1. Introduction
Telomeres are nucleoprotein structures at the end of eukaryotic chromosomes. In vertebrates, telomeres are composed by extended arrays of the hexanucleotide TTAGGG [
1] and by a specialized protein complex called shelterin [
2]. The main role of telomeres is to prevent chromosome ends from being recognized and processed as double-strand breaks. In normal somatic cells, telomeres shorten at each replication round, while in germ-line and stem cells the reverse transcriptase telomerase ensures DNA replication of chromosome ends by adding TTAGGG repeats to the 3’ end of telomeric DNA. In normal somatic cells, after several replication rounds, telomeres reach a critical length, resulting in the loss of their ability to maintain genome stability. Short telomeres induce a state called replicative senescence, which is characterized by irreversible arrest of the cell cycle and is responsible for a decline in tissue renewal capacity [
3,
4,
5,
6]. Senescence can be seen as a barrier to uncontrolled cell proliferation and tumor development. Cells escaping senescence enter in a condition called crisis that is characterized by genome instability and ultimately leads to apoptosis. Rare survivor cells can rescue telomere maintenance mechanisms which can lead to cell immortalization and cancer. In addition, noncoding RNA molecules transcribed from telomeres (telomeric-repeat-containing RNA (TERRA)) participate in the regulation of telomere function [
7,
8] and have been proposed as prognostic markers in different types of tumors [
9,
10,
11]. Therefore, telomeres together with telomerase, shelterin proteins and TERRA play crucial roles in the maintenance of genome integrity and stability.
Stretches of telomeric-like repeats are also located at internal sites [
12]. Given their position, they are called interstitial telomeric sequences (ITSs). Whereas the role of telomeres in genome stability and replication is well-defined, the function of ITSs remains unclear.
ITSs can be classified according to their sequence organization and localization [
13]. Het-ITSs are very extended blocks of telomeric-like repeats located at pericentromeric, terminal or intrachromosomal regions, generally coinciding with C-bands and easily detectable by cytogenetic analysis (fluorescence in situ hybridization (FISH)). They have been described in several vertebrate species [
14,
15,
16,
17,
18,
19] and in some insects [
20] and plants [
21] but are absent in other species such as human and mouse.
Although it has been proposed that Robertsonian fusion may be an important mechanism for ITS formation [
13], in the human genome the only “fusion” ITS is the one on chromosome 2q13 whose repeated units are organized in a head-to-head fashion [
22,
23].
Some ITSs, that we called “subtelomeric” [
13], are localized in regions immediately adjacent to the
bona fide telomeres and contain many degenerate units intermingled with other types of tandem and interspersed repeats.
In the present work we focused on “short-ITSs” which are stretches of telomeric-like repeats ranging in size from a few to a few hundred repeat units that are distributed at internal chromosomal sites. Although short-ITSs have been studied only in a few species, we hypothesized that they may be present in all species in which telomeres are maintained by telomerase [
13]. Short-ITSs, being too short to be efficiently visualized by FISH, can be found by sequence analysis of genomes. We previously studied the organization of short-ITSs in humans [
23,
24,
25] and other primates [
26,
27,
28] and in mouse and rat [
25]. We also showed that different types of ITSs can coexist: short and het-ITSs in Chinese hamster [
29,
30]; short, subtelomeric and fusion-ITSs in humans [
23]. In the horse, the absence of strong nonterminal telomeric FISH signals, that we now call het-ITSs, was first observed by de la Seña et al. [
31]. With regards to other Perissodactyls, large, detectable by FISH, ITSs are also absent in the donkey [
32] but present in five Hartmann’s Mountain zebra chromosomes [
33].
Being composed by the repetition of 6 bp long units, short-ITSs are a particular type of microsatellite. Whereas canonical microsatellites originate from progressive expansion of a few pre-existing repeated units through DNA polymerase slippage [
34], we showed that the mechanism of origin of ITSs during evolution is completely different [
25,
27]. Through a comparative analysis of human ITS loci and of their orthologous empty loci in other primates, we demonstrated that telomeric-like repeats appear suddenly during evolution, being introduced through a peculiar pathway of DNA double-strand break repair. We then proposed that telomerase may be directly involved in this pathway that should take place in the germ-line [
25,
27]. According to our model, the cytogenetic co-localization of short-ITSs and fragile sites that we previously observed in primates [
26,
28] and rodents [
29,
35] suggests they are not themselves prone to breakage but were rather inserted within DNA sites prone to breakage. Therefore, short-ITSs can be considered as ‘scars’ of DNA breaks that occurred at pre-existing fragile sites.
