Nutrition and Genetics in NAFLD: The Perfect Binomium
Abstract
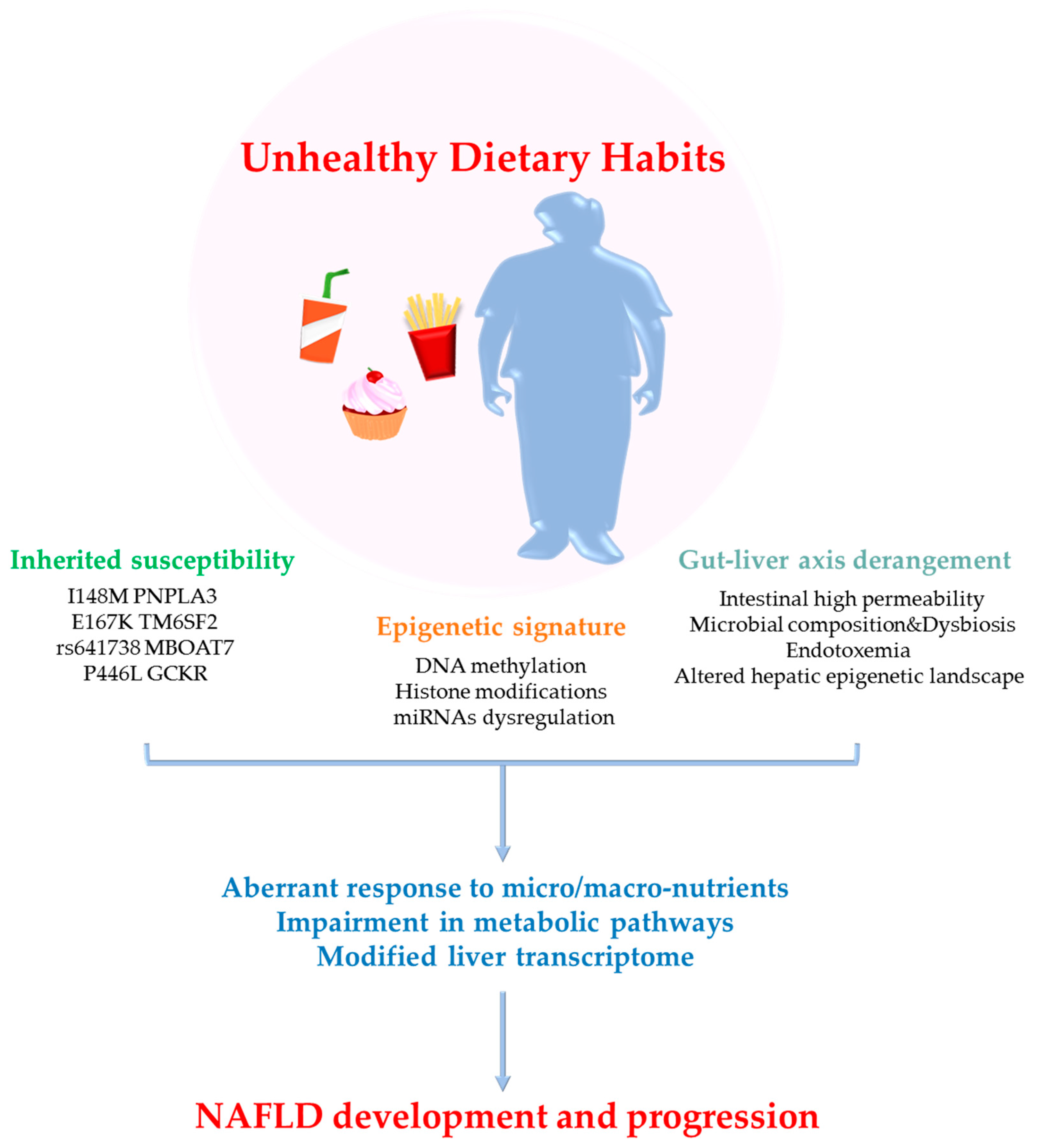
:1. Introduction
2. Methodological Approaches to Nutritional Genomics
3. Nutrigenetics: New Field to Customize Intervention Strategies
3.1. PNPLA3: An Appealing Genetic Sensor of Dietary Compounds
3.2. TM6SF2: Two Sides of the Same Coin
3.3. GCKR: A Potential Carbohydrates’ Modulator to Improve Liver Damage
3.4. MBOAT7 as a Novel High-Sensitive Converter of Nutritional Substrates
3.5. Other NAFLD Genetic Risk Factors Responsive to Diet
4. Nutriepigenomics: Is It Worth Looking into?
4.1. DNA Methylation: from NAFLD towards HCC
4.2. Histone Modifications in NAFLD: Dietary Control of Chromatin Reorganization
4.3. miRNAs-Diet Crosstalk in NAFLD: How Unhealthy Dietary Habits Act at Multiple Levels
5. Role of Gut Microbiota in NAFLD Pathogenesis and Possible Dietary-Based Strategies
Changes in Gut Microbiota Species Alters the Hepatic Epigenetic Landscape: A Tipping Point
6. Precision and Accuracy: The Dartboard of Nutrition
Nutri-Genetics/Epigenetics: A Smart Use of Nutrients in Ongoing Trials
7. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 5mC | 5-methylcytosine |
| αSMA | α smooth muscle actin |
| ACDRP | Amish Complex Disease Research Program |
| ACLY | ATP citrate lyase |
| ALA | α-lipoic acid |
| ALT | Alanine Aminotransferase |
| APO C3 | Apolipoprotein C3 |
| APOE | Apolipoprotein E |
| b3AR | Adrenergic receptor |
| BCAA | Branched-Chain Amino Acid |
| BPA | Bisphenol A |
| CASP1 | Caspase 1 |
| CBMN | Cytokinesis-block micronucleus assay |
| CCL4 | Carbon Tetrachloride |
| Cdkn1a | Cyclin Dependent Kinase Inhibitor 1A |
| ChDD | Choline-deficient diet |
| Col1A1 | Collagen Type I α1 |
| CPT1A | Carnitine Palmitoyltransferase 1A |
| CRISPR/Cas9 | Clustered regularly interspaced short palindromic repeats |
| Cyp7A1 | Cholesterol 7-α hydroxylase 1 |
| DHA | Docosahexaenoic acid |
| DNL | De novo lipogenesis |
| DNMTs | DNA methyltransferases |
| ELISA | Enzyme-linked immunosorbent assay |
| ER | Endothelial Reticulum |
| EPA | Eicosapentanoic acid |
| EZH2 | Enhancer of Zeste Homolog 2 |
| FASN | Fatty acid synthetase |
| FATP1 | Fatty acid transporter 1 |
| FA | Fatty Acid |
| FFA | Free Fatty Acid |
| FGF19 | Fibroblast Growth Factor 19 |
| FGFR2 | Fibroblast growth factor receptor 2 |
| FOXO1 | Forkhead box O1 |
| GCKR | Glucokinase Regulatory Protein |
| GIFT | Genetically Improved Farmed Tilapia |
| GSTM1 | Glutathione S-transferase Mu 1 |
| GSTT1 | Glutathione S-transferase theta 1 |
| GWAS | Genome-wide association study |
| H3K9 | Lysine 9 acetylation of histone H3 |
| H3K36 | Lysine 36 acetylation of histone H3 |
| H3Me2K4 | Lysine 4 dimethylation at histone H3 |
| HAT | Histone acetyltransferase |
| HCC | Hepatocellular Carcinoma |
| HDAC8 | Histone Deacetylase 8 |
| HFD | High Fat Diet |
| HFrD | high-fructose diet |
| HF/HS | High-Fat/High-Sucrose |
| HFS | High Fat Sucrose |
| HSCs | Hepatic Stellate Cells |
| HSD11B | 11β-hydroxysteroid dehydrogenase |
| iPOP | Integrated personal omic profile |
| IR | Insulin Resistance |
| IUGR | Intrauterine Grow Restriction |
| JHDM2A | Jumonji C (JmjC)-Domain-containing Histone Demethylase 2A |
| LDL | Low-density lipoprotein |
| LKB1/AMPK | Liver Kinase B1/AMP-activated Protein Kinase |
| LP | Low protein |
| LPIAT1 | Lyso-phosphatidylinositol acyl-transferase1 |
| LPL | Lipoprotein lipase activity |
| LPS | Lipopolysaccharides |
| LXRα | Liver X receptor alpha |
| Lyso-PI | Lyso-phosphatidylinositol |
| MAM | Mitochondria-associated membrane |
| MBOAT7 | Membrane Bound O-acyltransferase Domain-containing 7 |
| MCD | Methionine-Choline Deficient diet |
| MedDiet | Mediterranean diet |
| MetS | Metabolic Syndrome |
| miRNAs | microRNAs |
| MMP | Matrix Metalloproteinase |
| mtDNA | Mitochondrial DNA |
| MTHFR | Methylenetetrahydrofolate reductase |
| MT-ND6 | Mitochondrially encoded NADH dehydrogenase 6 |
| MTTP | Microsomal triglyceride transfer protein |
| MUFA | Monounsaturated fatty acid |
| Myd88 | Myeloid differentiation factor 88 |
| NAFLD | Nonalcoholic Fatty Liver Disease |
| NAS | NAFLD activity score |
| NASH | Nonalcoholic Steatohepatitis |
| NC | Normal Chow |
| NF-κB | Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B Cells |
| NIOR | Nutrient-induced insulin output ratio |
| NMR | Nuclear Magnetic Resonance |
| PA | Palmitic acid |
| PCG1-α | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha |
| PCSK7 | Proprotein Convertase Subtilisin/Kexin Type 7 |
| PCSK9 | Proprotein Convertase Subtilisin/Kexin Type 9 |
| PDGFα | Platelet-derived growth factor alpha |
| PMBCs | Peripheral blood mononuclear cells |
| PNPLA2 | Patatin-like Phospholipase Domain Protein 2 |
| PNPLA3 | Patatin-like Phospholipase Domain Protein 3 |
| PPARα | Peroxisome proliferator-activated receptor α |
| PPARγ 2 | Peroxisome proliferator-activated receptor γ2 |
| PPP1R3B | Protein Phosphatase 1 Regulatory subunit 3B |
| PUFA | Polyunsaturated fatty acids |
| RNAseq | RNA sequencing |
| ROS | Reactive Oxygen Species |
| SAM | S-adenyl methionine |
| SCD | Stearoyl-CoA Desaturase |
| SCFAs | Short Chain Fatty Acids |
| SFA | Saturated fatty acid |
| Sirt | Sirtuin |
| SNPs | Single nucleotide polymorphisms |
| SOD | Superoxide Dismutase |
| SREBP1c | Sterol Regulatory Element Binding Protein 1c |
| SULT1A1 | Sulfotransferase 1A1 |
| T2D | Type 2 Diabetes Mellitus |
| TA | Tannic acid |
| TG | Triglycerides |
| TGFβ1 | Transforming growth factor β1 |
| TLR4 | Toll-like receptor 4 |
| TM6SF2 | Transmembrane 6 Superfamily Member 2 |
| TMC4/MBOAT7 | Transmembrane Channel-like 4/Membrane Bound O-acyltransferase Domain-containing |
| TNFα | Tumor Necrosis Factor alpha |
| UCP-1 | Uncoupling Protein type I |
| VLDL | Very-low density lipoprotein |
| WNT | Wingless-related integration site |
References
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Day, C.P. From fat to inflammation. Gastroenterology 2006, 130, 207–210. [Google Scholar] [CrossRef]
- Wong, R.J.; Aguilar, M.; Cheung, R.; Perumpail, R.B.; Harrison, S.A.; Younossi, Z.M.; Ahmed, A. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology 2015, 148, 547–555. [Google Scholar] [CrossRef]
- Marchesini, G.; Brizi, M.; Bianchi, G.; Tomassetti, S.; Bugianesi, E.; Lenzi, M.; McCullough, A.J.; Natale, S.; Forlani, G.; Melchionda, N. Nonalcoholic fatty liver disease: A feature of the metabolic syndrome. Diabetes 2001, 50, 1844–1850. [Google Scholar] [CrossRef] [Green Version]
- Byrne, C.D.; Targher, G. NAFLD: A multisystem disease. J. Hepatol. 2015, 62 (Suppl. 1), S47–S64. [Google Scholar] [CrossRef] [Green Version]
- Dongiovanni, P.; Valenti, L. A Nutrigenomic Approach to Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2017, 18, 1534. [Google Scholar] [CrossRef] [Green Version]
- Dongiovanni, P.; Valenti, L. Genetics of nonalcoholic fatty liver disease. Metab. Clin. Exp. 2016, 65, 1026–1037. [Google Scholar] [CrossRef] [PubMed]
