FUT8 Alpha-(1,6)-Fucosyltransferase in Cancer
Abstract
:1. Glycosylation in Cancer
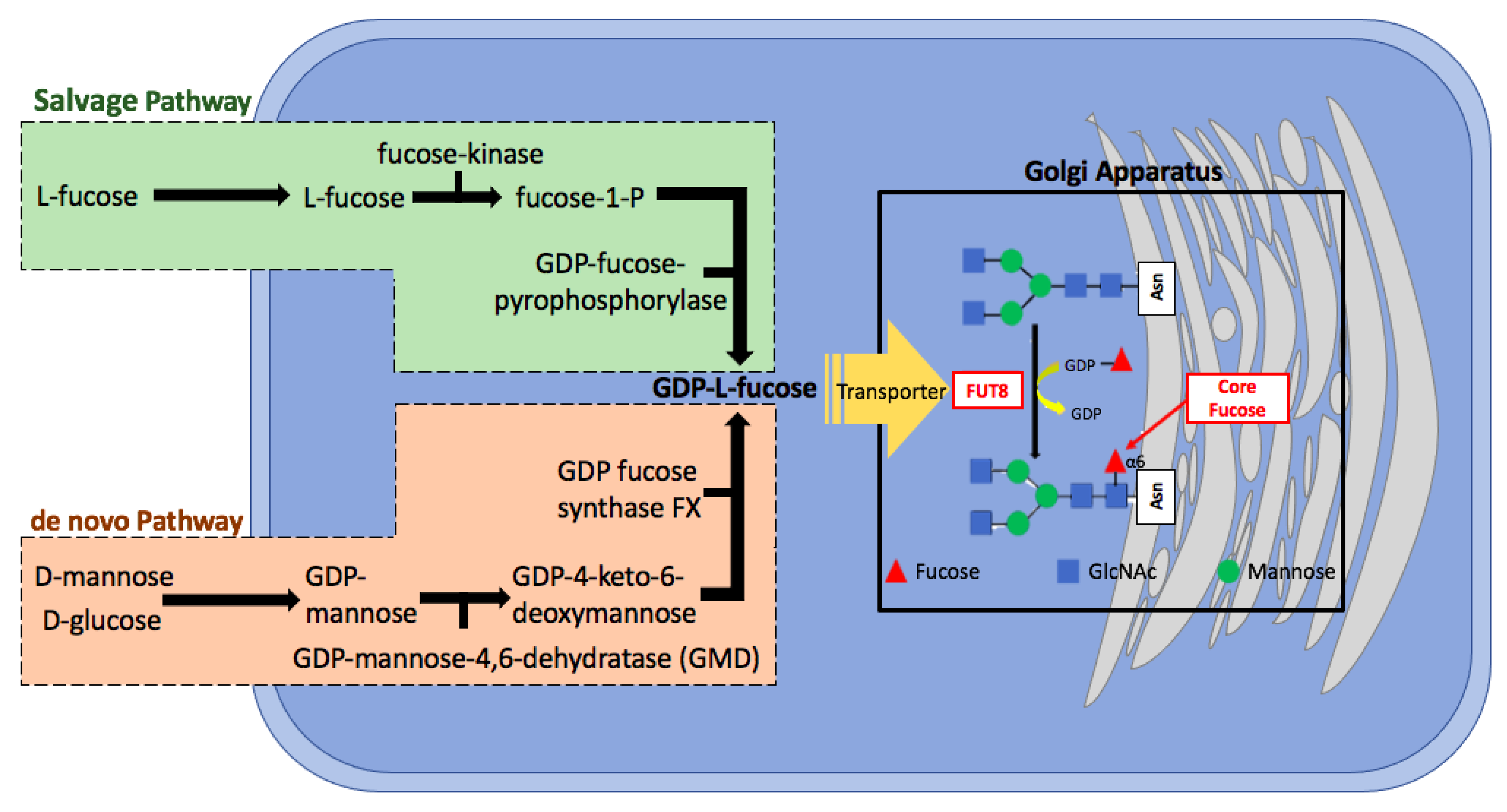
2. Fucosylation
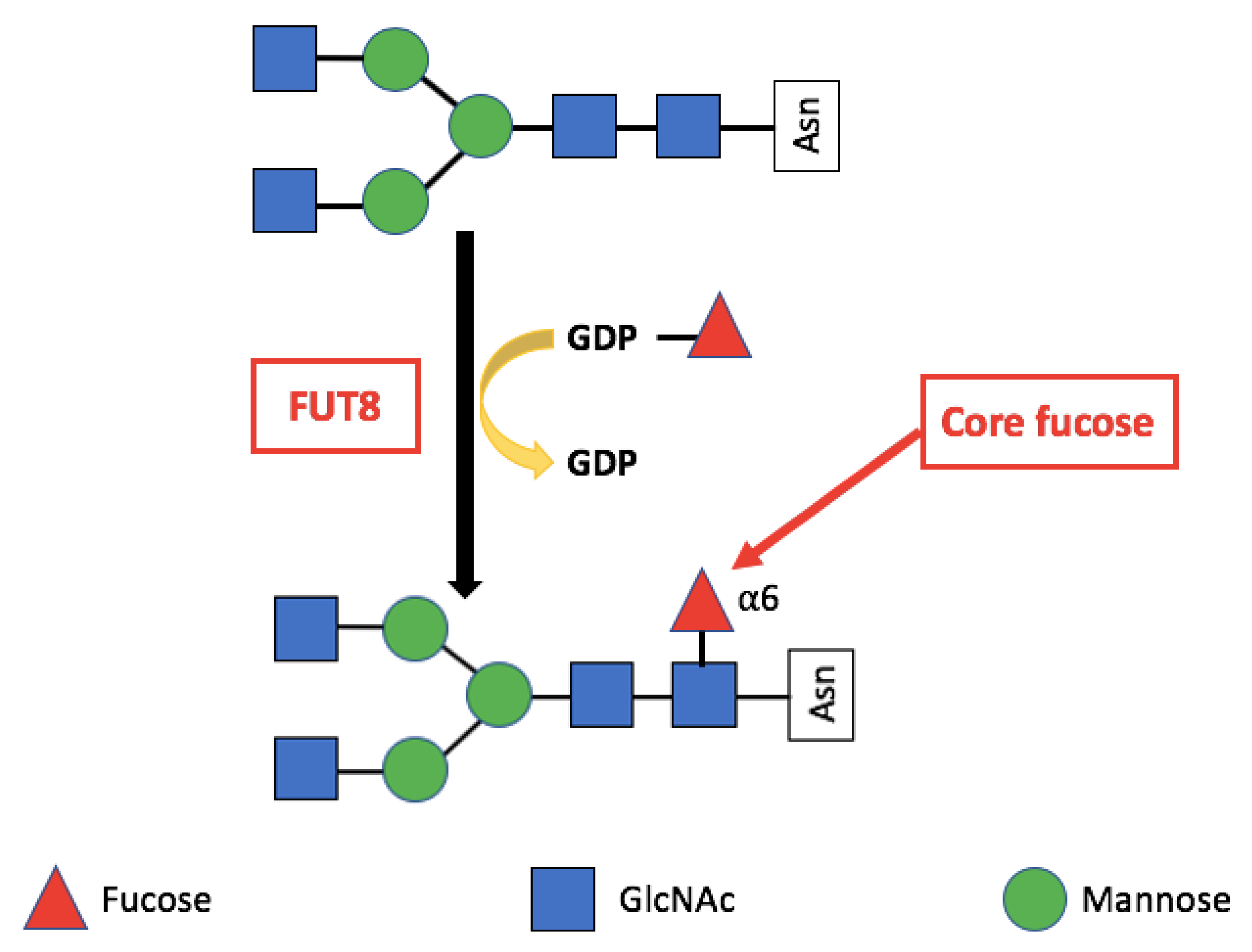
Core Fucosylation
3. Alpha-(1,6)-Fucosyltransferase (FUT8)
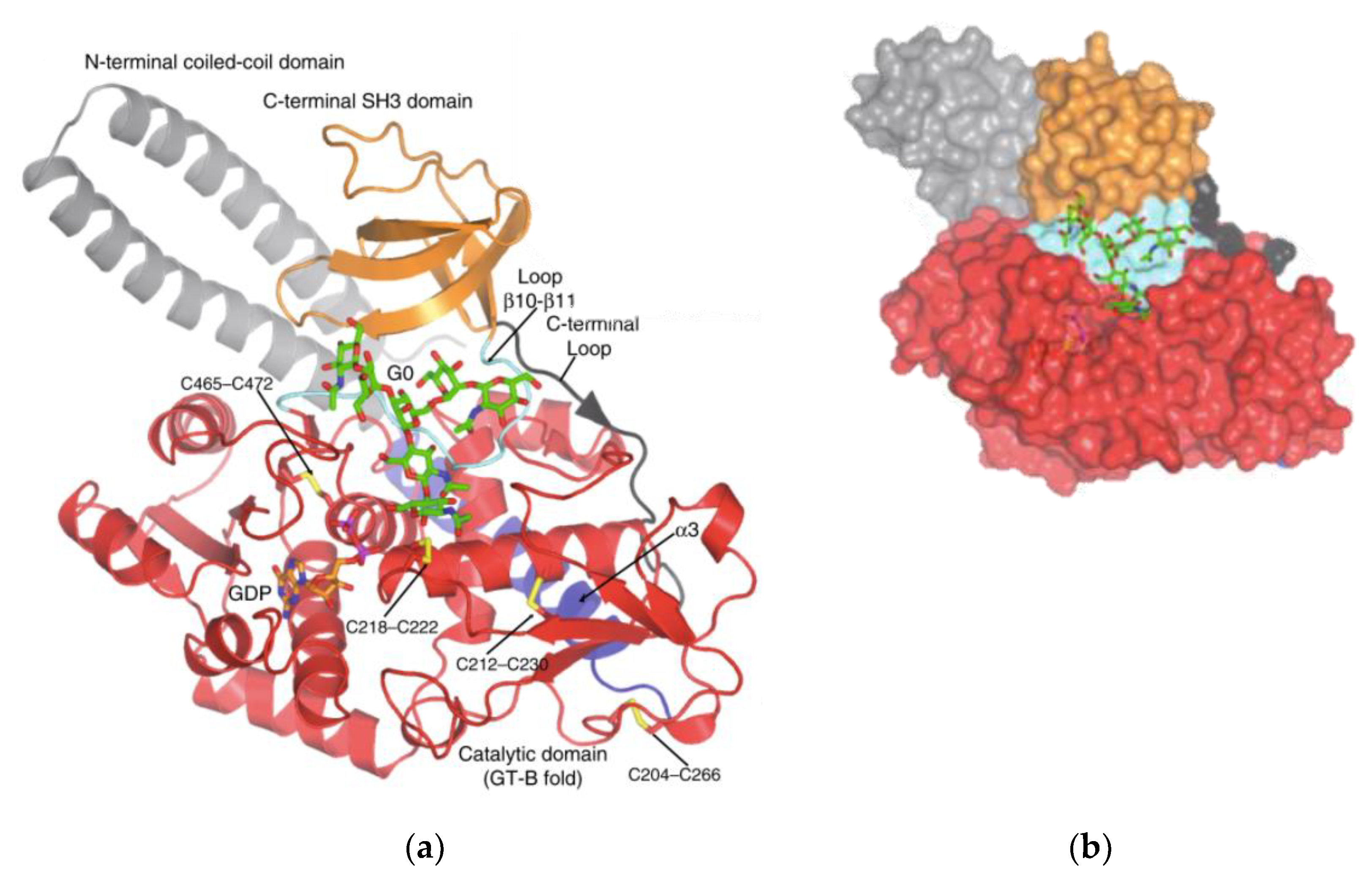
Structure
4. FUT8 Expression in Cancer
4.1. FUT8 Expression in Lung Cancer
4.2. FUT8 Expression in Liver Cancer
4.3. FUT8 Expression in Colorectal Cancer
4.4. FUT8 Expression in Ovarian Cancer
4.5. FUT8 Expression in Prostate Cancer
4.6. FUT8 Expression in Breast Cancer
4.7. FUT8 Expression in Thyroid Cancer
4.8. FUT8 Expression in Melanoma
4.9. FUT8 Expression in Pancreatic Cancer
5. Molecular Mechanisms of FUT8 in Cancer
5.1. Immune Evasion
5.2. Antibody-Dependent Cellular Cytotoxicity (ADCC)
5.3. Transforming Growth Factor Beta (TGF-β)
5.4. Epidermal Growth Factor (EGF)
5.5. α3β1. Integrin
5.6. E-Cadherin
6. Therapeutic Approaches
6.1. Afucosylated Antibodies Production Strategies
6.2. Therapeutic Afucosylated Antibody Drugs
6.3. Fucosylation Inhibitor-2-Fluorofucose
6.4. De novo and Salvage Pathway Inhibition of Cellular Fucosylation
7. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ER | Endoplasmic reticulum |
| GALNT | N-acetylgalactosaminyltransferase |
| GalNAc | N-acetylgalactosamine |
| FUT8 | α-(1,6)-fucosyltransferase |
| GLCNAc-TV or MGAT5 | N-acetylglucosaminyltransferase |
| GDP-β-l-fucose | Guanosine diphosphate |
| GMD | GDP-mannose 4,6-dehydrastase |
| FX protein | GDP-keto-6-deoxymannose 3,5-epimerase, 4-reductase |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| AFP | α-fetoprotein |
| HCC | Hepatocellular carcinoma |
| LCA | Lens culinaris agglutinin |
| PCa | Prostate Cancer |
| PSA | Prostate Specific Antigen |
| BPH | Benign Prostate Hyperplasia |
| HPLC | High-performance liquid chromatography |
| AAL | Aleuria aurantica lectin |
| AGP | α1-acid glycoprotein |
| CAIE | Crossed affinoimmunoelectrophoresis |
| fAGP | α1,3fucosylated AGP |
| RPN1 | Ribophorin I |
| FUTs | Fucosyltransferases |
| NSCLC | Nonsmall cell lung cancer |
| LEF-1 | Lymphoid enhancer binding factor 1 |
| CRC | Colorectal cancer |
| DFS | Disease free survival |
| EOC | Epithelial Ovarian Cancer |
| cDDP | Cisplatin |
| CTR1 | Copper transporter 1 |
| PrECs | Prostate epithelial cells |
| PrSCs | Prostate stromal cells |
| ADT | Androgen Deprivation Therapy |
| EV | Extracellular Vesicles |
| PDAC | Pancreatic ductal adenocarcinoma |
| ADCC | Antibody-dependent cellular cytotoxicity |
| PD-1 | Programmed cell death |
| TGF-β | Transforming growth factor- β1 |
| EGF | Epidermal growth factor |
| FcRs | Lymphocyte receptors |
| Fc | Constant region |
| IgG | Immunoglobulin G |
| CHO | Chinese hamster ovary |
| EMT | Epithelial-mesenchymal transition |
| E-box | Enhancer box |
| ZFNs | Zinc-finger nucleases |
| SGN-2FF | 2-fluorofucose |
| FDA | US Food and Drug Administration |
| NHL | Non-Hodgkin’s lymphoma |
| CTCL | T-cell lymphoma |
| HTLV1 | T-lymphotrophic virus 1 |
References
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Lowe, J.B. Biological roles of glycans. In Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Stanley, P.; Hart, G.W.; Aebi, M.; Darvill, A.G.; Kinoshita, T.; Packer, N.H.; Prestegard, J.H.; et al. Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Munkley, J.; Elliott, D.J. Hallmarks of glycosylation in cancer. Oncotarget 2016, 7, 35478–35489. [Google Scholar] [CrossRef] [Green Version]