Since ITS loci originated from the insertion of a telomeric repeat stretch at a DNA double-strand break site during evolution, we might expect that, for recently inserted ITS loci, empty ITS-less alleles are found in the same species, generating insertion polymorphism. Several studies demonstrated that transposable elements, such as Alu retrotransposons in humans, are characterized by insertion polymorphism [
36,
37,
38,
39,
40,
41,
42]. This type of transposable element polymorphism can be involved in gene expression modulation leading to phenotypic consequences [
43], including human and animal disease [
44,
45], and can be particularly informative for population genetics studies [
46,
47,
48]. Interestingly, we showed that the insertion of an ERE1 retroelement within the promoter of the horse myostatin gene greatly reduces its expression and improves racing performance in some breeds [
49]. We previously demonstrated that, in the horse, insertion polymorphism is particularly frequent for two types of sequences: (i) retrotransposons from the equine repetitive element 1 (ERE1) subfamily [
49] and (ii) nuclear sequences of mitochondrial origin (numt) [
50]. These observations, together with a number of molecular and cytogenetic comparative studies [
51,
52,
53,
54,
55,
56,
57,
58], support the hypothesis that the horse genome is in a stage of rapid evolution.
The first goal of the present work was to update the list of human ITSs and to investigate whether insertion polymorphism at ITS loci can be detected in the human population.
The second goal of this work was to identify ITS loci in the horse genome and to test whether, similarly to retrotransposons and numts, these loci are characterized by insertion polymorphism.
The third goal of the present work was to study the molecular mechanisms of ITS insertion through sequence comparison between ITSs and their corresponding empty loci.
3. Discussion
We previously classified interstitial telomeres according to their cytogenetic position and sequence organization as heterochromatic, short, fusion and subtelomeric [
13].
In previous studies, large blocks of telomeric-like repeats, corresponding to heterochromatic ITSs, could be detected by FISH in several metazoan and plant species [
14,
15,
16,
17,
18,
19,
20]. The application of the FISH technique revealed that this type of ITS is not present in the human genome, while allowing us to detect only a limited number of short-ITSs [
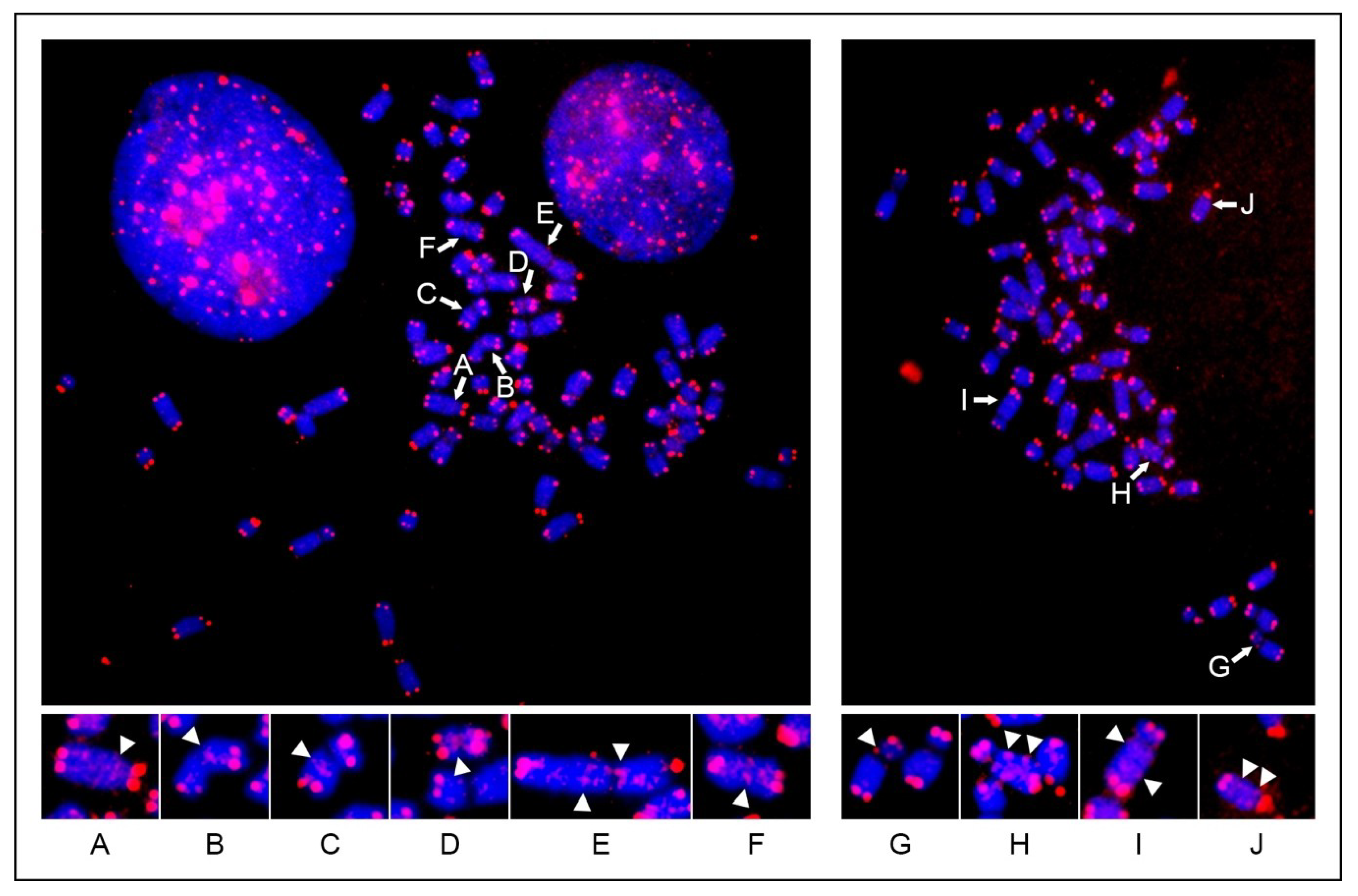