- Meroni, M.; Longo, M.; Erconi, V.; Valenti, L.; Gatti, S.; Fracanzani, A.L.; Dongiovanni, P. mir-101-3p Downregulation Promotes Fibrogenesis by Facilitating Hepatic Stellate Cell Transdifferentiation During Insulin Resistance. Nutrients 2019, 11, 2597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dongiovanni, P.; Meroni, M.; Longo, M.; Fargion, S.; Fracanzani, A.L. miRNA Signature in NAFLD: A Turning Point for a Non-Invasive Diagnosis. Int. J. Mol. Sci. 2018, 19, 3966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meroni, M.; Longo, M.; Dongiovanni, P. Alcohol or Gut Microbiota: Who Is the Guilty? Int. J. Mol. Sci. 2019, 20, 4568. [Google Scholar] [CrossRef] [Green Version]
- Meroni, M.; Longo, M.; Dongiovanni, P. The Role of Probiotics in Nonalcoholic Fatty Liver Disease: A New Insight into Therapeutic Strategies. Nutrients 2019, 11, 2642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hesketh, J. Personalised nutrition: How far has nutrigenomics progressed? Eur. J. Clin. Nutr. 2013, 67, 430–435. [Google Scholar] [CrossRef] [Green Version]
- Stover, P.J.; Caudill, M.A. Genetic and epigenetic contributions to human nutrition and health: Managing genome-diet interactions. J. Am. Diet. Assoc. 2008, 108, 1480–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.X. The coming of age of nutrigenetics and nutrigenomics. J. Nutr. Nutr. 2012, 5, I-II. [Google Scholar] [CrossRef] [PubMed]
- Arigony, A.L.; de Oliveira, I.M.; Machado, M.; Bordin, D.L.; Bergter, L.; Pra, D.; Henriques, J.A. The influence of micronutrients in cell culture: A reflection on viability and genomic stability. Biomed Res. Int. 2013, 2013, 597282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenech, M. Genome health nutrigenomics and nutrigenetics--diagnosis and nutritional treatment of genome damage on an individual basis. Food Chem. Toxicol. 2008, 46, 1365–1370. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Baghurst, P.; Luderer, W.; Turner, J.; Record, S.; Ceppi, M.; Bonassi, S. Low intake of calcium, folate, nicotinic acid, vitamin E, retinol, beta-carotene and high intake of pantothenic acid, biotin and riboflavin are significantly associated with increased genome instability--results from a dietary intake and micronucleus index survey in South Australia. Carcinogenesis 2005, 26, 991–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakajima, T.; Moriguchi, M.; Katagishi, T.; Sekoguchi, S.; Nishikawa, T.; Takashima, H.; Kimura, H.; Minami, M.; Itoh, Y.; Kagawa, K.; et al. Premature telomere shortening and impaired regenerative response in hepatocytes of individuals with NAFLD. Liver Int. 2006, 26, 23–31. [Google Scholar] [CrossRef]
- Ferguson, L.R.; De Caterina, R.; Görman, U.; Allayee, H.; Kohlmeier, M.; Prasad, C.; Choi, M.S.; Curi, R.; de Luis, D.A.; Gil, A.; et al. Guide and Position of the International Society of Nutrigenetics/Nutrigenomics on Personalised Nutrition: Part 1—Fields of Precision Nutrition. Lifestyle Genom. 2016, 9, 12–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stover, P.J. Physiology of folate and vitamin B12 in health and disease. Nutr. Rev. 2004, 62, S3–S12. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.; Arbour, L.; Tran, P.; Leclerc, D.; Sabbaghian, N.; Platt, R.; Gilfix, B.M.; Rosenblatt, D.S.; Gravel, R.A.; Forbes, P.; et al. Genetic polymorphisms in methylenetetrahydrofolate reductase and methionine synthase, folate levels in red blood cells, and risk of neural tube defects. Am. J. Med Genet. 1999, 84, 151–157. [Google Scholar] [CrossRef]
- Sun, M.Y.; Zhang, L.; Shi, S.L.; Lin, J.N. Associations between Methylenetetrahydrofolate Reductase (MTHFR) Polymorphisms and Non-Alcoholic Fatty Liver Disease (NAFLD) Risk: A Meta-Analysis. PLoS ONE 2016, 11, e0154337. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Stampfer, M.J.; Giovannucci, E.; Artigas, C.; Hunter, D.J.; Fuchs, C.; Willett, W.C.; Selhub, J.; Hennekens, C.H.; Rozen, R. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 1997, 57, 1098–1102. [Google Scholar] [PubMed]
- Esfahani, S.T.; Cogger, E.A.; Caudill, M.A. Heterogeneity in the prevalence of methylenetetrahydrofolate reductase gene polymorphisms in women of different ethnic groups. J. Am. Diet. Assoc. 2003, 103, 200–207. [Google Scholar] [CrossRef]
- Toomajian, C.; Ajioka, R.S.; Jorde, L.B.; Kushner, J.P.; Kreitman, M. A method for detecting recent selection in the human genome from allele age estimates. Genetics 2003, 165, 287–297. [Google Scholar]
- Brennan, L.; McNulty, B. New technology in nutrition research and practice. Proc. Nutr. Soc. 2017, 76, 173–174. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.; Kersten, S. Nutrigenomics: Goals and strategies. Nat. Rev. Genet. 2003, 4, 315–322. [Google Scholar] [CrossRef]
- Neeha, V.S.; Kinth, P. Nutrigenomics research: A review. J. Food Sci. Technol. 2013, 50, 415–428. [Google Scholar] [CrossRef] [Green Version]
- Roche, H.M. Nutrigenomics—New approaches for human nutrition research. J. Sci. Food Agric. 2006, 86, 1156–1163. [Google Scholar] [CrossRef]
- Pico, C.; Serra, F.; Rodriguez, A.M.; Keijer, J.; Palou, A. Biomarkers of Nutrition and Health: New Tools for New Approaches. Nutrients 2019, 11, 1092. [Google Scholar] [CrossRef] [Green Version]
- Xie, S.; Moya, C.; Bilgin, B.; Jayaraman, A.; Walton, S.P. Emerging affinity-based techniques in proteomics. Expert Rev. Proteom. 2009, 6, 573–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Li, D.; Dangott, L.J.; Wu, G. Proteomics and Its Role in Nutrition Research. J. Nutr. 2006, 136, 1759–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chirita-Emandi, A.; Niculescu, M. Chapter 7—Methods for Global Nutrigenomics and Precision Nutrition. In Principles of Nutrigenetics and Nutrigenomics; Caterina, R.D.E., Martinez, J.A., Kohlmeier, M., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 49–58. [Google Scholar] [CrossRef]
- Xie, Z.; Li, H.; Wang, K.; Lin, J.; Wang, Q.; Zhao, G.; Jia, W.; Zhang, Q. Analysis of transcriptome and metabolome profiles alterations in fatty liver induced by high-fat diet in rat. Metab. Clin. Exp. 2010, 59, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Mias, G.I.; Li-Pook-Than, J.; Jiang, L.; Lam, H.Y.; Chen, R.; Miriami, E.; Karczewski, K.J.; Hariharan, M.; Dewey, F.E.; et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012, 148, 1293–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dongiovanni, P.; Anstee, Q.M.; Valenti, L. Genetic predisposition in NAFLD and NASH: Impact on severity of liver disease and response to treatment. Curr. Pharm. Des. 2013, 19, 5219–5238. [Google Scholar] [CrossRef] [Green Version]
- Dongiovanni, P.; Romeo, S.; Valenti, L. Genetic Factors in the Pathogenesis of Nonalcoholic Fatty Liver and Steatohepatitis. Biomed Res. Int. 2015, 2015, 460190. [Google Scholar] [CrossRef]
- Schwimmer, J.B.; Celedon, M.A.; Lavine, J.E.; Salem, R.; Campbell, N.; Schork, N.J.; Shiehmorteza, M.; Yokoo, T.; Chavez, A.; Middleton, M.S.; et al. Heritability of nonalcoholic fatty liver disease. Gastroenterology 2009, 136, 1585–1592. [Google Scholar] [CrossRef] [Green Version]