- Scott, E.; Munkley, J. Glycans as Biomarkers in Prostate Cancer. Int. J. Mol. Sci. 2019, 20, 1389. [Google Scholar] [CrossRef] [Green Version]
- Hakomori, S. Glycosylation defining cancer malignancy: New wine in an old bottle. Proc. Natl. Acad. Sci. USA 2002, 99, 10231–10233. [Google Scholar] [CrossRef] [Green Version]
- Hakomori, S.-I. Aberrant Glycosylation In Tumors And Tumor-Associated Carbohydrate Antigens. Adv. Cancer Res. 1989, 257–331. [Google Scholar] [CrossRef]
- Meezan, E.; Wu, H.C.; Black, P.H.; Robbins, P.W. Comparative Studies on the Carbohydrate-containing membrane components of normal and virus-transformed mouse fibroblasts. II. Separation of glycoproteins and glycopeptides by Sephadex chromatography. Biochemistry 1969, 8, 2518–2524. [Google Scholar] [CrossRef] [PubMed]
- Reis, C.A.; Osorio, H.; Silva, L.; Gomes, C.; David, L. Alterations in glycosylation as biomarkers for cancer detection. J. Clin. Pathol. 2010, 63, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Woods, E.C.; Kai, F.; Barnes, J.M.; Pedram, K.; Pickup, M.W.; Hollander, M.J.; Weaver, V.M.; Bertozzi, C.R. A bulky glycocalyx fosters metastasis formation by promoting G1 cell cycle progression. eLife 2017, 6, e25752. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, E.; Schetters, S.T.T.; Van Kooyk, Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 2018, 18, 204–211. [Google Scholar] [CrossRef]
- Scott, E.; Elliott, D.J.; Munkley, J. Tumour associated glycans: A route to boost immunotherapy? Clin. Chim. Acta 2020, 502, 167–173. [Google Scholar] [CrossRef]
- Magalhães, A.; Duarte, H.O.; Reis, C.A. Aberrant Glycosylation in Cancer: A Novel Molecular Mechanism Controlling Metastasis. Cancer Cell 2017, 31, 733–735. [Google Scholar] [CrossRef] [Green Version]
- Powers, T.W.; Neely, B.A.; Shao, Y.; Tang, H.; Troyer, D.A.; Mehta, A.S.; Haab, B.B.; Drake, R.R. MALDI Imaging Mass Spectrometry Profiling of N-Glycans in Formalin-Fixed Paraffin Embedded Clinical Tissue Blocks and Tissue Microarrays. PLoS ONE 2014, 9, e106255. [Google Scholar] [CrossRef] [PubMed]
- Drake, R.R.; Powers, T.W.; Jones, E.E.; Bruner, E.; Mehta, A.S.; Angel, P.M. MALDI Mass Spectrometry Imaging of N-Linked Glycans in Cancer Tissues. In Advances in Cancer Research; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Becker, D.J.; Lowe, J.B. Fucose: Biosynthesis and biological function in mammals. Glycobiology 2003, 13, 41–53. [Google Scholar] [CrossRef]
- Stanley, P.; Taniguchi, N.; Aebi, M. N-glycans. In Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2017. [Google Scholar]
- Schneider, M.; Al-Shareffi, E.; Haltiwanger, R.S. Biological functions of fucose in mammals. Glycobiology 2017, 27, 601–618. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Wang, L.-X. Mammalian α-1,6-Fucosyltransferase (FUT8) Is the Sole Enzyme Responsible for theN-Acetylglucosaminyltransferase I-independent Core Fucosylation of High-mannoseN-Glycans. J. Biol. Chem. 2016, 291, 11064–11071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Inoue, S.; Gu, J.; Miyoshi, E.; Noda, K.; Li, W.; Mizuno-Horikawa, Y.; Nakano, M.; Asahi, M.; Takahashi, M.; et al. From The Cover: Dysregulation of TGF-β 1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc. Natl. Acad. Sci. USA 2005, 102, 15791–15796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, M.; Yang, D.; Dou, H.; Zhang, L. Fucosylation in cancer biology and its clinical applications. In Progress in Molecular Biology and Translational Science; Academic Press: Cambridge, MA, USA, 2019; pp. 93–119. [Google Scholar]