24]. In the present work, the same kind of analysis applied to horse metaphase spreads revealed that the general organization of interstitial telomeres in horses is similar to the one described in humans. As for the human situation, the horse short-ITSs were displayed as weak signals or remained largely undetected, due to the limited sensitivity of the FISH technique.
To compile a comprehensive list of short-ITSs in the human and horse genomes and to study their sequence organization, we analyzed the genome assemblies of the two species. It is worth mentioning that the number of ITSs that can be detected with this approach depends on the software, parameters used and coverage of the genome assembly. For instance, by BLAT search against the human genome version NCBI34/hg16, we previously found 83 ITSs composed by at least four repeats with less than one mismatch per repetition [
25]. In a successive work, in which we discovered that telomeric repeat factors 1 and 2 (TRF1 and TRF2), which are the two main telomere binding proteins involved in telomere structure and function, bind to a subset of interstitial telomeric repeats, a less stringent search of ITSs was carried out using the automatic RepeatMasker annotation [
69]. Following this search, we found 714 loci which included highly degenerate telomeric-like repeats. In that study we used pre-masked genome data from the software RepeatMasker, which tends to split long or degenerate repeat arrays into several shorter hits. In the present work, we used BLAST to carry out a search in the hg19/GRCh37 version of the human genome assembly, manually corrected overlapping hits and discarded degenerate repeat arrays. With this strategy, we identified 229 human ITSs containing at least four TTAGGG repeats with less than one mismatch per repeat. Using the same approach, we identified 142 short-ITSs in the horse reference genome. We have chosen to consider only stretches of at least four TTAGGG repeats with less than one mismatch per unit to avoid detection of short sequences possibly occurring in the genome by chance. A number of shorter and/or more degenerate ITSs are not included in our list. The choice of these parameters was arbitrary. In previous work, we demonstrated that, following their insertion, telomeric repeats undergo mutation during evolution; therefore, “young ITSs” are characterized by greater sequence conservation compared to “old ITSs” [
25]. Since we were interested in finding insertion polymorphism and in describing insertion mechanisms, we concentrated our analysis on well-conserved and not too short “young” ITSs. It is noteworthy that one ITS in the human genome, at chromosome 2q13, was derived by fusion between ancestral acrocentric chromosomes [
22], while we could not find any evidence of such ITS type in the horse.
