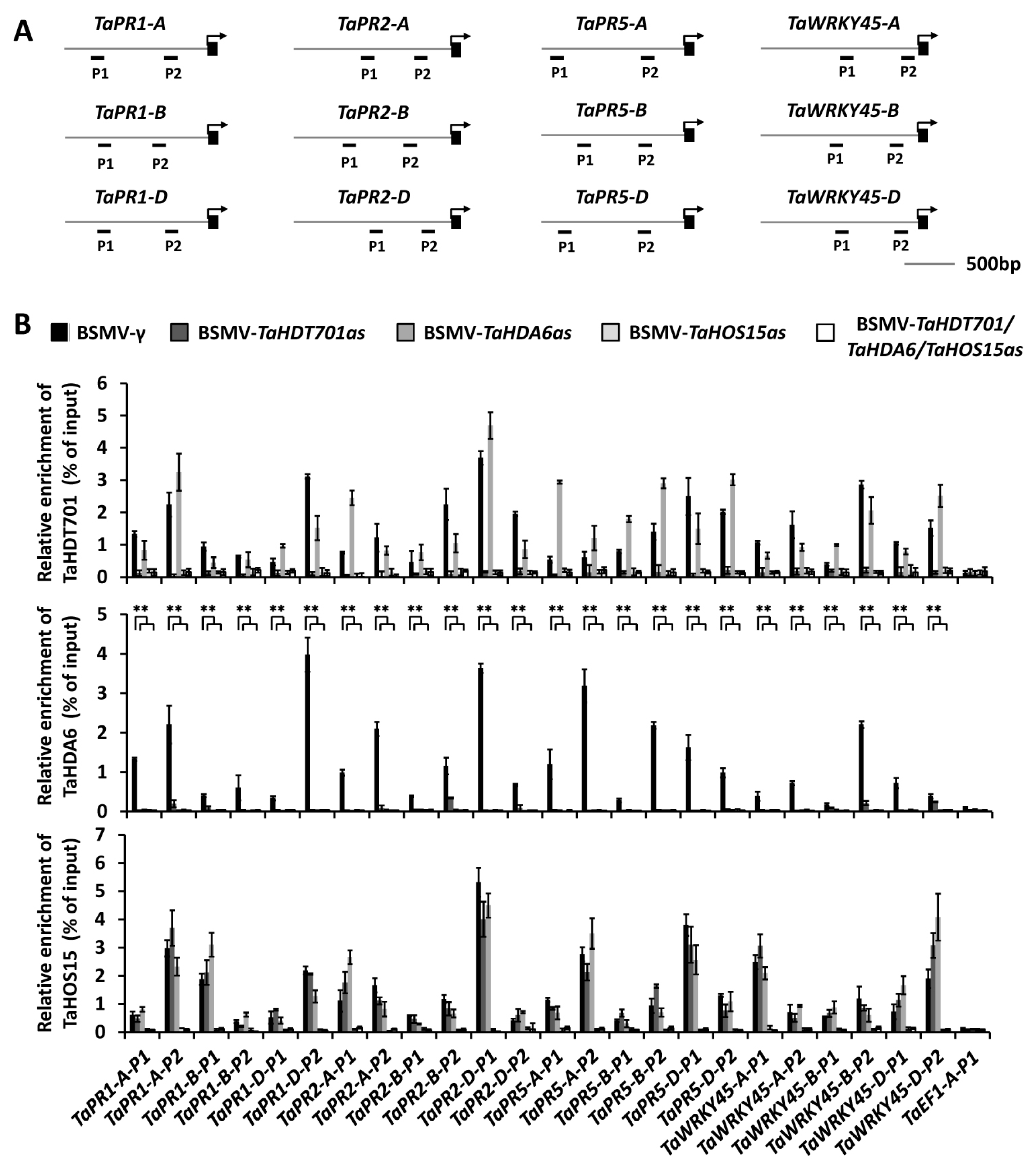
2.4. Down-Regulating TaHDT701 Impair The TaHDA6 Occupancy at Chromatin of Defense-Related Genes
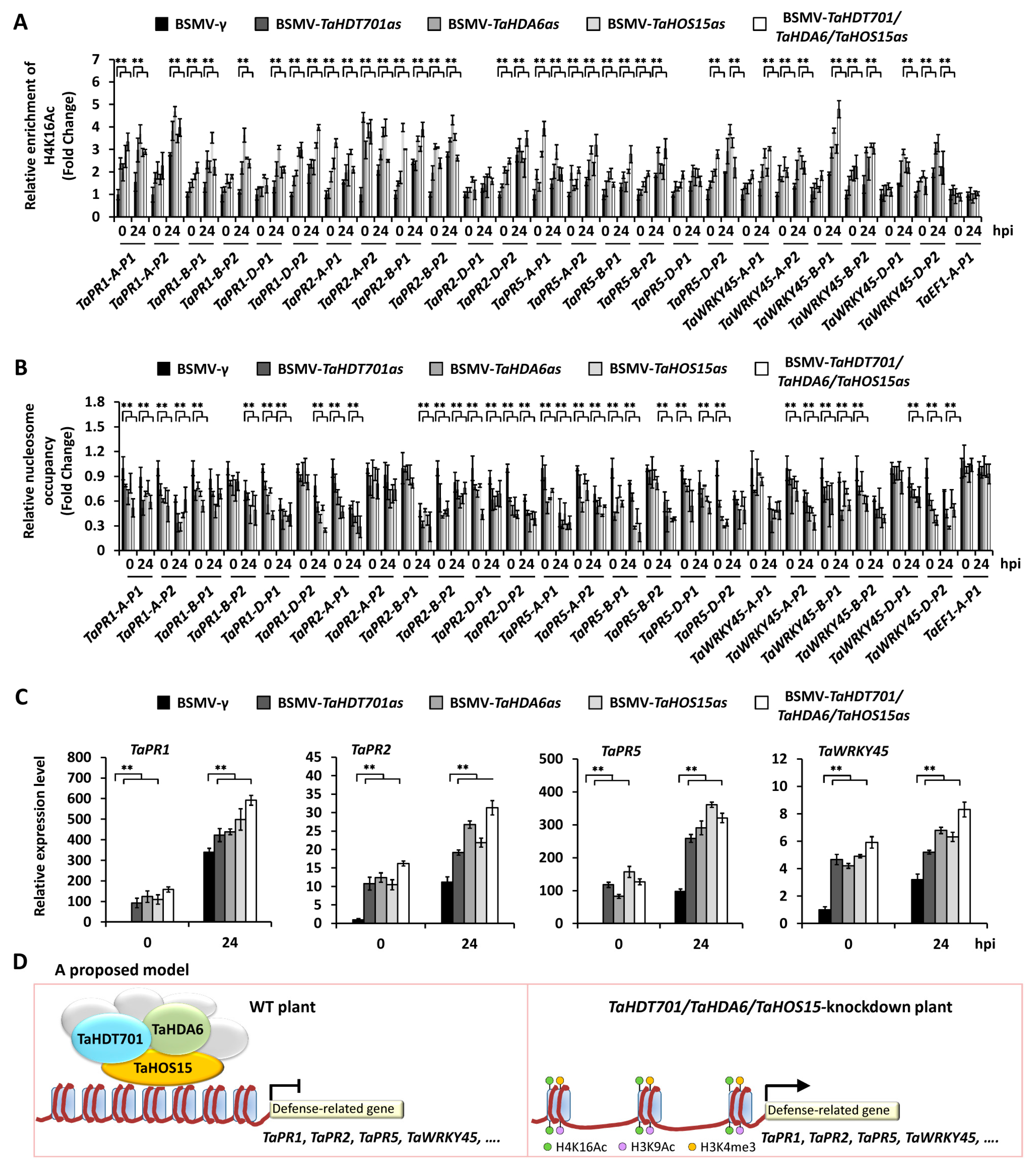
2.5. Silencing of TaHDT701, TaHDA6, and TaHOS15 Altered Chromatin State and Transcription Level of Defense-Related Genes under Bgt Infection
3. Discussion
4. Materials and Methods
4.1. Plant and Fungal Materials and Treatments
4.2. Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
4.3. BSMV-Mediated Gene-Silencing (BSMV-VIGS) Assays
4.4. Single-Cell Transient Gene-Silencing and Expression Assays
4.5. Yeast Two-Hybrid Analysis
4.6. GST Pull-Down Assays
4.7. Luciferase Complementation Imaging (LCI) Assays
4.8. Nuclear Co-Immunoprecipitation (Co-IP) Assays
4.9. Immunoprecipitation of Histone Deacetylase Activity
4.10. Chromatin Immunoprecipitation (ChIP) Assays
4.11. Nucleosome Occupancy Micrococcal Nuclease (MNase) Assay
4.12. Accession Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Bgt | Blumeria graminis f.sp. tritici |
| BSMV | barley stripe mosaic virus |
| BSMV-VIGS | barley stripe mosaic virus-induced gene-silencing |
| ChIP | chromatin immunoprecipitation |
| ChIP-qPCR | chromatin immunoprecipitation coupled with quantitative PCR |
| EF1 | elongation factor 1 |
| EV | empty vector |
| GST | glutathione S-transferase |
| H3K4Me3 | trimethylation of histone H3 lysine 4 |
| H3K9Ac | acetylation of histone H3 lysine 9 |
| H4K16Ac | acetylation of histone H4 lysine 16 |
| HD2 | type-II histone deacetylase |
| HDAC | histone deacetylase |
| HI% | haustorium index |
| hpi | hours post-inoculation |
| LCI | luciferase complementation imaging |
| MI% | microcolony index |
| MNase | micrococcal nuclease |
| qRT-PCR | quantitative reverse transcription-polymerase chain reaction |
| RPD3 | reduced potassium dependency protein 3 |
| TIGS | transient induced gene-silencing |
| WDR protein | WD40-repeat protein |
| Y2H | yeast two-hybrid. |
References
- Parlange, F.; Roffler, S.; Menardo, F.; Ben-David, R.; Bourras, S.; McNally, K.E.; Oberhaensli, S.; Stirnweis, D.; Buchmann, G.; Wicker, T.; et al. Genetic and molecular characterization of a locus involved in avirulence of Blumeria graminis f. sp. tritici on wheat Pm3 resistance alleles. Fungal Genet. Boil. 2015, 82, 181–192. [Google Scholar]