- Mehta, A.Y.; Cummings, R.D. GlycoGlyph: A glycan visualizing, drawing and naming application. Bioinformatics 2020, 36, 3613–3614. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Fucosyltransferase. 2020. Available online: https://www.proteinatlas.org/search/fucosyltransferase (accessed on 27 September 2020).
- Osumi, D.; Takahashi, M.; Miyoshi, E.; Yokoe, S.; Lee, S.H.; Noda, K.; Nakamori, S.; Gu, J.; Ikeda, Y.; Kuroki, Y.; et al. Core fucosylation of E-cadherin enhances cell-cell adhesion in human colon carcinoma WiDr cells. Cancer Sci. 2009, 100, 888–895. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Itoh, S.; Wang, X.; Isaji, T.; Miyoshi, E.; Kariya, Y.; Miyazaki, K.; Kawasaki, N.; Taniguchi, N.; Gu, J. Deletion of Core Fucosylation on α3β1 Integrin Down-regulates Its Functions. J. Biol. Chem. 2006, 281, 38343–38350. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Gu, J.; Ihara, H.; Miyoshi, E.; Honke, K.; Taniguchi, N. Core Fucosylation Regulates Epidermal Growth Factor Receptor-mediated Intracellular Signaling. J. Biol. Chem. 2006, 281, 2572–2577. [Google Scholar] [CrossRef] [Green Version]
- Tada, K.; Ohta, M.; Hidano, S.; Watanabe, K.; Hirashita, T.; Oshima, Y.; Fujnaga, A.; Nakanuma, H.; Masuda, T.; Endo, Y.; et al. Fucosyltransferase 8 plays a crucial role in the invasion and metastasis of pancreatic ductal adenocarcinoma. Surg. Today 2020, 50, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Shao, K.; Chen, Z.Y.; Gautam, S.; Deng, N.H.; Zhou, Y.; Wu, X.Z. Posttranslational modification of E-cadherin by core fucosylation regulates Src activation and induces epithelial–mesenchymal transition-like process in lung cancer cells. Glycobiology 2016, 26, 142–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, X.; Wang, N.; Pei, C.; Sun, L.; Sun, R.; Chen, J.; Liu, Y. Glycan-related gene expression signatures in human metastatic hepatocellular carcinoma cells. Exp. Ther. Med. 2012, 3, 415–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, C.-F.; Wu, M.-Y.; Lin, Y.-C.; Kannagi, R.; Yang, R.-B. FUT8 promotes breast cancer cell invasiveness by remodeling TGF-β receptor core fucosylation. Breast Cancer Res. 2017, 19, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Honma, R.; Kinoshita, I.; Miyoshi, E.; Tomaru, U.; Matsuno, Y.; Shimizu, Y.; Takeuchi, S.; Kobayashi, Y.; Kaga, K.; Taniguchi, N.; et al. Expression of Fucosyltransferase 8 Is Associated with an Unfavorable Clinical Outcome in Non-Small Cell Lung Cancers. Oncology 2015, 88, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, E.; Moriwaki, K.; Nakagawa, T. Biological Function of Fucosylation in Cancer Biology. J. Biochem. 2008, 143, 725–729. [Google Scholar] [CrossRef]
- A Norton, P.; Mehta, A.S. Expression of genes that control core fucosylation in hepatocellular carcinoma: Systematic review. World J. Gastroenterol. 2019, 25, 2947–2960. [Google Scholar] [CrossRef]
- Tonetti, M.; Sturla, L.; Bisso, A.; Benatti, U.; De Flora, A. Synthesis of GDP-l-fucose by the Human FX Protein. J. Biol. Chem. 1996, 271, 27274–27279. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, Y.; Fujii, J.; Inoue, S.; Uozumi, N.; Yanagidani, S.; Ikeda, Y.; Egashira, M.; Miyoshi, O.; Niikawa, N.; Taniguchi, N. Mapping of the α-1,6-fucosyltransferase gene, FUT8, to human chromosome 14q24.3. Cytogenet. Genome Res. 1999, 84, 58–60. [Google Scholar] [CrossRef]
- The Human Protein Atlas. FUT8. 2020. Available online: https://www.proteinatlas.org/ENSG00000033170-FUT8 (accessed on 28 April 2020).
