Characterization of Urine Stem Cell-Derived Extracellular Vesicles Reveals B Cell Stimulating Cargo
Abstract
:1. Introduction
2. Results
2.1. USC Isolation and Morphological Characterization
2.2. MSC Nature of the USCs
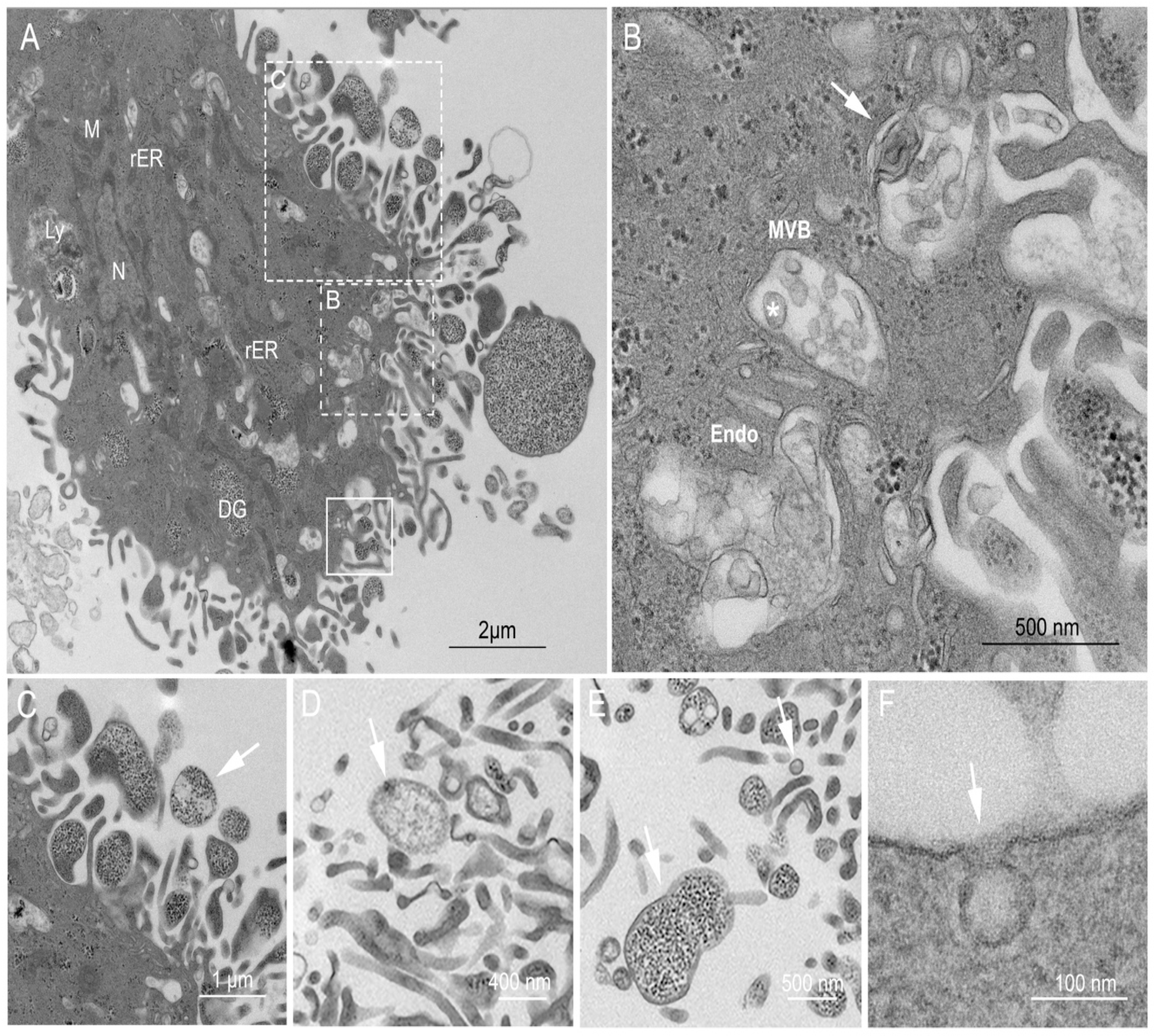
2.3. Ultrastructural Examination of the USCs
2.4. Characterization of Isolated EVs from USCs
2.5. USC–Derived EVs Activate T Cells without Inducing Proliferation
2.6. USC–Derived EVs Stimulate B Cells
2.7. Uptake of USC–Derived EVs by B Cells
2.8. USC–Derived EVs Are Carriers for B Cell Stimulants
2.9. USC Express Receptor for the B Cell Stimulant, BAFF
2.10. USC–Derived EVs Effect on Activated T Cells
3. Discussion
4. Materials and Methods
4.1. Isolation and Expansion of Human USCs
4.2. USCs Characterization
4.2.1. Cytochemical Staining
4.2.2. Immune Fluorescence Staining
4.2.3. Transmission Electron Microscopy (TEM)
4.3. MSCs Nature of the USCs
4.3.1. Phenotypical Characterization of USCs
4.3.2. USCs Differentiation
4.4. USCs Extracellular Vesicles Isolation
4.5. EV Characterization
4.5.1. Western Blot
4.5.2. Nanoparticle Tracking Analysis (NTA)
4.5.3. Transmission Electron Microscopy (TEM)
4.5.4. Immunogold Labelling
4.5.5. EVs Cytokine Content
4.6. PBMC Isolation
4.7. Co–Culture Studies
4.8. EV Uptake Study
4.9. Proliferation Study
4.10. Flow Cytometry
4.11. Antibody Quantification and Cytokines Assay
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Yin, K.; Wang, S.; Zhao, R.C. Exosomes from mesenchymal stem/stromal cells: A new therapeutic paradigm. Biomark. Res. 2019, 7, 8. [Google Scholar] [CrossRef] [Green Version]
- Margolis, L.; Sadovsky, Y. The biology of extracellular vesicles: The known unknowns. PLoS Biol. 2019, 17, e3000363. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar]
- Mianehsaz, E.; Mirzaei, H.R.; Mahjoubin-Tehran, M.; Rezaee, A.; Sahebnasagh, R.; Pourhanifeh, M.H.; Mirzaei, H.; Hamblin, M.R. Mesenchymal stem cell-derived exosomes: A new therapeutic approach to osteoarthritis? Stem Cell Res. 2019, 10, 340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phinney, D.G.; Pittenger, M.F. Concise review: MSC-derived exosomes for cell-free therapy. Stem Cells 2017, 35, 851–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baquir, B.; Hancock, R.E.W. Exosomes, your body’s answer to immune health. Ann. Transl. Med. 2017, 5, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dykes, I.M. Exosomes in Cardiovascular Medicine. Cardiol. Ther. 2017, 6, 225–237. [Google Scholar] [CrossRef]
- Baraniak, P.R.; McDevitt, T.C. Stem cell paracrine actions and tissue regeneration. Regen. Med. 2010, 5, 121–143. [Google Scholar] [CrossRef] [Green Version]
- Spees, J.L.; Lee, R.H.; Gregory, C.A. Mechanisms of mesenchymal stem/stromal cell function. Stem Cell Res. 2016, 7, 125. [Google Scholar] [CrossRef] [Green Version]