A comparative analysis between human and horse ITSs showed that six horse ITSs have been inserted in the genome of a common ancestor of Primates and Perissodactyla, more than 90 million years ago [
70]. It would be interesting to test whether the conservation of the telomeric repeat during such an extended evolutionary time may be related to any function. None of the human ITSs were inserted into exons of coding genes. This result is not surprising because such mutation would have inserted stop codons in both orientations.
The distribution of human and horse ITSs along chromosomes does not seem to be related to their size but is probably the result of random insertions. The fraction of human ITSs localized within introns (31%) is compatible with their random insertion in the genome since the fraction of human genome occupied by introns has been estimated to be between 26% and 38% [
71]. It will be interesting to test whether the presence of telomeric repeats within introns may affect splicing.
In previous studies, we demonstrated that short-ITSs were introduced in one step at a given time during the evolution of primate and rodent lineages [
25,
27]. Therefore, short-ITSs can be considered insertion sequences.
It is well-known that insertion sequences that were introduced recently during evolution can display insertion polymorphism [
36,
37,
38,
39,
40,
41,
42,
49,
50]. That is to say that the insertion-containing allele is not yet fixed, and the empty ancestral allele is also present in the population. Sequences showing insertion polymorphism have been used as markers for population genetic studies in many species including humans [
37,
42,
47,
49] and, in some cases, they have been associated to gene expression regulation [
43,
49]. To our knowledge, insertion polymorphism at short-ITS loci has not been described so far. Given the short length of these repeated arrays, their variation cannot be detected by FISH but only by sequence analysis. Indeed, only a fraction of short ITSs can be detected by FISH as faint signals whose frequency is related to the number of repeats at each locus (
Figure 1) [
24,
29]. On the contrary, variation of het-ITSs was described before through FISH experiments in plants [
72] and in PALA (N-(phosphonacetyl)-L-aspartate)-resistant CHO cells containing amplifications of the CAD (carbamyl-P-synthetase, aspartate transcarbamilase, dihydro-orotase) gene [
60].
Are short-ITSs inserted at random sites or within specific genomic regions? To answer this question, we analyzed the GC content of the regions surrounding human and horse short-ITSs. The analysis was carried out within windows of different length: 100 bp, 1 kb and 5 kb on each side of the telomeric repeat. The values varied greatly among different loci, ranging between 12% and 75%, and the average values corresponded to 41.6% and 41.9% in horse and human, respectively (data not shown). We could conclude that there is no preferential choice for ITS insertion based on GC content. In previous studies we showed that, in primates and rodents, ITS colocalize with fragile sites [
26,
28,
29,
35]. Although we do not know which genomic or epigenetic features may be related to the fragility of these sites, this correlation strongly supported the model of ITS insertion at DNA double-strand break sites.
In this work, we searched for the presence of ITS insertion polymorphism in the human population. Surprisingly, despite the large number of individuals analyzed, no ITS-less alleles were found, suggesting that this kind of polymorphism is not present, although we cannot exclude that very rare ITS-less alleles at some loci may exist.