- Taniguchi, N. Knockout Mice of α1,6 Fucosyltransferase (Fut 8). In Experimental Glycoscience; Springer: Berlin/Heidelberg, Germany, 2009; pp. 379–380. [Google Scholar]
- Malý, P.; Thall, A.D.; Petryniak, B.; E Rogers, C.; Smith, P.L.; Marks, R.M.; Kelly, R.J.; Gersten, K.M.; Cheng, G.; Saunders, T.L.; et al. The α(1,3)Fucosyltransferase Fuc-TVII Controls Leukocyte Trafficking through an Essential Role in L-, E-, and P-selectin Ligand Biosynthesis. Cell 1996, 86, 643–653. [Google Scholar] [CrossRef] [Green Version]
- Kudo, T.; Fujii, T.; Ikegami, S.; Inokuchi, K.; Takayama, Y.; Ikehara, Y.; Nishihara, S.; Togayachi, A.; Takahashi, S.; Tachibana, K.; et al. Mice lacking α1,3-fucosyltransferase IX demonstrate disappearance of Lewis x structure in brain and increased anxiety-like behaviors. Glycobiology 2007, 17, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ihara, H.; Ikeda, Y.; Toma, S.; Wang, X.; Suzuki, T.; Gu, J.; Miyoshi, E.; Tsukihara, T.; Honke, K.; Matsumoto, A.; et al. Crystal structure of mammalian α1,6-fucosyltransferase, FUT8. Glycobiology 2007, 17, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An Evolving Hierarchical Family Classification for Glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Qasba, P.K.; Ramakrishnan, B.; Boeggeman, E. Substrate-induced conformational changes in glycosyltransferases. Trends Biochem. Sci. 2005, 30, 53–62. [Google Scholar] [CrossRef] [PubMed]
- García-García, A.; Ceballos-Laita, L.; Serna, S.; Artschwager, R.; Reichardt, N.C.; Corzana, F.; Hurtado-Guerrero, R. Structural basis for substrate specificity and catalysis of α1,6-fucosyltransferase. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Tomida, S.; Takata, M.; Hirata, T.; Nagae, M.; Nakano, M.; Kizuka, Y. The SH3 domain in the fucosyltransferase FUT8 controls FUT8 activity and localization and is essential for core fucosylation. J. Biol. Chem. 2020, 295, 7992–8004. [Google Scholar] [CrossRef]
- Miyoshi, E.; Kawamoto, S.; Moriwaki, K.; Nakagawa, T.; Terao, M.; Shinzaki, S.; Yamane-Ohnuki, N.; Satoh, M.; Mehta, A.S.; Block, T.M. Overexpression of α1,6-fucosyltransferase in hepatoma enhances expression of Golgi phosphoprotein 2 in a fucosylation-independent manner. Int. J. Oncol. 2011, 39, 203–208. [Google Scholar] [CrossRef] [Green Version]
- Dube, D.H.; Bertozzi, C.R. Glycans in cancer and inflammation—Potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005, 4, 477–488. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Pinho, S.S.; Oliveira, P.; Cabral, J.; Carvalho, S.; Huntsman, D.; Gärtner, F.; Seruca, R.; Reis, C.A.; Oliveira, C. Loss and Recovery of Mgat3 and GnT-III Mediated E-cadherin N-glycosylation Is a Mechanism Involved in Epithelial-Mesenchymal-Epithelial Transitions. PLoS ONE 2012, 7, e33191. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-Y.; Jan, Y.-H.; Juan, Y.-H.; Yang, C.-J.; Huang, M.-S.; Yu, C.-J.; Yang, P.-C.; Hsiao, M.; Hsu, T.-L.; Wong, C.-H. Fucosyltransferase 8 as a functional regulator of nonsmall cell lung cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 630–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noda, K.; Miyoshi, E.; Uozumi, N.; Yanagidani, S.; Ikeda, Y.; Gao, C.-X.; Suzuki, K.; Yoshihara, H.; Yoshikawa, M.; Kawano, K.; et al. Gene expression of α1-6 fucosyltransferase in human hepatoma tissues: A possible implication for increased fucosylation of α-fetoprotein. Hepatology 1998, 28, 944–952. [Google Scholar] [CrossRef] [PubMed]
- Noda, M.; Okayama, H.; Kofunato, Y.; Chida, S.; Saito, K.; Tada, T.; Ashizawa, M.; Nakajima, T.; Aoto, K.; Kikuchi, T.; et al. Prognostic role of FUT8 expression in relation to p53 status in stage II and III colorectal cancer. PLoS ONE 2018, 13, e0200315. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Gao, S.; Song, X.; Dong, W.; Zhou, H.; Zhao, L.; Jia, L. Comprehensive N-glycan profiles of hepatocellular carcinoma reveal association of fucosylation with tumor progression and regulation of FUT8 by microRNAs. Oncotarget 2016, 7, 61199–61214. [Google Scholar] [CrossRef] [Green Version]
- Lv, X.; Song, J.; Xue, K.; Li, Z.; Li, M.; Zahid, D.; Cao, H.; Wang, L.; Song, W.; Ma, T.; et al. Core fucosylation of copper transporter 1 plays a crucial role in cisplatin-resistance of epithelial ovarian cancer by regulating drug uptake. Mol. Carcinog. 2019, 58, 794–807. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, J.; Li, Q.K.; Peskoe, S.B.; Zhang, B.; Choi, C.; A Platz, E.; Zhang, H. Overexpression of α (1,6) fucosyltransferase associated with aggressive prostate cancer. Glycobiology 2014, 24, 935–944. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, P.; Fontanals-Cirera, B.; Sokolova, E.; Jacob, S.; Vaiana, C.A.; Argibay, D.; Davalos, V.; McDermott, M.; Nayak, S.; Darvishian, F.; et al. A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis. Cancer Cell 2017, 31, 804–819.e7. [Google Scholar] [CrossRef] [Green Version]
- Ito, Y.; Miyauchi, A.; Yoshida, H.; Uruno, T.; Nakano, K.; Takamura, Y.; Miya, A.; Kobayashi, K.; Yokozawa, T.; Matsuzuka, F.; et al. Expression of α1,6-fucosyltransferase (FUT8) in papillary carcinoma of the thyroid: Its linkage to biological aggressiveness and anaplastic transformation. Cancer Lett. 2003, 200, 167–172. [Google Scholar] [CrossRef]
- Hoti, N.; Yang, S.; Hu, Y.; Shah, P.; Haffner, M.C.; Zhang, H. Overexpression of α (1,6) fucosyltransferase in the development of castration-resistant prostate cancer cells. Prostate Cancer Prostatic Dis. 2018, 21, 137–146. [Google Scholar] [CrossRef] [Green Version]
- Peracaula, R. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology 2003, 13, 457–470. [Google Scholar] [CrossRef] [Green Version]
- Yokobori, T.; Yazawa, S.; Asao, T.; Nakazawa, N.; Mogi, A.; Sano, R.; Kuwano, H.; Kaira, K.; Shirabe, K. Fucosylated α1-acid glycoprotein as a biomarker to predict prognosis following tumor immunotherapy of patients with lung cancer. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, S.; Asao, T.; Takahashi, J.; Yagihashi, Y.; Nishimura, T.; Saniabadi, A.R.; Poland, D.C.W.; Van Dijk, W.; Kuwano, H.; Kochibe, N.; et al. α1-Acid glycoprotein fucosylation as a marker of carcinoma progression and prognosis. Cancer 2004, 101, 2825–2836. [Google Scholar] [CrossRef] [PubMed]