- Lai, P.; Chen, X.; Guo, L.; Wang, Y.; Liu, X.; Liu, Y.; Zhou, T.; Huang, T.; Geng, S.; Luo, C.; et al. potent immunomodulatory role of exosomes derived from mesenchymal stromal cells in preventing cGVHD. J. Hematol. Oncol. 2018, 11, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shigemoto-Kuroda, T.; Oh, J.Y.; Kim, D.K.; Jeong, H.J.; Park, S.Y.; Lee, H.J.; Park, J.W.; Kim, T.W.; An, S.Y.; Prockop, D.J.; et al. MSC–derived Extracellular Vesicles Attenuate Immune Responses in Two Autoimmune Murine Models: Type 1 Diabetes and Uveoretinitis. Stem Cell Rep. 2017, 8, 1214–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.M.; Zhuansun, Y.X.; Chen, R.; Lin, L.; Lin, Y.; Li, J.G. Mesenchymal stem cell exosomes promote immunosuppression of regulatory T cells in asthma. Exp. Cell Res. 2018, 363, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Eirin, A.; Zhu, X.Y.; Puranik, A.S.; Tang, H.; McGurren, K.A.; van Wijnen, A.J.; Lerman, A.; Lerman, L.O. Mesenchymal stem cell-derived extracellular vesicles attenuate kidney inflammation. Kidney Int 2017, 92, 114–124. [Google Scholar] [CrossRef]
- Konala, V.B.R.; Mamidi, M.K.; Bhonde, R.; Das, A.K.; Pochampally, R.; Pal, R. The current landscape of the mesenchymal stromal cell secretome: A new paradigm for cell-free regeneration. Cytotherapy 2016, 18, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Galieva, L.R.; James, V.; Mukhamedshina, Y.O.; Rizvanov, A.A. Therapeutic Potential of Extracellular Vesicles for the Treatment of Nerve Disorders. Front. Neurosci. 2019, 13. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, S.W.; Wang, J.; Lee, C.J.; Liu, M.; Neelamegham, S.; Canty, J.M.; Nguyen, J. The microRNA regulatory landscape of MSC–derived exosomes: A systems view. Sci. Rep. 2018, 8, 1419. [Google Scholar] [CrossRef]
- Zhang, D.; Wei, G.; Li, P.; Zhou, X.; Zhang, Y. Urine–derived stem cells: A novel and versatile progenitor source for cell-based therapy and regenerative medicine. Genes Dis. 2014, 1, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; McNeill, E.; Tian, H.; Soker, S.; Andersson, K.-E.; Yoo, J.J.; Atala, A. Urine derived cells are a potential source for urological tissue reconstruction. J. Urol. 2008, 180, 2226–2233. [Google Scholar] [CrossRef]
- Pavathuparambil Abdul Manaph, N.; Al-Hawwas, M.; Bobrovskaya, L.; Coates, P.T.; Zhou, X.-F. Urine–derived cells for human cell therapy. Stem Cell Res. 2018, 9, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hass, R.; Kasper, C.; Böhm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue–derived MSC. Cell Commun. Signal. 2011, 9. [Google Scholar] [CrossRef] [Green Version]