- Zhu, X.; Yang, K.; Wei, X.; Zhang, Q.; Rong, W.; Du, L.; Ye, X.; Qi, L.; Zhang, Z. The wheat AGC kinase TaAGC1 is a positive contributor to host resistance to the necrotrophic pathogen Rhizoctonia cerealis. J. Exp. Bot. 2015, 66, 6591–6603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, S.; Wang, H.; Li, Y.; Kong, Z.; Tang, D. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 2017, 218, 298–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, S.; Kong, X.; Song, G.; Jia, M.; Guan, J.; Wang, F.; Qin, Z.; Wu, L.; Lan, X.; Li, A.; et al. DNA methylation dynamics during the interaction of wheat progenitor Aegilops tauschii with the obligate biotrophic fungus Blumeria graminis f. sp. tritici. New Phytol. 2018, 221, 1023–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Zhu, S.; Zhao, R.; Jiang, Z.; Ji, Y.; Ji, J.; Qiu, D.; Li, H.-J.; Bie, T. Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 2018, 11, 879–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koller, T.; Brunner, S.; Herren, G.; Hurni, S.; Keller, B. Pyramiding of transgenic Pm3 alleles in wheat results in improved powdery mildew resistance in the field. Theor. Appl. Genet. 2018, 131, 861–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, L.; Hu, P.; Liu, J.; Witek, K.; Zhou, S.; Xu, J.; Zhou, W.; Gao, L.; Huang, Z.; Zhang, R.; et al. Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 2018, 11, 874–878. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef] [Green Version]
- Adachi, H.; Tsuda, K. Convergence of cell-surface and intracellular immune receptor signalling. New Phytol. 2019, 221, 1676–1678. [Google Scholar] [CrossRef]
- Buscaill, P.; Rivas, S. Transcriptional control of plant defence responses. Curr. Opin. Plant Boil. 2014, 20, 35–46. [Google Scholar] [CrossRef]
- Jones, A.M.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez-Prado, J.S.; Piquerez, S.J.M.; Bendahmane, A.; Hirt, H.; Raynaud, C.; Benhamed, M. Modify the histone to win the battle: Chromatin dynamics in plant–pathogen interactions. Front. Plant Sci. 2018, 9, 355. [Google Scholar] [CrossRef]