- Asao, T.; Yazawa, S.; Nishimura, T.; Hayashi, T.; Shimaoka, H.; Saniabadi, A.R.; Kuwano, H. Development of a Novel System for Mass Spectrometric Analysis of Cancer-Associated Fucosylation in Plasmaα1-Acid Glycoprotein. BioMed Res. Int. 2013, 2013, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Yazawa, S.; Takahashi, R.; Yokobori, T.; Sano, R.; Mogi, A.; Saniabadi, A.R.; Kuwano, H.; Asao, T. Fucosylated Glycans in α1-Acid Glycoprotein for Monitoring Treatment Outcomes and Prognosis of Cancer Patients. PLoS ONE 2016, 11, e0156277. [Google Scholar] [CrossRef] [Green Version]
- The American Cancer Society. Lung Cancer. In The Cancer Atlas, 2nd ed.; The American Cancer Society: Atlanta, GA, USA, 2019. [Google Scholar]
- Zugazagoitia, J.; Enguita, A.B.; Nuñez, J.A.; Iglesias, L.; Ponce, S. The new IASLC/ATS/ERS lung adenocarcinoma classification from a clinical perspective: Current concepts and future prospects. J. Thorac. Dis. 2014, 6, S526–S536. [Google Scholar]
- Lin, H.; Wang, D.; Wu, T.; Dong, C.; Shen, N.; Sun, Y.; Sun, Y.; Xie, H.; Wang, N.; Shan, L. Blocking core fucosylation of TGF-β1 receptors downregulates their functions and attenuates the epithelial-mesenchymal transition of renal tubular cells. Am. J. Physiol. Physiol. 2011, 300, F1017–F1025. [Google Scholar] [CrossRef] [PubMed]
- Block, T.M.; Mehta, A.S.; Fimmel, C.J.; Jordan, R. Molecular viral oncology of hepatocellular carcinoma. Oncogene 2003, 22, 5093–5107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attwa, M.H. Guide for diagnosis and treatment of hepatocellular carcinoma. World J. Hepatol. 2015, 7, 1632–1651. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Liu, X.; Zhang, Y.; Li, T.; Zhan, C.; Huo, W.; He, A.; Yao, Y.; Jin, Y.; Qu, Y.; et al. Specific N-glycans of Hepatocellular Carcinoma Cell Surface and the Abnormal Increase of Core-α-1, 6-fucosylated Triantennary Glycan via N-acetylglucosaminyltransferases-IVa Regulation. Sci. Rep. 2015, 5, 16007. [Google Scholar] [CrossRef] [PubMed]
- Egashira, Y.; Suganuma, M.; Kataoka, Y.; Higa, Y.; Ide, N.; Morishita, K.; Kamada, Y.; Gu, J.; Fukagawa, K.; Miyoshi, E. Establishment and characterization of a fucosylated α-fetoprotein-specific monoclonal antibody: A potential application for clinical research. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Wang, Y.; Fukuda, T.; Isaji, T.; Lu, J.; Im, S.; Hang, Q.; Gu, W.; Hou, S.; Ohtsubo, K.; Gu, J. Loss of αl,6-fucosyltransferase inhibits chemical-induced hepatocellular carcinoma and tumorigenesis by down-regulating several cell signaling pathways. FASEB J. 2015, 29, 3217–3227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The American Cancer Society. Key Statistics for Colorectal Cancer; The American Cancer Society: Atlanta, GA, USA, 2020. [Google Scholar]
- Okagawa, Y.; Takada, K.; Arihara, Y.; Kikuchi, S.; Osuga, T.; Nakamura, H.; Kamihara, Y.; Hayasaka, N.; Usami, M.; Murase, K.; et al. Activated p53 with Histone Deacetylase Inhibitor Enhances l-Fucose-Mediated Drug Delivery through Induction of Fucosyltransferase 8 Expression in Hepatocellular Carcinoma Cells. PLoS ONE 2016, 11, e0168355. [Google Scholar] [CrossRef] [PubMed]
- Muinelo-Romay, L.; Villar-Portela, S.; Alvarez, E.C.; Gil-Martín, E.; Fern�Ndez-Briera, A. α(1,6)Fucosyltransferase expression is an independent prognostic factor for disease-free survival in colorectal carcinoma. Hum. Pathol. 2011, 42, 1740–1750. [Google Scholar] [CrossRef] [PubMed]
- Schwedler, C.; Kaup, M.; Weiz, S.; Hoppe, M.; Braicu, E.I.; Sehouli, J.; Hoppe, B.; Tauber, R.; Berger, M.; Blanchard, V. Identification of 34 N-glycan isomers in human serum by capillary electrophoresis coupled with laser-induced fluorescence allows improving glycan biomarker discovery. Anal. Bioanal. Chem. 2014, 406, 7185–7193. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Qin, W.; Qin, R.; Han, J.; Li, C.; Wang, Y.; Xu, C.-J. Lectin array and glycogene expression analyses of ovarian cancer cell line A2780 and its cisplatin-resistant derivate cell line A2780-cp. Clin. Proteom. 2017, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- The Global Cancer Observatory. Age standardized (World) incidence rates, prostate, all ages. Int. Agency Res. Cancer 2018, 876, 2018–2019. [Google Scholar]
- Bolla, M.; van Poppel, H. Management of Prostate Cancer, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2017; Volume 73. [Google Scholar]
- Fujita, K.; Hayashi, T.; Matsuzaki, K.; Nakata, W.; Masuda, M.; Kawashima, A.; Ujike, T.; Nagahara, A.; Tsuchiya, M.; Kobayashi, Y.; et al. Decreased fucosylated PSA as a urinary marker for high Gleason score prostate cancer. Oncotarget 2016, 7, 56643–56649. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.-L.; Yang, W.; Hu, Y.; Höti, N.; Liu, Y.; Shah, P.; Sun, S.; Clark, D.; Thomas, S.N.; Zhang, H. Site-Specific Fucosylation Analysis Identifying Glycoproteins Associated with Aggressive Prostate Cancer Cell Lines Using Tandem Affinity Enrichments of Intact Glycopeptides Followed by Mass Spectrometry. Anal. Chem. 2017, 89, 7623–7630. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.; Shimomura, M.; Uemura, M.; Nakata, W.; Sato, M.; Nagahara, A.; Nakai, Y.; Takamatsu, S.; Miyoshi, E.; Nonomura, N. Serum fucosylated haptoglobin as a novel prognostic biomarker predicting high-Gleason prostate cancer. Prostate 2014, 74, 1052–1058. [Google Scholar] [CrossRef]
- Shah, P.; Wang, X.; Yang, W.; Eshghi, S.T.; Sun, S.; Hoti, N.; Chen, L.; Yang, S.; Pasay, J.; Rubin, A.; et al. Integrated Proteomic and Glycoproteomic Analyses of Prostate Cancer Cells Reveal Glycoprotein Alteration in Protein Abundance and Glycosylation. Mol. Cell. Proteom. 2015, 14, 2753–2763. [Google Scholar] [CrossRef] [Green Version]
- Clark, D.J.; Schnaubelt, M.; Hoti, N.; Hu, Y.; Zhou, Y.; Gooya, M.; Zhang, H. Impact of Increased FUT8 Expression on the Extracellular Vesicle Proteome in Prostate Cancer Cells. J. Proteome Res. 2020, 19, 2195–2205. [Google Scholar] [CrossRef] [PubMed]
- World Cancer Research Fund. Diet, Nutrition, Physical Activity and Breast Cancer. 2018. Available online: https://www.wcrf.org/sites/default/files/Breast-cancer-report.pdf (accessed on 18 July 2020).