We have previously shown that, in the horse, insertion polymorphism is particularly frequent for numts and ERE1 transposable elements [
49,
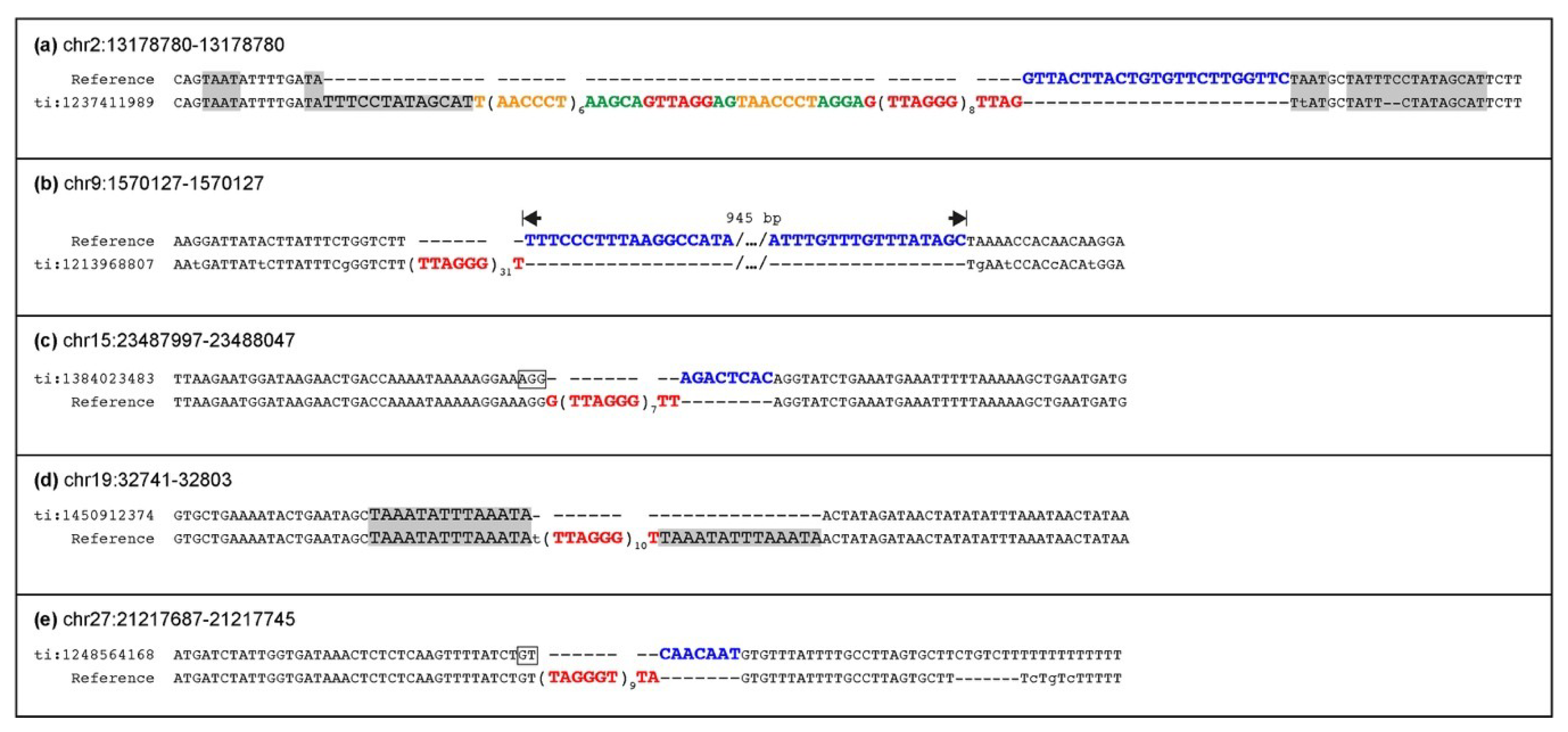
50]. We wondered whether loci polymorphic for ITS insertion could be detected as well. Indeed, as opposed to what we observed in humans, we found five ITS loci heterozygous for the presence of telomeric repeats in the genome of a single horse individual: the mare Twilight, who donated her DNA for the reference genome assembly. A PCR analysis of three loci heterozygous in Twilight in six horse breeds and in Przewalski’s horses confirmed that they are polymorphic. The ITS at chr15:23487997 is polymorphic both in
Equus caballus and in
Equus przewalskii, suggesting that the insertion of the telomeric repeat stretch pre-dates the separation of the domestic and Przewalski’s horse lineages. Therefore, this ITS was inserted in the genome of the common ancestor of the two horse lineages more than 0.5 million years ago [
51]. For the ITSs at chr2:13178780 and at chr19:32741, all analyzed individuals from Przewalski’s horse were homozygous for the ITS-less allele, suggesting that the insertion of telomeric repeats at these loci may have occurred in the domestic horse lineage very recently, after its separation from the Przewalski’s horse lineage. Alternatively, since the population of modern Przewalski’s horses derives from a few individuals [
73], the absence of the ITS may be due to genetic drift.