- Willner, I.R.; Waters, B.; Patil, S.R.; Reuben, A.; Morelli, J.; Riely, C.A. Ninety patients with nonalcoholic steatohepatitis: Insulin resistance, familial tendency, and severity of disease. Am. J. Gastroenterol. 2001, 96, 2957–2961. [Google Scholar] [CrossRef]
- Guerrero, R.; Vega, G.L.; Grundy, S.M.; Browning, J.D. Ethnic differences in hepatic steatosis: An insulin resistance paradox? Hepatology 2009, 49, 791–801. [Google Scholar] [CrossRef] [Green Version]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef] [Green Version]
- Pingitore, P.; Dongiovanni, P.; Motta, B.M.; Meroni, M.; Lepore, S.M.; Mancina, R.M.; Pelusi, S.; Russo, C.; Caddeo, A.; Rossi, G.; et al. PNPLA3 overexpression results in reduction of proteins predisposing to fibrosis. Hum. Mol. Genet. 2016, 25, 5212–5222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondul, A.; Mancina, R.M.; Merlo, A.; Dongiovanni, P.; Rametta, R.; Montalcini, T.; Valenti, L.; Albanes, D.; Romeo, S. PNPLA3 I148M Variant Influences Circulating Retinol in Adults with Nonalcoholic Fatty Liver Disease or Obesity. J. Nutr. 2015, 145, 1687–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dongiovanni, P.; Romeo, S.; Valenti, L. Hepatocellular carcinoma in nonalcoholic fatty liver: Role of environmental and genetic factors. World J. Gastroenterol. 2014, 20, 12945–12955. [Google Scholar] [CrossRef] [PubMed]
- BasuRay, S.; Smagris, E.; Cohen, J.C.; Hobbs, H.H. The PNPLA3 variant associated with fatty liver disease (I148M) accumulates on lipid droplets by evading ubiquitylation. Hepatology 2017, 66, 1111–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donati, B.; Motta, B.M.; Pingitore, P.; Meroni, M.; Pietrelli, A.; Alisi, A.; Petta, S.; Xing, C.; Dongiovanni, P.; del Menico, B.; et al. The rs2294918 E434K variant modulates patatin-like phospholipase domain-containing 3 expression and liver damage. Hepatology 2016, 63, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Li, J.Z.; Huang, Y.; Karaman, R.; Ivanova, P.T.; Brown, H.A.; Roddy, T.; Castro-Perez, J.; Cohen, J.C.; Hobbs, H.H. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J. Clin. Investig. 2012, 122, 4130–4144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dongiovanni, P.; Donati, B.; Fares, R.; Lombardi, R.; Mancina, R.M.; Romeo, S.; Valenti, L. PNPLA3 I148M polymorphism and progressive liver disease. World J. Gastroenterol. 2013, 19, 6969–6978. [Google Scholar] [CrossRef]
- Valenti, L.; Motta, B.M.; Soardo, G.; Iavarone, M.; Donati, B.; Sangiovanni, A.; Carnelutti, A.; Dongiovanni, P.; Rametta, R.; Bertelli, C.; et al. PNPLA3 I148M polymorphism, clinical presentation, and survival in patients with hepatocellular carcinoma. PLoS ONE 2013, 8, e75982. [Google Scholar] [CrossRef] [Green Version]
- Pirazzi, C.; Valenti, L.; Motta, B.M.; Pingitore, P.; Hedfalk, K.; Mancina, R.M.; Burza, M.A.; Indiveri, C.; Ferro, Y.; Montalcini, T.; et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum. Mol. Genet. 2014, 23, 4077–4085. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; He, S.; Li, J.Z.; Seo, Y.-K.; Osborne, T.F.; Cohen, J.C.; Hobbs, H.H. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc. Natl. Acad. Sci. USA 2010, 107, 7892–7897. [Google Scholar] [CrossRef] [Green Version]
- Baulande, S.; Lasnier, F.; Lucas, M.; Pairault, J. Adiponutrin, a transmembrane protein corresponding to a novel dietary- and obesity-linked mRNA specifically expressed in the adipose lineage. J. Biol. Chem. 2001, 276, 33336–33344. [Google Scholar] [CrossRef] [Green Version]
- Santoro, N.; Savoye, M.; Kim, G.; Marotto, K.; Shaw, M.M.; Pierpont, B.; Caprio, S. Hepatic fat accumulation is modulated by the interaction between the rs738409 variant in the PNPLA3 gene and the dietary omega6/omega3 PUFA intake. PLoS ONE 2012, 7, e37827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Cohen, J.C.; Hobbs, H.H. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J. Biol. Chem. 2011, 286, 37085–37093. [Google Scholar] [CrossRef] [Green Version]
- Simopoulos, A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008, 233, 674–688. [Google Scholar] [CrossRef]
- Perttilä, J.; Huaman-Samanez, C.; Caron, S.; Tanhuanpää, K.; Staels, B.; Yki-Järvinen, H.; Olkkonen, V.M. PNPLA3 is regulated by glucose in human hepatocytes, and its I148M mutant slows down triglyceride hydrolysis. Am. J. Physiol. Endocrinol. Metab. 2012, 302, E1063–E1069. [Google Scholar] [CrossRef]
- Scorletti, E.; West, A.L.; Bhatia, L.; Hoile, S.P.; McCormick, K.G.; Burdge, G.C.; Lillycrop, K.A.; Clough, G.F.; Calder, P.C.; Byrne, C.D. Treating liver fat and serum triglyceride levels in NAFLD, effects of PNPLA3 and TM6SF2 genotypes: Results from the WELCOME trial. J. Hepatol. 2015, 63, 1476–1483. [Google Scholar] [CrossRef] [Green Version]
- Nobili, V.; Bedogni, G.; Donati, B.; Alisi, A.; Valenti, L. The I148M variant of PNPLA3 reduces the response to docosahexaenoic acid in children with non-alcoholic fatty liver disease. J. Med. Food 2013, 16, 957–960. [Google Scholar] [CrossRef] [Green Version]
- Mancina, R.M.; Matikainen, N.; Maglio, C.; Söderlund, S.; Lundbom, N.; Hakkarainen, A.; Rametta, R.; Mozzi, E.; Fargion, S.; Valenti, L.; et al. Paradoxical dissociation between hepatic fat content and de novo lipogenesis due to PNPLA3 sequence variant. J. Clin. Endocrinol. Metab. 2015, 100, E821–E825. [Google Scholar] [CrossRef] [Green Version]
- Davis, J.N.; Lê, K.-A.; Walker, R.W.; Vikman, S.; Spruijt-Metz, D.; Weigensberg, M.J.; Allayee, H.; Goran, M.I. Increased hepatic fat in overweight Hispanic youth influenced by interaction between genetic variation in PNPLA3 and high dietary carbohydrate and sugar consumption. Am. J. Clin. Nutr. 2010, 92, 1522–1527. [Google Scholar] [CrossRef] [Green Version]
- Sevastianova, K.; Kotronen, A.; Gastaldelli, A.; Perttilä, J.; Hakkarainen, A.; Lundbom, J.; Suojanen, L.; Orho-Melander, M.; Lundbom, N.; Ferrannini, E.; et al. Genetic variation in PNPLA3 (adiponutrin) confers sensitivity to weight loss-induced decrease in liver fat in humans. Am. J. Clin. Nutr. 2011, 94, 104–111. [Google Scholar] [CrossRef]
- Nobili, V.; Liccardo, D.; Bedogni, G.; Salvatori, G.; Gnani, D.; Bersani, I.; Alisi, A.; Valenti, L.; Raponi, M. Influence of dietary pattern, physical activity, and I148M PNPLA3 on steatosis severity in at-risk adolescents. Genes Nutr. 2014, 9, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzuillo, P.; Grandone, A.; Perrone, L.; del Giudice, E.M. Weight loss allows the dissection of the interaction between abdominal fat and PNPLA3 (adiponutrin) in the liver damage of obese children. J. Hepatol. 2013, 59, 1143–1144. [Google Scholar] [CrossRef] [Green Version]
- Kozlitina, J.; Smagris, E.; Stender, S.; Nordestgaard, B.G.; Zhou, H.H.; Tybjærg-Hansen, A.; Vogt, T.F.; Hobbs, H.H.; Cohen, J.C. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014, 46, 352–356. [Google Scholar] [CrossRef] [Green Version]