- Marquez-Curtis, L.A.; Janowska-Wieczorek, A.; McGann, L.E.; Elliott, J.A.W. Mesenchymal stromal cells derived from various tissues: Biological, clinical and cryopreservation aspects. Cryobiology 2015, 71, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Bieback, K.; Kern, S.; Kocaömer, A.; Ferlik, K.; Bugert, P. Comparing mesenchymal stromal cells from different human tissues: Bone marrow, adipose tissue and umbilical cord blood. Bio-Med. Mater. Eng. 2008, 18, 71–76. [Google Scholar]
- Al-Nbaheen, M.; Vishnubalaji, R.; Ali, D.; Bouslimi, A.; Al-Jassir, F.; Megges, M.; Prigione, A.; Adjaye, J.; Kassem, M.; Aldahmash, A. Human Stromal (Mesenchymal) Stem Cells from Bone Marrow, Adipose Tissue and Skin Exhibit Differences in Molecular Phenotype and Differentiation Potential. Stem Cell Rev. Rep. 2013, 9, 32–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.Z.; Liu, Y.M.; Niu, X.; Yin, J.Y.; Hu, B.; Guo, S.C.; Fan, Y.; Wang, Y.; Wang, N.S. Exosomes secreted by human urine–derived stem cells could prevent kidney complications from type I diabetes in rats. Stem Cell Res. 2016, 7, 24. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-Y.; Rao, S.-S.; Ren, L.; Hu, X.-K.; Tan, Y.-J.; Hu, Y.; Luo, J.; Liu, Y.-W.; Yin, H.; Huang, J.; et al. Exosomal DMBT1 from human urine–derived stem cells facilitates diabetic wound repair by promoting angiogenesis. Theranostics 2018, 8, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Li, Q.; Niu, X.; Zhang, G.; Ling, X.; Zhang, J.; Wang, Y.; Deng, Z. Extracellular Vesicles Secreted by Human Urine-Derived Stem Cells Promote Ischemia Repair in a Mouse Model of Hind-Limb Ischemia. Cell. Physiol. Biochem. 2018, 47, 1181–1192. [Google Scholar] [CrossRef]
- Wu, R.; Huang, C.; Wu, Q.; Jia, X.; Liu, M.; Xue, Z.; Qiu, Y.; Niu, X.; Wang, Y. Exosomes secreted by urine–derived stem cells improve stress urinary incontinence by promoting repair of pubococcygeus muscle injury in rats. Stem Cell Res. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Di Trapani, M.; Bassi, G.; Midolo, M.; Gatti, A.; Takam Kamga, P.; Cassaro, A.; Carusone, R.; Adamo, A.; Krampera, M. Differential and transferable modulatory effects of mesenchymal stromal cell-derived extracellular vesicles on T, B and NK cell functions. Sci. Rep. 2016, 6, 24120. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.; Chung, H.; Moon, J.H.; Kang, S.J.; Park, C.-G. Mesenchymal stem cell-derived exosomes suppress proliferation of T cells by inducing cell cycle arrest through p27kip1/Cdk2 signaling. Immunol. Lett. 2020, 225, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Huang, Y.; Han, J.; Yu, L.; Li, Y.; Lu, Z.; Li, H.; Liu, Z.; Shi, C.; Duan, F. Immunomodulatory effects of mesenchymal stromal cells–derived exosome. Immunol. Res. 2016, 64, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Blazquez, R.; Sanchez-Margallo, F.M.; de la Rosa, O.; Dalemans, W.; Álvarez, V.; Tarazona, R.; Casado, J.G. Immunomodulatory potential of human adipose mesenchymal stem cells derived exosomes on in vitro stimulated T cells. Front. Immunol. 