- Yue, L.; Han, C.; Li, Z.; Li, X.; Liu, D.; Liu, S.; Yu, H. Fucosyltransferase 8 expression in breast cancer patients: A high throughput tissue microarray analysis. Histol. Histopathol. 2015, 31, 547–855. [Google Scholar] [PubMed]
- Chanrion, M.; Negre, V.; Fontaine, H.; Salvetat, N.; Bibeau, F.; Mac Grogan, G.; Mauriac, L.; Katsaros, D.; Molina, F.; Theillet, C.; et al. A Gene Expression Signature that Can Predict the Recurrence of Tamoxifen-Treated Primary Breast Cancer. Clin. Cancer Res. 2008, 14, 1744–1752. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Yau, C.; Gray, J.W.; Chew, K.; Dairkee, S.H.; Moore, D.H.; Eppenberger, U.; Eppenberger-Castori, S.; Benz, C.C. Enhanced NFκB and AP-1 transcriptional activity associated with antiestrogen resistant breast cancer. BMC Cancer 2007, 7, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Sieuwerts, A.M.; McGreevy, M.; Casey, G.; Cufer, T.; Paradiso, A.; Harbeck, N.; Span, P.N.; Hicks, D.G.; Crowe, J.; et al. The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res. Treat. 2008, 116, 303–309. [Google Scholar] [CrossRef]
- Aldinger, K.A.; Samaan, N.A.; Ibanez, M.; Hill, C.S. Anaplastic carcinoma of the thyroid.A review of 84 cases of spindle and giant cell carcinoma of the thyroid. Cancer 1978, 41, 2267–2275. [Google Scholar] [CrossRef]
- Miyoshi, E.; Ito, Y.; Miyoshi, Y. Involvement of Aberrant Glycosylation in Thyroid Cancer. J. Oncol. 2010, 2010, 816595. [Google Scholar] [CrossRef] [Green Version]
- I Riker, A.; Enkemann, S.A.; Fodstad, O.; Liu, S.; Ren, S.; Morris, C.; Xi, Y.; Howell, P.; Metge, B.; Samant, R.; et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genom. 2008, 1, 13. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Shen, S.S.; Hoshida, Y.; Subramanian, A.; Ross, K.; Brunet, J.-P.; Wagner, S.N.; Ramaswamy, S.; Mesirov, J.P.; Hynes, R.O. Gene Expression Changes in an Animal Melanoma Model Correlate with Aggressiveness of Human Melanoma Metastases. Mol. Cancer Res. 2008, 6, 760–769. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, K.; Ohta, M.; Yada, K.; Komori, Y.; Iwashita, Y.; Kashima, K.; Inomata, M. Fucosylation is associated with the malignant transformation of intraductal papillary mucinous neoplasms: A lectin microarray-based study. Surg. Today 2016, 46, 1217–1223. [Google Scholar] [CrossRef]
- Okuyama, N.; Ide, Y.; Nakano, M.; Nakagawa, T.; Yamanaka, K.; Moriwaki, K.; Murata, K.; Ohigashi, H.; Yokoyama, S.; Eguchi, H.; et al. Fucosylated haptoglobin is a novel marker for pancreatic cancer: A detailed analysis of the oligosaccharide structure and a possible mechanism for fucosylation. Int. J. Cancer 2006, 118, 2803–2808. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, E.; Nakano, M. Fucosylated haptoglobin is a novel marker for pancreatic cancer: Detailed analyses of oligosaccharide structures. Proteomics 2008, 8, 3257–3262. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Chikuma, S.; Kondo, T.; Hibino, S.; Machiyama, H.; Yokosuka, T.; Nakano, M.; Yoshimura, A. Blockage of Core Fucosylation Reduces Cell-Surface Expression of PD-1 and Promotes Anti-tumor Immune Responses of T Cells. Cell Rep. 2017, 20, 1017–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, K.; Yokote, H.; Arao, T.; Maegawa, M.; Tanaka, K.; Fujita, Y.; Shimizu, C.; Hanafusa, T.; Fujiwara, Y.; Nishio, K. N-Glycan fucosylation of epidermal growth factor receptor modulates receptor activity and sensitivity to epidermal growth factor receptor tyrosine kinase inhibitor. Cancer Sci. 2008, 99, 1611–1617. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.; Shi, B.Z.; Yuan, Y.F.; Wu, X.Z. The expression of core fucosylated E-cadherin in cancer cells and lung cancer patients: Prognostic implications. Cell Res. 2004, 14, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.; Shi, B.; Geng, F.; Zhang, C.; Wu, W.; Wu, X.Z. E-cadherin core fucosylation regulates nuclear β-catenin accumulation in lung cancer cells. Glycoconj. J. 2008, 25, 843–850. [Google Scholar] [CrossRef]
- Shinkawa, T.; Nakamura, K.; Yamane, N.; Shoji-Hosaka, E.; Kanda, Y.; Sakurada, M.; Uchida, K.; Anazawa, H.; Satoh, M.; Yamasaki, M.; et al. The Absence of Fucose but Not the Presence of Galactose or BisectingN-Acetylglucosamine of Human IgG1 Complex-type Oligosaccharides Shows the Critical Role of Enhancing Antibody-dependent Cellular Cytotoxicity. J. Biol. Chem. 2002, 278, 3466–3473. [Google Scholar] [CrossRef] [Green Version]
- Vidarsson, G.; Dekkers, G.; Rispens, T. IgG Subclasses and Allotypes: From Structure to Effector Functions. Front. Immunol. 2014, 5, 520. [Google Scholar] [CrossRef] [Green Version]
- Clynes, R.A.; Towers, T.L.; Presta, L.G.; Ravetch, J.V. Inhibitory Fc receptors modulate in vivo cytoxicity against tumor targets. Nat. Med. 2000, 6, 443–446. [Google Scholar] [CrossRef]
- Imai-Nishiya, H.; Mori, K.; Inoue, M.; Wakitani, M.; Iida, S.; Shitara, K.; Satoh, M. Double knockdown of alpha1,6-fucosyltransferase (FUT8) and GDP-mannose 4,6-dehydratase (GMD) in antibody-producing cells: A new strategy for generating fully non-fucosylated therapeutic antibodies with enhanced ADCC. BMC Biotechnol. 2007, 7, 84. [Google Scholar] [CrossRef] [Green Version]
- Roy, G.; Martin, T.; Barnes, A.; Wang, J.; Jimenez, R.B.; Rice, M.; Li, L.; Feng, H.; Zhang, S.; Chaerkady, R.; et al. A novel bicistronic gene design couples stable cell line selection with a fucose switch in a designer CHO host to produce native and afucosylated glycoform antibodies. mAbs 2018, 10, 416–430. [Google Scholar] [CrossRef] [Green Version]
- Niwa, R. Enhancement of the Antibody-Dependent Cellular Cytotoxicity of Low-Fucose IgG1 Is Independent of Fc RIIIa Functional Polymorphism. Clin. Cancer Res. 2004, 10, 6248–6255. [Google Scholar] [CrossRef] [Green Version]
- Niwa, R.; Natsume, A.; Uehara, A.; Wakitani, M.; Iida, S.; Uchida, K.; Satoh, M.; Shitara, K. IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. J. Immunol. Methods 2005, 306, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Niwa, R.; Sakurada, M.; Kobayashi, Y.; Uehara, A.; Matsushima, K.; Ueda, R.; Nakamura, K.; Shitara, K. Enhanced Natural Killer Cell Binding and Activation by Low-Fucose IgG1 Antibody Results in Potent Antibody-Dependent Cellular Cytotoxicity Induction at Lower Antigen Density. Clin. Cancer Res. 2005, 11, 2327–2336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niwa, R.; Shoji-Hosaka, E.; Sakurada, M.; Shinkawa, T.; Uchida, K.; Nakamura, K.; Matsushima, K.; Ueda, R.; Hanai, N.; Shitara, K. Defucosylated Chimeric Anti-CC Chemokine Receptor 4 IgG1 with Enhanced Antibody-Dependent Cellular Cytotoxicity Shows Potent Therapeutic Activity to T-Cell Leukemia and Lymphoma. Cancer Res. 2004, 64, 2127–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahmani, A.; Delisle, J.-S. TGF-β in T Cell Biology: Implications for Cancer Immunotherapy. Cancers 2018, 10, 194. [Google Scholar] [CrossRef] [Green Version]
- Thiery, J.P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Puisieux, A.; Brabletz, T.; Caramel, J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 2014, 16, 488–494. [Google Scholar] [CrossRef]
- Nieto, M.A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002, 3, 155–166. [Google Scholar] [CrossRef]
- Yamane-Ohnuki, N.; Kinoshita, S.; Inoue-Urakubo, M.; Kusunoki, M.; Iida, S.; Nakano, R.; Wakitani, M.; Niwa, R.; Sakurada, M.; Uchida, K.; et al. Establishment ofFUT8 knockout Chinese hamster ovary cells: An ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol. Bioeng. 2004, 87, 614–622. [Google Scholar] [CrossRef]
- Pereira, N.A.; Chan, K.F.; Lin, P.C.; Song, Z. The “less-is-more” in therapeutic antibodies: Afucosylated anti-cancer antibodies with enhanced antibody-dependent cellular cytotoxicity. mAbs 2018, 10, 693–711. [Google Scholar] [CrossRef] [PubMed]
- Malphettes, L.; Freyvert, Y.; Chang, J.; Liu, P.-Q.; Chan, E.; Miller, J.C.; Zhou, Z.; Nguyen, T.; Tsai, C.; Snowden, A.W.; et al. Highly efficient deletion of FUT8 in CHO cell lines using zinc-finger nucleases yields cells that produce completely nonfucosylated antibodies. Biotechnol. Bioeng. 2010, 106, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Loos, A.; Steinkellner, H. IgG-Fc glycoengineering in non-mammalian expression hosts. Arch. Biochem. Biophys. 2012, 526, 167–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okeley, N.M.; Alley, S.C.; Anderson, M.E.; Boursalian, T.E.; Burke, P.J.; Emmerton, K.M.; Jeffrey, S.C.; Klussman, K.; Law, C.-L.; Sussman, D.; et al. Development of orally active inhibitors of protein and cellular fucosylation. Proc. Natl. Acad. Sci. USA 2013, 110, 5404–5409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasdaska, J.R.; Sherwood, S.; Regan, J.T.; Dickey, L.F. An afucosylated anti-CD20 monoclonal antibody with greater antibody-dependent cellular cytotoxicity and B-cell depletion and lower complement-dependent cytotoxicity than rituximab. Mol. Immunol. 2012, 50, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; DiLillo, D.J.; Bournazos, S.; Giddens, J.P.; Ravetch, J.V.; Wang, L.-X. Modulating IgG effector function by Fc glycan engineering. Proc. Natl. Acad. Sci. USA 2017, 114, 3485–3490. [Google Scholar] [CrossRef] [Green Version]