- Luukkonen, P.K.; Zhou, Y.; Nidhina Haridas, P.A.; Dwivedi, O.P.; Hyötyläinen, T.; Ali, A.; Juuti, A.; Leivonen, M.; Tukiainen, T.; Ahonen, L.; et al. Impaired hepatic lipid synthesis from polyunsaturated fatty acids in TM6SF2 E167K variant carriers with NAFLD. J. Hepatol. 2017, 67, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Dongiovanni, P.; Petta, S.; Maglio, C.; Fracanzani, A.L.; Pipitone, R.; Mozzi, E.; Motta, B.M.; Kaminska, D.; Rametta, R.; Grimaudo, S.; et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology 2015, 61, 506–514. [Google Scholar] [CrossRef]
- Goffredo, M.; Caprio, S.; Feldstein, A.E.; D’Adamo, E.; Shaw, M.M.; Pierpont, B.; Savoye, M.; Zhao, H.; Bale, A.E.; Santoro, N. Role of TM6SF2 rs58542926 in the pathogenesis of nonalcoholic pediatric fatty liver disease: A multiethnic study. Hepatology 2016, 63, 117–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smagris, E.; Gilyard, S.; BasuRay, S.; Cohen, J.C.; Hobbs, H.H. Inactivation of Tm6sf2, a Gene Defective in Fatty Liver Disease, Impairs Lipidation but Not Secretion of Very Low Density Lipoproteins. J. Biol. Chem. 2016, 291, 10659–10676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Hare, E.A.; Yang, R.; Yerges-Armstrong, L.M.; Sreenivasan, U.; McFarland, R.; Leitch, C.C.; Wilson, M.H.; Narina, S.; Gorden, A.; Ryan, K.A.; et al. TM6SF2 rs58542926 impacts lipid processing in liver and small intestine. Hepatology 2017, 65, 1526–1542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musso, G.; Cipolla, U.; Cassader, M.; Pinach, S.; Saba, F.; De Michieli, F.; Paschetta, E.; Bongiovanni, D.; Framarin, L.; Leone, N.; et al. TM6SF2 rs58542926 variant affects postprandial lipoprotein metabolism and glucose homeostasis in NAFLD. J. Lipid Res. 2017, 58, 1221–1229. [Google Scholar] [CrossRef] [Green Version]
- Krawczyk, M.; Stachowska, E.; Milkiewicz, P.; Lammert, F.; Milkiewicz, M. Reduction of Caloric Intake Might Override the Prosteatotic Effects of the PNPLA3 p.I148M and TM6SF2 p.E167K Variants in Patients with Fatty Liver: Ultrasound-Based Prospective Study. Digestion 2016, 93, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Dongiovanni, P.; Petta, S.; Mannisto, V.; Mancina, R.M.; Pipitone, R.; Karja, V.; Maggioni, M.; Kakela, P.; Wiklund, O.; Mozzi, E.; et al. Statin use and non-alcoholic steatohepatitis in at risk individuals. J. Hepatol. 2015, 63, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Beer, N.L.; Tribble, N.D.; McCulloch, L.J.; Roos, C.; Johnson, P.R.; Orho-Melander, M.; Gloyn, A.L. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum. Mol. Genet. 2009, 18, 4081–4088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santoro, N.; Caprio, S.; Pierpont, B.; Van Name, M.; Savoye, M.; Parks, E.J. Hepatic De Novo Lipogenesis in Obese Youth Is Modulated by a Common Variant in the GCKR Gene. J. Clin. Endocrinol. Metab. 2015, 100, E1125–E1132. [Google Scholar] [CrossRef] [Green Version]
- Valenti, L.; Alisi, A.; Nobili, V. Unraveling the genetics of fatty liver in obese children: Additive effect of P446L GCKR and I148M PNPLA3 polymorphisms. Hepatology 2012, 55, 661–663. [Google Scholar] [CrossRef]
- Kaliora, A.C.; Kalafati, I.P.; Gioxari, A.; Diolintzi, A.; Kokkinos, A.; Dedoussis, G.V. A modified response of NAFLD patients with non-significant fibrosis in nutritional counseling according to GCKR rs1260326. Eur. J. Nutr. 2018, 57, 2227–2235. [Google Scholar] [CrossRef]
- Santoro, N.; Zhang, C.K.; Zhao, H.; Pakstis, A.J.; Kim, G.; Kursawe, R.; Dykas, D.J.; Bale, A.E.; Giannini, C.; Pierpont, B.; et al. Variant in the glucokinase regulatory protein (GCKR) gene is associated with fatty liver in obese children and adolescents. Hepatology 2012, 55, 781–789. [Google Scholar] [CrossRef] [Green Version]
- Nettleton, J.A.; McKeown, N.M.; Kanoni, S.; Lemaitre, R.N.; Hivert, M.F.; Ngwa, J.; van Rooij, F.J.; Sonestedt, E.; Wojczynski, M.K.; Ye, Z.; et al. Interactions of dietary whole-grain intake with fasting glucose- and insulin-related genetic loci in individuals of European descent: A meta-analysis of 14 cohort studies. Diabetes Care 2010, 33, 2684–2691. [Google Scholar] [CrossRef] [Green Version]
- Sotos-Prieto, M.; Guillén, M.; Vicente Sorli, J.; Portolés, O.; Guillem-Saiz, P.; Ignacio Gonzalez, J.; Qi, L.; Corella, D. Relevant associations of the glucokinase regulatory protein/glucokinase gene variation with TAG concentrations in a high-cardiovascular risk population: Modulation by the Mediterranean diet. Br. J. Nutr. 2013, 109, 193–201. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-J.; Jang, H.B.; Kim, H.-J.; Ahn, Y.; Hong, K.-W.; Cho, S.B.; Kang, J.H.; Park, S.I. The dietary monounsaturated to saturated fatty acid ratio modulates the genetic effects of GCKR on serum lipid levels in children. Clin. Chim. Acta Int. J. Clin. Chem. 2015, 450, 155–161. [Google Scholar] [CrossRef]
- Buch, S.; Stickel, F.; Trépo, E.; Way, M.; Herrmann, A.; Nischalke, H.D.; Brosch, M.; Rosendahl, J.; Berg, T.; Ridinger, M.; et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat. Genet. 2015, 47, 1443–1448. [Google Scholar] [CrossRef]
- Mancina, R.M.; Dongiovanni, P.; Petta, S.; Pingitore, P.; Meroni, M.; Rametta, R.; Boren, J.; Montalcini, T.; Pujia, A.; Wiklund, O.; et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology 2016, 150, 1219–1230. [Google Scholar] [CrossRef] [Green Version]
- Donati, B.; Dongiovanni, P.; Romeo, S.; Meroni, M.; McCain, M.; Miele, L.; Petta, S.; Maier, S.; Rosso, C.; De Luca, L.; et al. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci. Rep. 2017, 7, 4492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thabet, K.; Chan, H.L.Y.; Petta, S.; Mangia, A.; Berg, T.; Boonstra, A.; Brouwer, W.P.; Abate, M.L.; Wong, V.W.; Nazmy, M.; et al. The membrane-bound O-acyltransferase domain-containing 7 variant rs641738 increases inflammation and fibrosis in chronic hepatitis B. Hepatology 2017, 65, 1840–1850. [Google Scholar] [CrossRef]
- Thabet, K.; Asimakopoulos, A.; Shojaei, M.; Romero-Gomez, M.; Mangia, A.; Irving, W.L.; Berg, T.; Dore, G.J.; Gronbaek, H.; Sheridan, D.; et al. MBOAT7 rs641738 increases risk of liver inflammation and transition to fibrosis in chronic hepatitis C. Nat. Commun. 2016, 7, 12757. [Google Scholar] [CrossRef] [Green Version]
- Zarini, S.; Hankin, J.A.; Murphy, R.C.; Gijon, M.A. Lysophospholipid acyltransferases and eicosanoid biosynthesis in zebrafish myeloid cells. Prostaglandins Other Lipid Mediat. 2014, 113–115, 52–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meroni, M.; Dongiovanni, P.; Longo, M.; Carli, F.; Baselli, G.; Rametta, R.; Pelusi, S.; Badiali, S.; Maggioni, M.; Gaggini, M.; et al. Mboat7 down-regulation by hyper-insulinemia induces fat accumulation in hepatocytes. EBioMedicine 2020, 52, 102658. [Google Scholar] [CrossRef] [Green Version]
- Miele, L.; Dall’armi, V.; Cefalo, C.; Nedovic, B.; Arzani, D.; Amore, R.; Rapaccini, G.; Gasbarrini, A.; Ricciardi, W.; Grieco, A.; et al. A case-control study on the effect of metabolic gene polymorphisms, nutrition, and their interaction on the risk of non-alcoholic fatty liver disease. Genes Nutr. 2014, 9, 383. [Google Scholar] [CrossRef] [Green Version]