2014, 5, 556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Fattore, A.; Luciano, R.; Pascucci, L.; Goffredo, B.M.; Giorda, E.; Scapaticci, M.; Fierabracci, A.; Muraca, M. Immunoregulatory effects of mesenchymal stem cell-derived extracellular vesicles on T lymphocytes. Cell Transpl. 2015, 24, 2615–2627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plumas, J.; Chaperot, L.; Richard, M.-J.; Molens, J.-P.; Bensa, J.-C.; Favrot, M.-C. Mesenchymal stem cells induce apoptosis of activated T cells. Leukemia 2005, 19, 1597–1604. [Google Scholar] [CrossRef] [PubMed]
- Rosado, M.M.; Bernardo, M.E.; Scarsella, M.; Conforti, A.; Giorda, E.; Biagini, S.; Cascioli, S.; Rossi, F.; Guzzo, I.; Vivarelli, M.; et al. Inhibition of B-cell proliferation and antibody production by mesenchymal stromal cells is mediated by T cells. Stem Cells Dev. 2015, 24, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.; Yang, Q.; Wang, Q.; Shi, C.; Wang, D.; Armato, U.; Prà, I.D.; Chiarini, A. Mesenchymal stromal cells–exosomes: A promising cell-free therapeutic tool for wound healing and cutaneous regeneration. Burn. Trauma 2019, 7, 38. [Google Scholar] [CrossRef]
- Andaloussi, S.E.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef]
- Riazifar, M.; Mohammadi, M.R.; Pone, E.J.; Yeri, A.; Lasser, C.; Segaliny, A.I.; McIntyre, L.L.; Shelke, G.V.; Hutchins, E.; Hamamoto, A. Stem cell-derived exosomes as nanotherapeutics for autoimmune and neurodegenerative disorders. ACS Nano 2019, 13, 6670–6688. [Google Scholar] [CrossRef]
- Kordelas, L.; Rebmann, V.; Ludwig, A.; Radtke, S.; Ruesing, J.; Doeppner, T.; Epple, M.; Horn, P.; Beelen, D.; Giebel, B. MSC–derived exosomes: A novel tool to treat therapy–refractory graft–versus–host disease. Leukemia 2014, 28, 970–973. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, S.; Liu, G.; Shi, Y.; Wu, R.; Yang, B.; He, T.; Fan, Y.; Lu, X.; Zhou, X.; Liu, H.; et al. Multipotential differentiation of human urine–derived stem cells: Potential for therapeutic applications in urology. Stem Cells 2013, 31, 1840–1856. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Sun, A.; Zou, Y.; Ge, J. Inducible metabolic adaptation promotes mesenchymal stem cell therapy for ischemia: A hypoxia–induced and glycogen–based energy prestorage strategy. Arter. Thromb. Vasc. Biol. 2014, 34, 870–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muhič, M.; Vardjan, N.; Chowdhury, H.H.; Zorec, R.; Kreft, M. Insulin and insulin–like growth factor 1 (IGF–1) modulate cytoplasmic glucose and glycogen levels but not glucose transport across the membrane in astrocytes. J. Biol. Chem. 2015, 290, 11167–11176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef]
- Tang, Y.-T.; Huang, Y.-Y.; Zheng, L.; Qin, S.-H.; Xu, X.-P.; An, T.-X.; Xu, Y.; Wu, Y.-S.; Hu, X.-M.; Ping, B.-H. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int. J. Mol. Med. 2017, 40, 834–844. [Google Scholar] [CrossRef] [Green Version]