- Mössner, E.; Brünker, P.; Moser, S.; Püntener, U.; Schmidt, C.; Herter, S.; Grau, R.; Gerdes, C.; Nopora, A.; Van Puijenbroek, E.; et al. Increasing the efficacy of CD20 antibody therapy through the engineering of a new type II anti-CD20 antibody with enhanced direct and immune effector cell–mediated B-cell cytotoxicity. Blood 2010, 115, 4393–4402. [Google Scholar] [CrossRef]
- Cartron, G.; De Guibert, S.; Dilhuydy, M.-S.; Morschhauser, F.; Leblond, V.; Dupuis, J.; Mahe, B.; Bouabdallah, R.; Lei, G.; Wenger, M.; et al. Obinutuzumab (GA101) in relapsed/refractory chronic lymphocytic leukemia: Final data from the phase 1/2 GAUGUIN study. Blood 2014, 124, 2196–2202. [Google Scholar] [CrossRef] [Green Version]
- Goede, V.; Fischer, K.; Engelke, A.; Schlag, R.; Lepretre, S.; Montero, L.F.C.; Montillo, M.; Fegan, C.; Asikanius, E.; Humphrey, K.; et al. Obinutuzumab as frontline treatment of chronic lymphocytic leukemia: Updated results of the CLL11 study. Leukemia 2015, 29, 1602–1604. [Google Scholar] [CrossRef]
- Goede, V.; Fischer, K.; Busch, R.; Engelke, A.; Eichhorst, B.; Wendtner, C.M.; Chagorova, T.; De La Serna, J.; Dilhuydy, M.-S.; Illmer, T.; et al. Obinutuzumab plus Chlorambucil in Patients with CLL and Coexisting Conditions. N. Engl. J. Med. 2014, 370, 1101–1110. [Google Scholar] [CrossRef] [Green Version]
- Cardarelli, P.M.; Rao-Naik, C.; Chen, S.; Huang, H.; Pham, A.; Moldovan-Loomis, M.-C.; Pan, C.; Preston, B.; Passmore, D.; Liu, J.; et al. A nonfucosylated human antibody to CD19 with potent B-cell depletive activity for therapy of B-cell malignancies. Cancer Immunol. Immunother. 2010, 59, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Ward, E.; Mittereder, N.; Kuta, E.; Sims, G.P.; Bowen, M.A.; Dall’Acqua, W.; Tedder, T.; Kiener, P.; Coyle, A.J.; Wu, H.; et al. A glycoengineered anti-CD19 antibody with potent antibody-dependent cellular cytotoxicity activity in vitro and lymphoma growth inhibition in vivo. Br. J. Haematol. 2011, 155, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.; Wang, Y.; Gallagher, S.; Mittereder, N.; Kuta, E.; Damschroder, M.M.; Woods, R.; Rowe, D.C.; Cheng, L.; Cook, K.; et al. B-Cell Depletion In Vitro and In Vivo with an Afucosylated Anti-CD19 Antibody. J. Pharmacol. Exp. Ther. 2010, 335, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Matlawska-Wasowska, K.; Ward, E.S.; Stevens, S.; Wang, Y.; Herbst, R.; Winter, S.S.; Wilson, B.S. Macrophage and NK-mediated killing of precursor-B acute lymphoblastic leukemia cells targeted with a-fucosylated anti-CD19 humanized antibodies. Leukemia 2013, 27, 1263–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breton, C.; Nahimana, A.; Aubry, D.; Macoin, J.; Moretti, P.; Bertschinger, M.; Hou, S.; Duchosal, M.A.; Back, J. A novel anti-CD19 monoclonal antibody (GBR 401) with high killing activity against B cell malignancies. J. Hematol. Oncol. 2014, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Gerdes, C.A.; Nicolini, V.G.; Herter, S.; Van Puijenbroek, E.; Lang, S.; Roemmele, M.; Moessner, E.; Freytag, O.; Friess, T.; Ries, C.H.; et al. GA201 (RG7160): A Novel, Humanized, Glycoengineered Anti-EGFR Antibody with Enhanced ADCC and Superior In Vivo Efficacy Compared with Cetuximab. Clin. Cancer Res. 2013, 19, 1126–1138. [Google Scholar] [CrossRef] [Green Version]
- Grugan, K.D.; Dorn, K.; Jarantow, S.W.; Bushey, B.S.; Pardinas, J.R.; Laquerre, S.; Moores, S.L.; Chiu, M.L. Fc-mediated activity of EGFR x c-Met bispecific antibody JNJ-61186372 enhanced killing of lung cancer cells. mAbs 2017, 9, 114–126. [Google Scholar] [CrossRef] [Green Version]
- Schanzer, J.M.; Wartha, K.; Croasdale, R.; Moser, S.; Künkele, K.-P.; Ries, C.; Scheuer, W.; Duerr, H.; Pompiati, S.; Pollmann, J.; et al. A Novel Glycoengineered Bispecific Antibody Format for Targeted Inhibition of Epidermal Growth Factor Receptor (EGFR) and Insulin-like Growth Factor Receptor Type I (IGF-1R) Demonstrating Unique Molecular Properties. J. Biol. Chem. 2014, 289, 18693–18706. [Google Scholar] [CrossRef] [Green Version]
- Duvic, M.; Pinter-Brown, L.C.; Foss, F.M.; Sokol, L.; Jorgensen, J.L.; Challagundla, P.; Dwyer, K.M.; Zhang, X.; Kurman, M.R.; Ballerini, R.; et al. Phase 1/2 study of mogamulizumab, a defucosylated anti-CCR4 antibody, in previously treated patients with cutaneous T-cell lymphoma. Blood 2015, 125, 1883–1889. [Google Scholar] [CrossRef]
- Beck, A.; Reichert, J.M. Marketing approval of mogamulizumab. mAbs 2012, 4, 419–425. [Google Scholar] [CrossRef] [Green Version]
- Sawas, A.; Farber, C.M.; Schreeder, M.T.; Khalil, M.Y.; Mahadevan, D.; Deng, C.; Amengual, J.E.; Nikolinakos, P.G.; Kolesar, J.M.; Kuhn, J.G.; et al. A phase 1/2 trial of ublituximab, a novel anti-CD20 monoclonal antibody, in patients with B-cell non-Hodgkin lymphoma or chronic lymphocytic leukaemia previously exposed to rituximab. Br. J. Haematol. 2017, 177, 243–253. [Google Scholar] [CrossRef]
- Eisner, F.; Pichler, M.; Goletz, S.; Stoeger, H.; Samonigg, H. A glyco-engineered anti-HER2 monoclonal antibody (TrasGEX) induces a long-lasting remission in a patient with HER2 overexpressing metastatic colorectal cancer after failure of all available treatment options. J. Clin. Pathol. 2015, 68, 1044.1–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neff-LaFord, H.; Grilley-Olson, J.E.; Smith, D.C.; Curti, B.; Goel, S.; Kuzel, T.M.; Markovic, S.N.; Rixe, O.; Bajor, D.L.; Gajewski, T.F.; et al. Abstract 5535: SEA-CD40 is a non-fucosylated anti-CD40 antibody with potent pharmacodynamic activity in preclinical models and patients with advanced solid tumors. Immunology 2020, 80, 5535. [Google Scholar]
- Grilley-Olson, J.E.; Curti, B.D.; Smith, D.C.; Goel, S.; Gajewski, T.; Markovic, S.; Rixe, O.; Bajor, D.L.; Gutierrez, M.; Kuzel, T.; et al. SEA-CD40, a non-fucosylated CD40 agonist: Interim results from a phase 1 study in advanced solid tumors. J. Clin. Oncol. 2018, 36, 3093. [Google Scholar] [CrossRef]
- Coveler, A.L.; Bajor, D.L.; Masood, A.; Yilmaz, E.; Shields, A.F.; Javle, M.M.; Paluri, R.K.; Vaccaro, G.M.; Zalupski, M.; Grilley-Olson, J.E.; et al. Phase I study of SEA-CD40, gemcitabine, nab-paclitaxel, and pembrolizumab in patients with metastatic pancreatic ductal adenocarcinoma (PDAC). J. Clin. Oncol. 2020, 38, TPS4671. [Google Scholar] [CrossRef]
- Pijnenborg, J.; Rossing, E.; Noga, M.; Titulaer, W.; Veizaj, R.; Lefeber, D.J.; Boltje, T. Fluorinated Mannosides Inhibit Cellular Fucosylation. ChemRxiv 2020. Preprint. [Google Scholar] [CrossRef]
- Okeley, N.M.; Heiser, R.A.; Zeng, W.; Hengel, S.M.; Wall, J.; Haughney, P.C.; Yap, T.A.; Robert, F.; Sanborn, R.E.; Burris, H.; et al. Abstract 5551: SGN-2FF: A small-molecule inhibitor of fucosylation modulates immune cell activity in preclinical models and demonstrates pharmacodynamic activity in early phase 1 analysis. Clin. Trials 2018, 78, 5551. [Google Scholar] [CrossRef]
- Pijnenborg, J.F.; Visser, E.A.; Noga, M.; Rossing, E.; Veizaj, R.; Lefeber, D.J.; Büll, C.; Boltje, T.J. Cellular fucosylation inhibitors based on fluorinated fucose-1-phosphates. ChemRxiv 2020. Preprint. [Google Scholar] [CrossRef]
- Zhou, Y.; Fukuda, T.; Hang, Q.; Hou, S.; Isaji, T.; Kameyama, A.; Gu, J. Inhibition of fucosylation by 2-fluorofucose suppresses human liver cancer HepG2 cell proliferation and migration as well as tumor formation. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Lim, J.; Chun, J.N.; Lee, S.; Kim, T.M.; Kim, D.; Kim, S.; Bae, D.; Bae, S.; So, I.; et al. Altered expression of fucosylation pathway genes is associated with poor prognosis and tumor metastasis in non-small cell lung cancer. Int. J. Oncol. 2020, 56, 559–567. [Google Scholar] [CrossRef] [Green Version]
- U.S. National Library of Medicine. ClinicalTrials.gov. 2020. Available online: https://clinicaltrials.gov/ct2/home (accessed on 10 October 2020).