- Stachowska, E.; Ryterska, K.; Maciejewska, D.; Banaszczak, M.; Milkiewicz, P.; Milkiewicz, M.; Gutowska, I.; Ossowski, P.; Kaczorowska, M.; Jamioł-Milc, D.; et al. Nutritional Strategies for the Individualized Treatment of Non-Alcoholic Fatty Liver Disease (NAFLD) Based on the Nutrient-Induced Insulin Output Ratio (NIOR). Int. J. Mol. Sci. 2016, 17, 1192. [Google Scholar] [CrossRef] [Green Version]
- Dongiovanni, P.; Meroni, M.; Baselli, G.; Mancina, R.M.; Ruscica, M.; Longo, M.; Rametta, R.; Cespiati, A.; Pelusi, S.; Ferri, N.; et al. PCSK7 gene variation bridges atherogenic dyslipidemia with hepatic inflammation in NAFLD patients. J. Lipid Res. 2019, 60, 1144–1153. [Google Scholar] [CrossRef]
- Huang, T.; Huang, J.; Qi, Q.; Li, Y.; Bray, G.A.; Rood, J.; Sacks, F.M.; Qi, L. PCSK7 genotype modifies effect of a weight-loss diet on 2-year changes of insulin resistance: The POUNDS LOST trial. Diabetes Care 2015, 38, 439–444. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.; Pertsemlidis, A.; Kotowski, I.K.; Graham, R.; Garcia, C.K.; Hobbs, H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005, 37, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Ruscica, M.; Ferri, N.; Macchi, C.; Meroni, M.; Lanti, C.; Ricci, C.; Maggioni, M.; Fracanzani, A.L.; Badiali, S.; Fargion, S.; et al. Liver fat accumulation is associated with circulating PCSK9. Ann. Med. 2016, 48, 384–391. [Google Scholar] [CrossRef]
- Trinder, M.; Francis, G.A.; Brunham, L.R. Association of Monogenic vs Polygenic Hypercholesterolemia With Risk of Atherosclerotic Cardiovascular Disease. JAMA Cardiol. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costet, P.; Cariou, B.; Lambert, G.; Lalanne, F.; Lardeux, B.; Jarnoux, A.L.; Grefhorst, A.; Staels, B.; Krempf, M. Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatory element-binding protein 1c. J. Biol. Chem. 2006, 281, 6211–6218. [Google Scholar] [CrossRef] [Green Version]
- Dongiovanni, P.; Meroni, M.; Mancina, R.M.; Baselli, G.; Rametta, R.; Pelusi, S.; Mannisto, V.; Fracanzani, A.L.; Badiali, S.; Miele, L.; et al. Protein phosphatase 1 regulatory subunit 3B gene variation protects against hepatic fat accumulation and fibrosis in individuals at high risk of nonalcoholic fatty liver disease. Hepatol. Commun. 2018, 2, 666–675. [Google Scholar] [CrossRef] [Green Version]
- Stender, S.; Smagris, E.; Lauridsen, B.K.; Kofoed, K.F.; Nordestgaard, B.G.; Tybjaerg-Hansen, A.; Pennacchio, L.A.; Dickel, D.E.; Cohen, J.C.; Hobbs, H.H. Relationship between genetic variation at PPP1R3B and levels of liver glycogen and triglyceride. Hepatology 2018, 67, 2182–2195. [Google Scholar] [CrossRef] [Green Version]
- Mehta, M.B.; Shewale, S.V.; Sequeira, R.N.; Millar, J.S.; Hand, N.J. Hepatic protein phosphatase 1 regulatory subunit 3B (Ppp1r3b) promotes hepatic glycogen synthesis and thereby regulates fasting energy homeostasis. J. Biol. Chem. 2017, 292, 10444–10454. [Google Scholar] [CrossRef] [Green Version]
- Remely, M.; Stefanska, B.; Lovrecic, L.; Magnet, U.; Haslberger, A.G. Nutriepigenomics: The role of nutrition in epigenetic control of human diseases. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 328–333. [Google Scholar] [CrossRef]
- Barker, D.J. Developmental origins of adult health and disease. J. Epidemiol. Community Health 2004, 58, 114–115. [Google Scholar] [CrossRef] [Green Version]
- Goyal, R.; Goyal, D.; Leitzke, A.; Gheorghe, C.P.; Longo, L.D. Brain renin-angiotensin system: Fetal epigenetic programming by maternal protein restriction during pregnancy. Reprod. Sci. 2010, 17, 227–238. [Google Scholar] [CrossRef]
- Wankhade, U.D.; Zhong, Y.; Kang, P.; Alfaro, M.; Chintapalli, S.V.; Thakali, K.M.; Shankar, K. Enhanced offspring predisposition to steatohepatitis with maternal high-fat diet is associated with epigenetic and microbiome alterations. PLoS ONE 2017, 12, e0175675. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.; Pan, Y.X. Pathophysiological basis for compromised health beyond generations: Role of maternal high-fat diet and low-grade chronic inflammation. J. Nutr. Biochem. 2015, 26, 1–8. [Google Scholar] [CrossRef]
- Murphy, S.K.; Yang, H.; Moylan, C.A.; Pang, H.; Dellinger, A.; Abdelmalek, M.F.; Garrett, M.E.; Ashley-Koch, A.; Suzuki, A.; Tillmann, H.L.; et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology 2013, 145, 1076–1087. [Google Scholar] [CrossRef] [Green Version]
- Pirola, C.J.; Gianotti, T.F.; Burgueno, A.L.; Rey-Funes, M.; Loidl, C.F.; Mallardi, P.; Martino, J.S.; Castano, G.O.; Sookoian, S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut 2013, 62, 1356–1363. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordero, P.; Milagro, F.I.; Campion, J.; Martinez, J.A. Maternal methyl donors supplementation during lactation prevents the hyperhomocysteinemia induced by a high-fat-sucrose intake by dams. Int. J. Mol. Sci. 2013, 14, 24422–24437. [Google Scholar] [CrossRef] [Green Version]
- Mikael, L.G.; Pancer, J.; Wu, Q.; Rozen, R. Disturbed one-carbon metabolism causing adverse reproductive outcomes in mice is associated with altered expression of apolipoprotein AI and inflammatory mediators PPARalpha, interferon-gamma, and interleukin-10. J. Nutr. 2012, 142, 411–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.J.; Zhang, H.W.; Zhou, J.Y.; Liu, Y.; Yang, Y.; Chen, X.L.; Zhu, C.H.; Zheng, R.D.; Ling, W.H.; Zhu, H.L. Betaine attenuates hepatic steatosis by reducing methylation of the MTTP promoter and elevating genomic methylation in mice fed a high-fat diet. J. Nutr. Biochem. 2014, 25, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Takaya, J.; Yamanouchi, S.; Kaneko, K. A Calcium-Deficient Diet in Rat Dams during Gestation and Nursing Affects Hepatic 11β-hydroxysteroid dehydrogenase-1 Expression in the Offspring. PLoS ONE 2014, 9, e84125. [Google Scholar] [CrossRef] [PubMed]
- Takaya, J.; Iharada, A.; Okihana, H.; Kaneko, K. Magnesium deficiency in pregnant rats alters methylation of specific cytosines in the hepatic hydroxysteroid dehydrogenase-2 promoter of the offspring. Epigenetics 2011, 6, 573–578. [Google Scholar] [CrossRef] [Green Version]
- Dudley, K.J.; Sloboda, D.M.; Connor, K.L.; Beltrand, J.; Vickers, M.H. Offspring of mothers fed a high fat diet display hepatic cell cycle inhibition and associated changes in gene expression and DNA methylation. PLoS ONE 2011, 6, e21662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pogribny, I.P.; Shpyleva, S.I.; Muskhelishvili, L.; Bagnyukova, T.V.; Jill James, S.; Beland, F.A. Role of DNA damage and alterations in cytosine DNA methylation in rat liver carcinogenesis induced by a methyl-deficient diet. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2009, 669, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Zeybel, M.; Hardy, T.; Robinson, S.M.; Fox, C.; Anstee, Q.M.; Ness, T.; Masson, S.; Mathers, J.C.; French, J.; White, S.; et al. Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin. Epigenetics 2015, 7, 25. [Google Scholar] [CrossRef] [Green Version]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Inagaki, T.; Tachibana, M.; Magoori, K.; Kudo, H.; Tanaka, T.; Okamura, M.; Naito, M.; Kodama, T.; Shinkai, Y.; Sakai, J. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells 2009, 14, 991–1001. [Google Scholar] [CrossRef]