Our results underline a striking difference between the human and horse genomes in terms of ITS insertion polymorphism. In humans, such polymorphism is either absent or very rare, while our data strongly suggest that it is extremely frequent in the horse. As mentioned above, in the horse, insertion polymorphism is also very frequent for ERE1 retrotransposons and numts. All together, these findings provide further evidence to the notion that the horse genome is in a stage of rapid evolution. In line with this hypothesis is our discovery of an evolutionary new centromere, totally devoid of satellite tandem repeats, on horse chromosome 11 [
52,
58]. Therefore, different molecular mechanisms, such as transposition, DNA double-strand break repair and centromere repositioning contribute to the great plasticity of the horse genome in the current evolutionary stage.
We previously described polymorphism of human ITS loci due to variable number of tandem repeats [
63]. VNTR polymorphism is also present at horse ITSs, indicating that this peculiar type of microsatellite can be unstable and that, similarly to microsatellites with shorter units, they may be useful polymorphic markers for linkage analysis and parentage testing.
Mechanisms of ITS Insertion in the Horse Genome
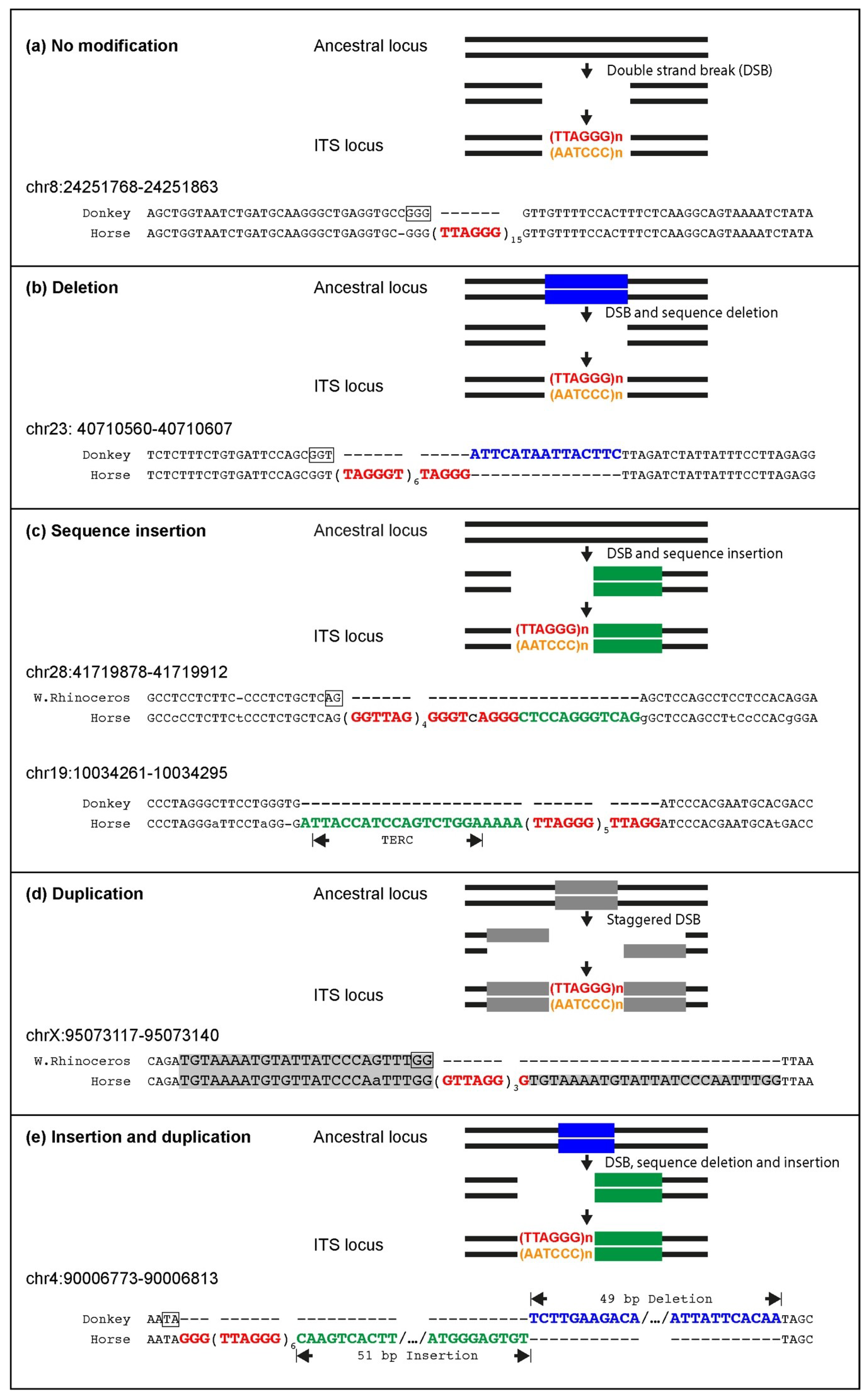
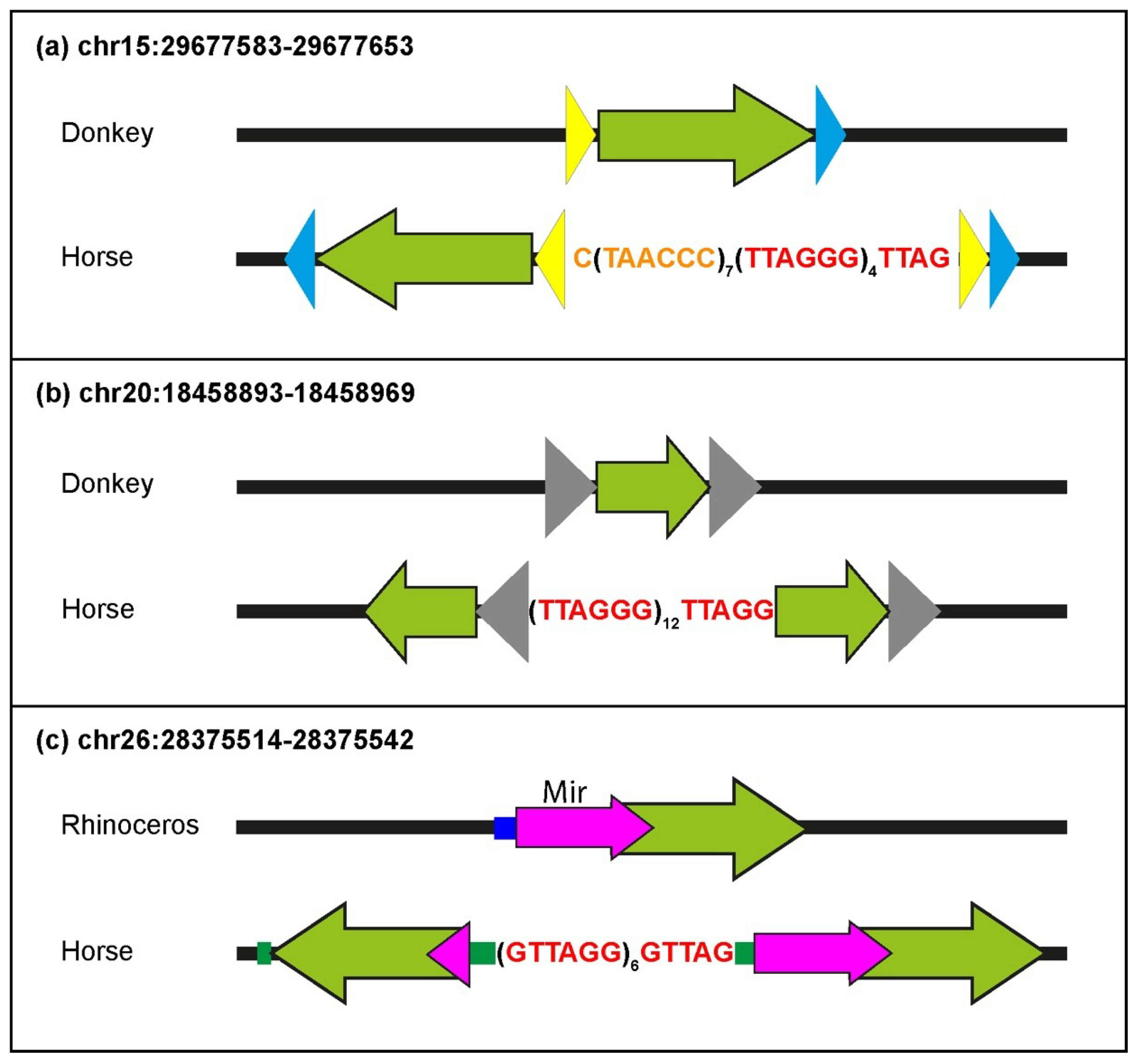
Several ITSs were inserted in the horse genome within target sequences that are well-conserved in the orthologous position of the donkey or rhinoceros genome. Therefore, similarly to primates and rodents, ITS insertions have also occurred in one step in equids [
25,
27]. In the present work, the comparison between ITSs and ITS-less ancestral orthologous loci allowed us to demonstrate that the insertion sites underwent modifications that are typical of the nonhomologous end-joining pathway, supporting our previous hypothesis that they are generated in the course of evolution during the repair of DNA double-strand breaks [