- Eleftheriadis, T.; Pissas, G.; Liakopoulos, V.; Stefanidis, I. Cytochrome c as a Potentially Clinical Useful Marker of Mitochondrial and Cellular Damage. Front. Immunol. 2016, 7, 279. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Mun, J.Y. Structural analysis of exosomes using different types of Electron microscopy. Appl. Microsc. 2017, 47, 171–175. [Google Scholar] [CrossRef] [Green Version]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus–like vesicles, and apoptotic bodies. J. Neuro Oncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [Green Version]
- McKelvey, K.J.; Powell, K.L.; Ashton, A.W.; Morris, J.M.; McCracken, S.A. Exosomes: Mechanisms of Uptake. J. Circ. Biomark. 2015, 4, 7. [Google Scholar] [CrossRef] [Green Version]
- Gonda, A.; Kabagwira, J.; Senthil, G.N.; Wall, N.R. Internalization of Exosomes through Receptor-Mediated Endocytosis. Mol. Cancer Res. 2019, 17, 337–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrell, C.R.; Jovicic, N.; Djonov, V.; Arsenijevic, N.; Volarevic, V. Mesenchymal Stem Cell-Derived Exosomes and Other Extracellular Vesicles as New Remedies in the Therapy of Inflammatory Diseases. Cells 2019, 8, 1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laman, J.D.; Claassen, E.; Noelle, R.J. Functions of CD40 and its ligand, gp39 (CD40L). Crit. Rev. Immunol. 1996, 16, 59. [Google Scholar] [CrossRef]
- Thompson, N.; Isenberg, D.A.; Jury, E.C.; Ciurtin, C. Exploring BAFF: Its expression, receptors and contribution to the immunopathogenesis of Sjögren’s syndrome. Rheumatology 2016, 55, 1548–1555. [Google Scholar] [CrossRef] [Green Version]
- Jourdan, M.; Cren, M.; Robert, N.; Bolloré, K.; Fest, T.; Duperray, C.; Guilloton, F.; Hose, D.; Tarte, K.; Klein, B. IL–6 supports the generation of human long–lived plasma cells in combination with either APRIL or stromal cell-soluble factors. Leukemia 2014, 28, 1647–1656. [Google Scholar] [CrossRef]
- Schlom, J.; Jochems, C.; Gulley, J.L.; Huang, J. The role of soluble CD40L in immunosuppression. Oncoimmunology 2013, 2, e22546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howland, K.C.; Ausubel, L.J.; London, C.A.; Abbas, A.K. The Roles of CD28 and CD40 Ligand in T Cell Activation and Tolerance. J. Immunol. 2000, 164, 4465–4470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khare, D.; Or, R.; Resnick, I.; Barkatz, C.; Almogi-Hazan, O.; Avni, B. Mesenchymal Stromal Cell-Derived Exosomes Affect mRNA Expression and Function of B–Lymphocytes. Front. Immunol. 2018, 9, 3053. [Google Scholar] [CrossRef] [Green Version]
- Traggiai, E.; Volpi, S.; Schena, F.; Gattorno, M.; Ferlito, F.; Moretta, L.; Martini, A. Bone marrow-derived mesenchymal stem cells induce both polyclonal expansion and differentiation of B cells isolated from healthy donors and systemic lupus erythematosus patients. Stem Cells 2008, 26, 562–569. [Google Scholar] [CrossRef]