- Sohi, G.; Marchand, K.; Revesz, A.; Arany, E.; Hardy, D.B. Maternal protein restriction elevates cholesterol in adult rat offspring due to repressive changes in histone modifications at the cholesterol 7alpha-hydroxylase promoter. Mol. Endocrinol. 2011, 25, 785–798. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.C.; Guarente, L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 2014, 25, 138–145. [Google Scholar] [CrossRef]
- Pazienza, V.; Borghesan, M.; Mazza, T.; Sheedfar, F.; Panebianco, C.; Williams, R.; Mazzoccoli, G.; Andriulli, A.; Nakanishi, T.; Vinciguerra, M. SIRT1-metabolite binding histone macroH2A1.1 protects hepatocytes against lipid accumulation. Aging 2014, 6, 35–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Friso, S.; Choi, S.-W. Epigenetic mechanisms underlying the link between non-alcoholic fatty liver diseases and nutrition. Nutrients 2014, 6, 3303–3325. [Google Scholar] [CrossRef] [Green Version]
- Hirschey, M.D.; Shimazu, T.; Jing, E.; Grueter, C.A.; Collins, A.M.; Aouizerat, B.; Stancakova, A.; Goetzman, E.; Lam, M.M.; Schwer, B.; et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell 2011, 44, 177–190. [Google Scholar] [CrossRef] [Green Version]
- Chung, M.Y.; Song, J.H.; Lee, J.; Shin, E.J.; Park, J.H.; Lee, S.H.; Hwang, J.T.; Choi, H.K. Tannic acid, a novel histone acetyltransferase inhibitor, prevents non-alcoholic fatty liver disease both in vivo and in vitro model. Mol. Metab. 2019, 19, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wong, V.W.S.; Wong, G.L.H.; Yang, W.; Sun, H.; Shen, J.; Tong, J.H.M.; Go, M.Y.Y.; Cheung, Y.S.; Lai, P.B.S.; et al. Histone Deacetylase HDAC8 Promotes Insulin Resistance and β-Catenin Activation in NAFLD-Associated Hepatocellular Carcinoma. Cancer Res. 2015, 75, 4803–4816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Baffy, G. MicroRNAs in Nonalcoholic Fatty Liver Disease. J. Clin. Med. 2015, 4, 1977–1988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panera, N.; Gnani, D.; Crudele, A.; Ceccarelli, S.; Nobili, V.; Alisi, A. MicroRNAs as controlled systems and controllers in non-alcoholic fatty liver disease. World J. Gastroenterol. 2014, 20, 15079–15086. [Google Scholar] [CrossRef]
- Meroni, M.; Longo, M.; Rametta, R.; Dongiovanni, P. Genetic and Epigenetic Modifiers of Alcoholic Liver Disease. Int. J. Mol. Sci. 2018, 19, 3857. [Google Scholar] [CrossRef] [Green Version]
- Del Campo, J.A.; Gallego-Durán, R.; Gallego, P.; Grande, L. Genetic and Epigenetic Regulation in Nonalcoholic Fatty Liver Disease (NAFLD). Int. J. Mol. Sci. 2018, 19, 911. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.; Deiters, A. MicroRNA miR-122 as a therapeutic target for oligonucleotides and small molecules. Curr. Med. Chem. 2013, 20, 3629–3640. [Google Scholar] [CrossRef]
- Qiang, J.; Tao, Y.F.; Bao, J.W.; Chen, D.J.; Li, H.X.; He, J.; Xu, P. High Fat Diet-Induced miR-122 Regulates Lipid Metabolism and Fat Deposition in Genetically Improved Farmed Tilapia (GIFT, Oreochromis niloticus) Liver. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Long, J.-K.; Dai, W.; Zheng, Y.-W.; Zhao, S.-P. miR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease. Mol. Med. 2019, 25, 26. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.; Li, M.; Wan, X.; Jin, X.; Chen, S.; Yu, C.; Li, Y. Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease. Sci. Rep. 2015, 5, 13729. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhu, K.; Yu, W.; Wang, H.; Liu, L.; Wu, Q.; Li, S.; Guo, J. MiR-181b regulates steatosis in nonalcoholic fatty liver disease via targeting SIRT1. Biochem. Biophys. Res. Commun. 2017, 493, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Dongiovanni, P.; Crudele, A.; Panera, N.; Romito, I.; Meroni, M.; De Stefanis, C.; Palma, A.; Comparcola, D.; Fracanzani, A.L.; Miele, L.; et al. beta-Klotho gene variation is associated with liver damage in children with NAFLD. J. Hepatol. 2020, 72, 411–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bala, S.; Marcos, M.; Kodys, K.; Csak, T.; Catalano, D.; Mandrekar, P.; Szabo, G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J. Biol. Chem. 2011, 286, 1436–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csak, T.; Bala, S.; Lippai, D.; Kodys, K.; Catalano, D.; Iracheta-Vellve, A.; Szabo, G. MicroRNA-155 Deficiency Attenuates Liver Steatosis and Fibrosis without Reducing Inflammation in a Mouse Model of Steatohepatitis. PLoS ONE 2015, 10, e0129251. [Google Scholar] [CrossRef] [Green Version]
- Bonci, D. MicroRNA-21 as therapeutic target in cancer and cardiovascular disease. Recent Pat. Cardiovasc. Drug Discov. 2010, 5, 156–161. [Google Scholar] [CrossRef]
- Zhao, J.; Tang, N.; Wu, K.; Dai, W.; Ye, C.; Shi, J.; Zhang, J.; Ning, B.; Zeng, X.; Lin, Y. MiR-21 simultaneously regulates ERK1 signaling in HSC activation and hepatocyte EMT in hepatic fibrosis. PLoS ONE 2014, 9, e108005. [Google Scholar] [CrossRef] [Green Version]
- Tryndyak, V.P.; Marrone, A.K.; Latendresse, J.R.; Muskhelishvili, L.; Beland, F.A.; Pogribny, I.P. MicroRNA changes, activation of progenitor cells and severity of liver injury in mice induced by choline and folate deficiency. J. Nutr. Biochem. 2016, 28, 83–90. [Google Scholar] [CrossRef]
- Tryndyak, V.P.; Latendresse, J.R.; Montgomery, B.; Ross, S.A.; Beland, F.A.; Rusyn, I.; Pogribny, I.P. Plasma microRNAs are sensitive indicators of inter-strain differences in the severity of liver injury induced in mice by a choline- and folate-deficient diet. Toxicol. Appl. Pharmacol. 2012, 262, 52–59. [Google Scholar] [CrossRef] [Green Version]
- Hillman, E.T.; Lu, H.; Yao, T.; Nakatsu, C.H. Microbial Ecology along the Gastrointestinal Tract. Microbes Environ. 2017, 32, 300–313. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Medina, M.; Denizot, J.; Dreux, N.; Robin, F.; Billard, E.; Bonnet, R.; Darfeuille-Michaud, A.; Barnich, N. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 2014, 63, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Denizot, J.; Thevenot, J.; Martinez-Medina, M.; Massier, S.; Sauvanet, P.; Bernalier-Donadille, A.; Denis, S.; Hofman, P.; Bonnet, R.; et al. Western diet induces a shift in microbiota composition enhancing susceptibility to Adherent-Invasive E. coli infection and intestinal inflammation. Sci. Rep. 2016, 6, 19032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hills, R.D., Jr.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef] [Green Version]
- Brahe, L.K.; Astrup, A.; Larsen, L.H. Is butyrate the link between diet, intestinal microbiota and obesity-related metabolic diseases? Obes. Rev. 2013, 14, 950–959. [Google Scholar] [CrossRef]