- Tsuda, K.; Somssich, I. Transcriptional networks in plant immunity. New Phytol. 2015, 206, 932–947. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Wang, G.-L. Chromatin versus pathogens: The function of epigenetics in plant immunity. Front. Plant Sci. 2015, 6, 675. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Liu, Y.; Wang, X.; Chang, C. Insight into the role of epigenetic processes in abiotic and biotic stress response in wheat and barley. Int. J. Mol. Sci. 2020, 21, 1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinas, N.A.; Saze, H.; Saijo, Y. Epigenetic control of defense signaling and priming in plants. Front. Plant Sci. 2016, 7, 205. [Google Scholar] [CrossRef] [PubMed]
- Kurdistani, S.K.; Grunstein, M. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Boil. 2003, 4, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Bellizzi, M.D.R.; Ning, Y.; Meyers, B.C.; Wang, G.-L. HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 2012, 24, 3783–3794. [Google Scholar] [CrossRef] [Green Version]
- Pandey, R. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef] [Green Version]
- Mehdi, S.; Derkacheva, M.; Ramström, M.; Kralemann, L.; Bergquist, J.; Hennig, L. The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. Plant Cell 2015, 28, 42–54. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.; Wang, Y.-Y.; Liu, X.; Yang, S.; Lu, Q.; Cui, Y.; Wu, K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012, 63, 3297–3306. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lim, C.J.; Shen, M.; Park, H.J.; Cha, J.-Y.; Iniesto, E.; Rubio, V.; Mengiste, T.; Zhu, J.-K.; Bressan, R.A.; et al. Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. USA 2018, 115, E5400–E5409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.-M.; Song, H.-R.; Han, S.-K.; Han, M.; Kim, C.-Y.; Park, J.; Lee, Y.; Jeon, J.-S.; Noh, Y.-S.; Noh, B. HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 2012, 71, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, Q.; Wu, Z.; Wang, H.; Han, S.; Jin, Y.; Zhou, J.; Zhang, Z.; Jiang, J.; Shen, Y.; et al. HISTONE DEACETYLASE 6 represses pathogen defense responses in Arabidopsis thaliana. Plant Cell Environ. 2017, 40, 2972–2986. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Zhang, L.; Zhou, C.; Yu, C.