- Järvå, M.A.; Dramicanin, M.; Lingford, J.P.; Mao, R.; John, A.; Jarman, K.E.; Grinter, R.; Goddard-Borger, E.D. Structural basis of substrate recognition and catalysis by fucosyltransferase 8. J. Biol. Chem. 2020, 295, 6677–6688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihara, H.; Okada, T.; Taniguchi, N.; Ikeda, Y. Involvement of the α-helical and Src homology 3 domains in the molecular assembly and enzymatic activity of human α1,6-fucosyltransferase, FUT8. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129596. [Google Scholar] [CrossRef] [PubMed]
- Boruah, B.M.; Kadirvelraj, R.; Liu, L.; Ramiah, A.; Li, C.; Zong, G.; Bosman, G.P.; Yang, J.-Y.; Wang, L.-X.; Boons, G.-J.; et al. Characterizing human α-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases. J. Biol. Chem. 2020, 295, 17027–17045. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Hartke, R.; Li, C.; Yang, Q.; Liu, J.O.; Wang, L.-X. Synthetic Fluorinated l-Fucose Analogs Inhibit Proliferation of Cancer Cells and Primary Endothelial Cells. ACS Chem. Biol. 2020. [Google Scholar] [CrossRef]
- Takei, J.; Ohishi, T.; Kaneko, M.K.; Harada, H.; Kawada, M.; Kato, Y. A defucosylated anti-PD-L1 monoclonal antibody 13-mG2a-f exerts antitumor effects in mouse xenograft models of oral squamous cell carcinoma. Biochem. Biophys. Rep. 2020, 24, 100801. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, Q.; Huang, H.; Chen, X.; Huang, T.; Li, W.; Zhang, J.; Liu, Y. Caveolin-1 upregulates Fut8 expression by activating the Wnt/β-catenin pathway to enhance HCC cell proliferative and invasive ability. Cell Biol. Int. 2020, 44. [Google Scholar] [CrossRef]
- Li, F.; Zhao, S.; Cui, Y.; Guo, T.; Qiang, J.; Xie, Q.; Yu, W.; Guo, W.; Deng, W.; Gu, C.; et al. 6-Fucosyltransferase (FUT8) regulates the cancer-promoting capacity of cancer-associated fibroblasts (CAFs) by modifying EGFR core fucosylation (CF) in non-small cell lung cancer (NSCLC). Am. J. Cancer Res. 2020, 10, 816. [Google Scholar]
- Höti, N.; Lih, T.-S.; Pan, J.; Zhou, Y.; Yang, G.; Deng, A.; Chen, L.; Dong, M.; Yang, R.-B.; Tu, C.-F.; et al. A Comprehensive Analysis of FUT8 Overexpressing Prostate Cancer Cells Reveals the Role of EGFR in Castration Resistance. Cancers 2020, 12, 468. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Xia, M.; Deng, X.; Ke, Q.; Zhang, C.; Peng, F.; Dong, X.; Luo, Z. A lectin-based glycomic approach identifies FUT8 as a driver of radioresistance in oesophageal squamous cell carcinoma. Cell. Oncol. 2020, 43, 695–707. [Google Scholar] [CrossRef]
- Zhang, N.; Li, M.; Xu, X.; Zhang, Y.; Liu, Y.; Zhao, M.; Li, P.; Chen, J.; Fukuda, T.; Gu, J.; et al. Loss of core fucosylation enhances the anticancer activity of cytotoxic T lymphocytes by increasing PD-1 degradation. Eur. J. Immunol. 2020, 50, 1820–1833. [Google Scholar] [CrossRef]
| Common Name(s) | Abbreviation | Subcellular Location [23] | Representative Major Products |
|---|---|---|---|
| H blood group α2fucosyltransferase | FUT1 | Membrane |  |
| Secretor (Se) blood group α2fucosyltransferase | FUT2 | Plasma membrane Cytosol |  |
| Fuc-TII α3/4fucosyltransferase Lewis blood group fucosyltransferase | FUT3 | Intracellular membrane (different isoforms) | Sialyl-Lewis Structures Lewis Structures  |
| Fuc-TIV α3fucosyltransferase ELAM-1 ligand fucosyltransferase | FUT4 | Vesicles |  |
| Fuc-TV α3fucosyltransferase | FUT5 | Membrane |  |
| Fuc-VI α3fucosyltransferase | FUT6 | Golgi apparatus |  |
| Fuc-VII α3fucosyltransferase | FUT7 | Golgi apparatus |  |
| Fuc-VIII α3fucosyltransferase | FUT8 | Golgi apparatus Cytosol |  |
| Fuc-TIX α3fucosyltransferase | FUT9 | Nucleoplasm Endoplasmic reticulum Golgi apparatus |  |
| Fuc-TX α3fucosyltransferase | FUT10 | Nucleoplasm Endoplasmic reticulum Golgi apparatus | Unknown |
| Fuc-TXI α3fucosyltransferase | FUT11 | Nuclear membrane Golgi apparatus | Unknown |
| Protein O-fucosyltransferase 1 | POFUT1/FUT12 | Centrosome |  |
| Protein O-fucosyltransferase 2 | POFUT2/FUT13 | Intracellular |  |
| Lung Cancer [20,28,49] |
|
| Liver Cancer [50,52] |
|
| Colorectal Cancer [24] |
|
| Ovarian Cancer [53] |
|
| Prostate Cancer [5,57] |
|
| Breast Cancer [30] |
|
| Thyroid Cancer [56] |
|
| Melanoma [55] |
|
| Pancreatic Cancer [27] |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bastian, K.; Scott, E.; Elliott, D.J.; Munkley, J. FUT8 Alpha-(1,6)-Fucosyltransferase in Cancer. Int. J. Mol. Sci. 2021, 22, 455. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22010455
Bastian K, Scott E, Elliott DJ, Munkley J. FUT8 Alpha-(1,6)-Fucosyltransferase in Cancer. International Journal of Molecular Sciences. 2021; 22(1):455. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22010455
Chicago/Turabian StyleBastian, Kayla, Emma Scott, David J. Elliott, and Jennifer Munkley. 2021. "FUT8 Alpha-(1,6)-Fucosyltransferase in Cancer" International Journal of Molecular Sciences 22, no. 1: 455. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22010455