25,
27]. Deletions of short sequences are the most frequent modifications occurring at the break site during the insertion of ITSs, but random sequence addition also occurred. At a few horse ITS loci, direct duplications of target sequences occurred that are likely resulting from the repair of staggered double-strand DNA breaks. Deletions of sequences flanking the break site were indeed the most frequent modifications observed at junctions produced by the repair of double-strand breaks (DSB) induced in experimental systems, while additions and duplications were also observed [
74,
75,
76]. During DSB repair, sequence modifications of the broken ends seem to be often necessary to provide the correct substrate for the final ligation reaction and are operated by specific enzymes such as the Mre11, Exo1 and Artemis nucleases, polynucleotide kinases and template-independent DNA polymerases [
77]. Interestingly, the insertion of telomeric repeats in the horse genome was frequently accompanied by complex modifications of the target sequence involving combinations of deletions, additions, inversions and duplications. Such complex rearrangements were not observed in our previous analysis of rodents and primates, further confirming the great plasticity of the horse genome. In one ITS (chr19:10034261,
Figure 3c), the telomeric repeat stretch was inserted together with a sequence retrotranscribed from a region of the telomerase RNA component (TERC) far away from the telomeric template. Our previous observation of 14 mouse ITS loci with a similar sequence arrangement, called TERC-ITS [