- Lindstedt, G.; Lindstedt, S.; Gustafsson, B.E. Mucus in Intestinal Contents of Germfree Rats. J. Exp. Med. 1965, 121, 201–213. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Fouts, D.E.; Starkel, P.; Hartmann, P.; Chen, P.; Llorente, C.; DePew, J.; Moncera, K.; Ho, S.B.; Brenner, D.A.; et al. Intestinal REG3 Lectins Protect against Alcoholic Steatohepatitis by Reducing Mucosa-Associated Microbiota and Preventing Bacterial Translocation. Cell Host Microbe 2016, 19, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Hritz, I.; Mandrekar, P.; Velayudham, A.; Catalano, D.; Dolganiuc, A.; Kodys, K.; Kurt-Jones, E.; Szabo, G. The critical role of toll-like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88. Hepatology 2008, 48, 1224–1231. [Google Scholar] [CrossRef]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef]
- Wagnerberger, S.; Spruss, A.; Kanuri, G.; Volynets, V.; Stahl, C.; Bischoff, S.C.; Bergheim, I. Toll-like receptors 1-9 are elevated in livers with fructose-induced hepatic steatosis. Br. J. Nutr. 2012, 107, 1727–1738. [Google Scholar] [CrossRef] [Green Version]
- Jin, R.; Willment, A.; Patel, S.S.; Sun, X.; Song, M.; Mannery, Y.O.; Kosters, A.; McClain, C.J.; Vos, M.B. Fructose induced endotoxemia in pediatric nonalcoholic Fatty liver disease. Int. J. Hepatol. 2014, 2014, 560620. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmer, J.; Lange, B.; Frick, J.S.; Sauer, H.; Zimmermann, K.; Schwiertz, A.; Rusch, K.; Klosterhalfen, S.; Enck, P. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur. J. Clin. Nutr. 2012, 66, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Aries, V.; Crowther, J.S.; Drasar, B.S.; Hill, M.J.; Williams, R.E. Bacteria and the aetiology of cancer of the large bowel. Gut 1969, 10, 334–335. [Google Scholar] [CrossRef] [Green Version]
- Conlon, M.A.; Bird, A.R. The impact of diet and lifestyle on gut microbiota and human health. Nutrients 2014, 7, 17–44. [Google Scholar] [CrossRef]
- Chassard, C.; Lacroix, C. Carbohydrates and the human gut microbiota. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Bae, S.C. Histone deacetylase inhibitors: Molecular mechanisms of action and clinical trials as anti-cancer drugs. Am. J. Transl. Res. 2011, 3, 166–179. [Google Scholar]
- Qin, Y.; Wade, P.A. Crosstalk between the microbiome and epigenome: Messages from bugs. J. Biochem. 2018, 163, 105–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krautkramer, K.A.; Kreznar, J.H.; Romano, K.A.; Vivas, E.I.; Barrett-Wilt, G.A.; Rabaglia, M.E.; Keller, M.P.; Attie, A.D.; Rey, F.E.; Denu, J.M. Diet-Microbiota Interactions Mediate Global Epigenetic Programming in Multiple Host Tissues. Mol. Cell 2016, 64, 982–992. [Google Scholar] [CrossRef] [Green Version]
- Celis-Morales, C.; Livingstone, K.M.; Marsaux, C.F.M.; Macready, A.L.; Fallaize, R.; O’Donovan, C.B.; Woolhead, C.; Forster, H.; Walsh, M.C.; Navas-Carretero, S.; et al. Effect of personalized nutrition on health-related behaviour change: Evidence from the Food4Me European randomized controlled trial. Int. J. Epidemiol. 2016, 46, 578–588. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Lopez, O.; Milagro, F.I.; Allayee, H.; Chmurzynska, A.; Choi, M.S.; Curi, R.; De Caterina, R.; Ferguson, L.R.; Goni, L.; Kang, J.X.; et al. Guide for Current Nutrigenetic, Nutrigenomic, and Nutriepigenetic Approaches for Precision Nutrition Involving the Prevention and Management of Chronic Diseases Associated with Obesity. J. Nutr. Nutr. 2017, 10, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Arpon, A.; Riezu-Boj, J.I.; Milagro, F.I.; Marti, A.; Razquin, C.; Martinez-Gonzalez, M.A.; Corella, D.; Estruch, R.; Casas, R.; Fito, M.; et al. Adherence to Mediterranean diet is associated with methylation changes in inflammation-related genes in peripheral blood cells. J. Physiol. Biochem. 2016, 73, 445–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yubero-Serrano, E.M.; Gonzalez-Guardia, L.; Rangel-Zuñiga, O.; Delgado-Casado, N.; Delgado-Lista, J.; Perez-Martinez, P.; Garcia-Rios, A.; Caballero, J.; Marin, C.; Gutierrez-Mariscal, F.M.; et al. Postprandial antioxidant gene expression is modified by Mediterranean diet supplemented with coenzyme Q(10) in elderly men and women. Age 2013, 35, 159–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez-Mariscal, F.M.; Perez-Martinez, P.; Delgado-Lista, J.; Yubero-Serrano, E.M.; Camargo, A.; Delgado-Casado, N.; Cruz-Teno, C.; Santos-Gonzalez, M.; Rodriguez-Cantalejo, F.; Castaño, J.P.; et al. Mediterranean diet supplemented with coenzyme Q10 induces postprandial changes in p53 in response to oxidative DNA damage in elderly subjects. Age 2012, 34, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, C.F.; Nonino, C.B.; de Oliveira, B.A.; Pinhel, M.A.; Mansego, M.L.; Milagro, F.I.; Zulet, M.A.; Martinez, J.A. DNA Methylation and Hydroxymethylation Levels in Relation to Two Weight Loss Strategies: Energy-Restricted Diet or Bariatric Surgery. Obes. Surg. 2016, 26, 603–611. [Google Scholar] [CrossRef]
- Garcia-Caraballo, S.C.; Comhair, T.M.; Verheyen, F.; Gaemers, I.; Schaap, F.G.; Houten, S.M.; Hakvoort, T.B.M.; Dejong, C.H.C.; Lamers, W.H.; Koehler, S.E. Prevention and reversal of hepatic steatosis with a high-protein diet in mice. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2013, 1832, 685–695. [Google Scholar] [CrossRef] [Green Version]
- Rahmanabadi, A.; Mahboob, S.; Amirkhizi, F.; Hosseinpour-Arjmand, S.; Ebrahimi-Mameghani, M. Oral alpha-lipoic acid supplementation in patients with non-alcoholic fatty liver disease: Effects on adipokines and liver histology features. Food Funct. 2019, 10, 4941–4952. [Google Scholar] [CrossRef]
- Kapoor, S. Hepato-protective effects of alpha lipoic acid besides its role in preventing fatty liver disease. Liver Int. 2013, 33, 162–163. [Google Scholar] [CrossRef]
- Stoner, M.W.; Thapa, D.; Zhang, M.; Gibson, G.A.; Calderon, M.J.; St Croix, C.M.; Scott, I. α-Lipoic acid promotes α-tubulin hyperacetylation and blocks the turnover of mitochondria through mitophagy. Biochem. J. 2016, 473, 1821–1830. [Google Scholar] [CrossRef]
- Rudkowska, I.; Caron-Dorval, D.; Verreault, M.; Couture, P.; Deshaies, Y.; Barbier, O.; Vohl, M.-C. PPARα L162V polymorphism alters the potential of n-3 fatty acids to increase lipoprotein lipase activity. Mol. Nutr. Food Res. 2010, 54, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Grenier, E.; Ziv, E.; Delvin, E.; Leduc, L.; Spahis, S.; Lafond, J.; Levy, E. n-3 fatty acids on utero programming of insulin resistance, NASH and hyperlipidemia in Psammomys obesus. FASEB J. 2008, 22 (Suppl. 1), 687.5. [Google Scholar] [CrossRef]
- Boque, N.; de la Iglesia, R.; de la Garza, A.L.; Milagro, F.I.; Olivares, M.; Banuelos, O.; Soria, A.C.; Rodriguez-Sanchez, S.; Martinez, J.A.; Campion, J. Prevention of diet-induced obesity by apple polyphenols in Wistar rats through regulation of adipocyte gene expression and DNA methylation patterns. Mol. Nutr. Food Res. 2013, 57, 1473–1478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gracia, A.; Elcoroaristizabal, X.; Fernandez-Quintela, A.; Miranda, J.; Bediaga, N.G.; de Pancorbo, M.M.; Rimando, A.M.; Portillo, M.P. Fatty acid synthase methylation levels in adipose tissue: Effects of an obesogenic diet and phenol compounds. Genes Nutr. 2014, 9, 411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morimoto, T.; Sunagawa, Y.; Kawamura, T.; Takaya, T.; Wada, H.; Nagasawa, A.; Komeda, M.; Fujita, M.; Shimatsu, A.; Kita, T.; et al. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. J. Clin. Investig. 2008, 118, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Huang, R.; Xiong, Y.L.; Wu, C. Protective effects of curcumin against liver fibrosis through modulating DNA methylation. Chin. J. Nat. Med. 2016, 14, 255–264. [Google Scholar] [CrossRef]