-W.; Chaikam, V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 2008, 59, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhang, L.; Duan, J.; Miki, B.; Wu, K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 2005, 17, 1196–1204. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Gao, F.; Wu, J.; Dai, J.; Wei, C.; Li, Y. Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol. 2010, 51, 1291–1299. [Google Scholar] [CrossRef]
- Latrasse, D.; Jégu, T.; Li, H.; De Zelicourt, A.; Raynaud, C.; Legras, S.; Gust, A.; Samajova, O.; Veluchamy, A.; Rayapuram, N.; et al. MAPK-triggered chromatin reprogramming by histone deacetylase in plant innate immunity. Genome Boil. 2017, 18, 131. [Google Scholar] [CrossRef]
- Liu, J.; Zhi, P.; Wang, X.; Fan, Q.; Chang, C. Wheat WD40-repeat protein TaHOS15 functions in a histone deacetylase complex to fine-tune defense responses to Blumeria graminis f.sp. tritici. J. Exp. Bot. 2018, 70, 255–268. [Google Scholar] [CrossRef]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.; Wang, Y.-Y.; Liu, X.; Yang, S.; Wu, K. HD2 proteins interact with RPD3-type histone deacetylases. Plant Signal. Behav. 2012, 7, 608–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.J.; Baek, D.; Cha, J.-Y.; Liao, X.; Kang, S.-H.; McClung, C.R.; Lee, S.Y.; Yun, D.-J.; Kim, W.-Y. HOS15 interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell 2019, 31, 37–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourque, S.; Jeandroz, S.; Grandperret, V.; Lehotai, N.; Aimé, S.; Soltis, D.; Miles, N.; Melkonian, M.; Deyholos, M.; Leebens-Mack, J.; et al. The evolution of HD2 proteins in green plants. Trends Plant Sci. 2016, 21, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-T.; Luo, M.; Wang, Y.-Y.; Wu, K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010, 61, 3345–3353. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.-W.; Tai, R.; Wang, S.-C.; Yang, P.; Luo, M.; Yang, S.; Cheng, K.; Wang, W.-C.; Cheng, Y.-S.; Wu, K. HISTONE DEACETYLASE6 acts in concert with histone methyltransferases SUVH4, SUVH5, and SUVH6 to regulate transposon silencing. Plant Cell 2017, 29, 1970–1983. [Google Scholar] [CrossRef] [Green Version]