25], provided a strong indication that the telomerase enzyme may be involved in the generation of interstitial telomeres. Having also found a TERC-ITS in the horse genome corroborates this interpretation. Further evidence supporting the involvement of telomerase in ITS insertion is the observation that, in a highly significant number of ITS loci, nucleotides in register with the telomeric repeat sequence were exposed at the break site that occurred in the ancestral sequence. In this scenario, ITS insertion represents one of the noncanonical and controversial roles of telomerase that have been recently proposed [
78]. An alternative mechanism that may account for the generation of some ITSs relies on the introduction of retrotranscribed telomeric RNA into DNA double-strand break sites. These two proposed pathways may be activated in different conditions.
To test whether ITSs can be introduced at DNA double-strand break sites in somatic cells in culture, we previously set up an experimental system based on the induction of site-specific breaks by the I-SceI endonuclease [
75]. We analyzed about 350,000 junctions generated by the repair of these breaks but never observed the insertion of a telomeric repeat stretch, suggesting that, in this system, this event is very rare or does not occur at all. In a successive work, Onozawa and colleagues [
76] transfected total RNA into cultured human cancer cells in which double-strand breaks were induced at I-SceI sites and showed that sequences retrotranscribed from the RNA could be introduced at the break site. At four of these insertions they found telomeric repeats and suggested that these sequences may have been retrotranscribed from telomerase RNA. We can now suggest that the telomeric repeats observed in this experimental system may have been retrotranscribed from TERRA, the family of RNA molecules transcribed from telomeres. It is tempting to postulate that also in vivo some ITSs may have been generated in the germ-line by a DNA repair pathway involving the insertion of DNA fragments retrotranscribed from TERRA molecules.