- Rahmani, S.; Asgary, S.; Askari, G.; Keshvari, M.; Hatamipour, M.; Feizi, A.; Sahebkar, A. Treatment of Non-alcoholic Fatty Liver Disease with Curcumin: A Randomized Placebo-controlled Trial. Phytother. Res. 2016, 30, 1540–1548. [Google Scholar] [CrossRef]
- Bruno, R.S.; Dugan, C.E.; Smyth, J.A.; DiNatale, D.A.; Koo, S.I. Green tea extract protects leptin-deficient, spontaneously obese mice from hepatic steatosis and injury. J. Nutr. 2008, 138, 323–331. [Google Scholar] [CrossRef] [Green Version]
- Park, H.J.; DiNatale, D.A.; Chung, M.Y.; Park, Y.K.; Lee, J.Y.; Koo, S.I.; O’Connor, M.; Manautou, J.E.; Bruno, R.S. Green tea extract attenuates hepatic steatosis by decreasing adipose lipogenesis and enhancing hepatic antioxidant defenses in ob/ob mice. J. Nutr. Biochem. 2011, 22, 393–400. [Google Scholar] [CrossRef]
- Bose, M.; Lambert, J.D.; Ju, J.; Reuhl, K.R.; Shapses, S.A.; Yang, C.S. The major green tea polyphenol, (-)-epigallocatechin-3-gallate, inhibits obesity, metabolic syndrome, and fatty liver disease in high-fat-fed mice. J. Nutr. 2008, 138, 1677–1683. [Google Scholar] [CrossRef]
- Ueno, T.; Torimura, T.; Nakamura, T.; Sivakumar, R.; Nakayama, H.; Otabe, S.; Yuan, X.; Yamada, K.; Hashimoto, O.; Inoue, K.; et al. Epigallocatechin-3-gallate improves nonalcoholic steatohepatitis model mice expressing nuclear sterol regulatory element binding protein-1c in adipose tissue. Int. J. Mol. Med. 2009, 24, 17–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzu, N.; Bahcecioglu, I.H.; Dagli, A.F.; Ozercan, I.H.; Ustundag, B.; Sahin, K. Epigallocatechin gallate attenuates experimental non-alcoholic steatohepatitis induced by high fat diet. J. Gastroenterol. Hepatol. 2008, 23 (8 Pt 2), e465–e470. [Google Scholar] [CrossRef]
- Nakamoto, K.; Takayama, F.; Mankura, M.; Hidaka, Y.; Egashira, T.; Ogino, T.; Kawasaki, H.; Mori, A. Beneficial Effects of Fermented Green Tea Extract in a Rat Model of Non-alcoholic Steatohepatitis. J. Clin. Biochem. Nutr. 2009, 44, 239–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, M.Y.; Park, H.J.; Manautou, J.E.; Koo, S.I.; Bruno, R.S. Green tea extract protects against nonalcoholic steatohepatitis in ob/ob mice by decreasing oxidative and nitrative stress responses induced by proinflammatory enzymes. J. Nutr. Biochem. 2012, 23, 361–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, L.F.; Cogliati, B.; Otton, R. Green Tea Prevents NAFLD by Modulation of miR-34a and miR-194 Expression in a High-Fat Diet Mouse Model. Oxidative Med. Cell. Longev. 2019, 2019, 4168380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Clinical Trial Start-End Date | Status | Study Type | Interventions | Conditions | Objectives | Locations |
|---|---|---|---|---|---|---|
| NCT00760513 11/09-11/18 | Completed (n = 103) | Interventional Randomized Phase IV | Omega-3-acid (OMACOR) 4 gr/d oral capsule for 18 months versus placebo | Biopsy-verified NASH or Ultrasound-verified NAFLD | Identify the effects of n-3 fatty acids on fibrosis and cardiovascular risk biomarkers in NAFLD patients | Southampton General Hospital Southampton, Hants, United Kingdom |
| NCT00885313 03/09–03/11 | Completed (n = 60) | Interventional Randomized Phase II | DHA 250 or 500mg/kg/d oral for 24 months vs. placebo | NAFLD and obesity in children | Evaluate the effects of DHA on Children With NAFLD | Bambino Gesù Hospital and Research Institute Rome, Italy |
| NCT00697580 05/059–06/07 | Completed (n = 104) | Interventional Randomized | Behavioral: Nutrition changes and physical exercise | NAFLD and obesity in adolescents | Investigate the Intra-Abdominal Fat, insulin sensitivity and Risk of Disease in Adolescents | Veronica Atkins Lifestyle Intervention Laboratory Los Angeles, CA (USA) |
| NCT00072995 09/03–09/07 | Completed (n = 811) | Interventional | Four Diets Differing in Macronutrient Composition and Low in Saturated Fat | Cardiovascular disease, NAFLD, obesity | Preventing Obesity Using Novel Dietary Strategies | Pennington Biomedical Research Center, Louisiana State University, Baton Rouge, LA (USA). Harvard University School of Public Health, Boston, MA (USA) |
| NCT01530139 02/12–02/16 | Completed (n = 1607) | Interventional Randomized | Three personalized diets (Food4Me) versus control diet | Adult individuals | Personalization of dietary advices based on biochemical and/or genetic information | Universities of Newcastle; College Dublin; Maastricht; Oslo; Navarra; Harokopio; Reading; München; Newcastle Upon-Tyne and National Food and Nutrition Institute |
| NCT02663388 01/16–12/19 | Recruiting (n = 20,000) * | Observational | Gastric bypass, sleeve, gastric banding | Cardiovascular disease, NAFLD, obesity, MetS, T2D | Evaluate DNA Methylation on severe obesity-related complications before and after bariatric surgery | University of Lorraine, CHU Nancy Vandoeuvre Les Nancy, France |
| NCT03183193 06/16–12/19 | Recruiting (n = 120) * | Interventional Randomized | FLiO diet versus control diet | NAFLD and obesity | Nutritional/lifestyle interventions to identify non-invasive biomarkers to early diagnosis of NAFLD in future obese people | Centre for Nutrition Research, University of Navarra Pamplona, Navarra, Spain |
| NCT03135873 03/17–12/19 | Recruiting (n = 52) * | Interventional Randomized (Phase I) | Mastiha vs. placebo | Obese NAFLD | Explore gene-diet interactions and correlate genetic/epigenetic markers with metabolome and gut microbiota | Harokopio University, Athens, Attica, Greece |
| NCT02837367 06/19–12/19 | Recruiting (n = 600) * | Interventional Randomized | Administration of supplements containing methyl-donors | Obesity and dyslipidemia | Explore the genetic signature involved in the donation of methyl groups and unsaturated omega-6/3 fatty acid metabolism | Clinica II Pediatrie Bega Timisoara, Timis, Romania. Spitalul Judetean Timisoara; Centrul de Diabet Timisoara, Timis, Romania |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meroni, M.; Longo, M.; Rustichelli, A.; Dongiovanni, P. Nutrition and Genetics in NAFLD: The Perfect Binomium. Int. J. Mol. Sci. 2020, 21, 2986. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21082986
Meroni M, Longo M, Rustichelli A, Dongiovanni P. Nutrition and Genetics in NAFLD: The Perfect Binomium. International Journal of Molecular Sciences. 2020; 21(8):2986. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21082986
Chicago/Turabian StyleMeroni, Marica, Miriam Longo, Alice Rustichelli, and Paola Dongiovanni. 2020. "Nutrition and Genetics in NAFLD: The Perfect Binomium" International Journal of Molecular Sciences 21, no. 8: 2986. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21082986