- Mozgova, I.; Wildhaber, T.; Liu, Q.; Mansour, E.A.; L’Haridon, F.; Métraux, J.-P.; Gruissem, W.; Hofius, D.; Hennig, L. Chromatin assembly factor CAF-1 represses priming of plant defence response genes. Nat. Plants 2015, 1, 15127. [Google Scholar] [CrossRef]
- Yuan, C.; Li, C.; Yan, L.; Jackson, A.O.; Liu, Z.; Han, C.; Yu, J.; Li, D. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 2011, 6, e26468. [Google Scholar] [CrossRef]
- Kong, L.; Chang, C. Suppression of wheat TaCDK8/TaWIN1 interaction negatively affects germination of Blumeria graminis f.sp. tritici by interfering with very-long-chain aldehyde biosynthesis. Plant Mol. Biol. 2018, 96, 165–178. [Google Scholar]
- Taunton, J.; Hassig, C.A.; Schreiber, S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996, 272, 408–411. [Google Scholar] [CrossRef]
- Ding, Y.; Ndamukong, I.; Xu, Z.; Lapko, H.; Fromm, M.; Avramova, Z. ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet. 2012, 8, e1003111. [Google Scholar] [CrossRef] [Green Version]
- Shu, H.; Gruissem, W.; Hennig, L. Measuring Arabidopsis chromatin accessibility using DNase I-polymerase chain reaction and DNase I-chip assays. Plant Physiol. 2013, 162, 1794–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhi, P.; Kong, L.; Liu, J.; Zhang, X.; Wang, X.; Li, H.; Sun, M.; Li, Y.; Chang, C. Histone Deacetylase TaHDT701 Functions in TaHDA6-TaHOS15 Complex to Regulate Wheat Defense Responses to Blumeria graminis f.sp. tritici. Int. J. Mol. Sci. 2020, 21, 2640. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072640
Zhi P, Kong L, Liu J, Zhang X, Wang X, Li H, Sun M, Li Y, Chang C. Histone Deacetylase TaHDT701 Functions in TaHDA6-TaHOS15 Complex to Regulate Wheat Defense Responses to Blumeria graminis f.sp. tritici. International Journal of Molecular Sciences. 2020; 21(7):2640. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072640
Chicago/Turabian StyleZhi, Pengfei, Lingyao Kong, Jiao Liu, Xiaona Zhang, Xiaoyu Wang, Haoyu Li, Maokai Sun, Yan Li, and Cheng Chang. 2020. "Histone Deacetylase TaHDT701 Functions in TaHDA6-TaHOS15 Complex to Regulate Wheat Defense Responses to Blumeria graminis f.sp. tritici" International Journal of Molecular Sciences 21, no. 7: 2640. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21072640