- Rasmusson, I.; Le Blanc, K.; Sundberg, B.; Ringdén, O. Mesenchymal Stem Cells Stimulate Antibody Secretion in Human B Cells. Scand. J. Immunol. 2007, 65, 336–343. [Google Scholar] [CrossRef]
- Ji, Y.R.; Yang, Z.X.; Han, Z.-B.; Meng, L.; Liang, L.; Feng, X.M.; Yang, S.G.; Chi, Y.; Chen, D.D.; Wang, Y.W. Mesenchymal stem cells support proliferation and terminal differentiation of B cells. Cell. Physiol. Biochem. 2012, 30, 1526–1537. [Google Scholar] [CrossRef] [PubMed]
- Asari, S.; Itakura, S.; Ferreri, K.; Liu, C.-P.; Kuroda, Y.; Kandeel, F.; Mullen, Y. Mesenchymal stem cells suppress B-cell terminal differentiation. Exp. Hematol. 2009, 37, 604–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abumaree, M.; Al Jumah, M.; Pace, R.A.; Kalionis, B. Immunosuppressive properties of mesenchymal stem cells. Stem Cell Rev. Rep. 2012, 8, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Franquesa, M.; Hoogduijn, M.J.; Bestard, O.; Grinyó, J.M. Immunomodulatory effect of mesenchymal stem cells on B cells. Front. Immunol. 2012, 3, 212. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Wu, M.; Yuan, Y.; Wang, Z.Z.; Jiang, H.; Chen, T. Priming of Toll–like receptor 4 pathway in mesenchymal stem cells increases expression of B cell activating factor. Biochem. Biophys. Res. Commun. 2014, 448, 212–217. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Wu, X.-R.; Liu, H.-S.; Liu, X.-H.; Liu, G.-H.; Zheng, X.-B.; Hu, T.; Liang, Z.-X.; He, X.-W.; Wu, X.-J.; et al. Immunomodulatory Effect of Urine–derived Stem Cells on Inflammatory Bowel Diseases via Downregulating Th1/Th17 Immune Responses in a PGE2–dependent Manner. J. Crohn’s Colitis 2019, 14, 654–668. [Google Scholar] [CrossRef]
- Bifari, F.; Lisi, V.; Mimiola, E.; Pasini, A.; Krampera, M. Immune modulation by mesenchymal stem cells. Transfus. Med. Hemother. 2008, 35, 194–204. [Google Scholar] [CrossRef] [Green Version]
- Lai, P.; Weng, J.; Guo, L.; Chen, X.; Du, X. Novel insights into MSC–EVs therapy for immune diseases. Biomark. Res. 2019, 7, 6. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Liao, J.; Su, X.; Li, W.; Bi, Z.; Wang, J.; Su, Q.; Huang, H.; Wei, Y.; Gao, Y.; et al. Human urine–derived stem cells protect against renal ischemia/reperfusion injury in a rat model via exosomal miR–146a–5p which targets IRAK1. Theranostics 2020, 10, 9561–9578. [Google Scholar] [CrossRef]
- Domenis, R.; Cifù, A.; Quaglia, S.; Pistis, C.; Moretti, M.; Vicario, A.; Parodi, P.C.; Fabris, M.; Niazi, K.R.; Soon-Shiong, P.; et al. Pro inflammatory stimuli enhance the immunosuppressive functions of adipose mesenchymal stem cells–derived exosomes. Sci. Rep. 2018, 8, 13325. [Google Scholar] [CrossRef]
- Martin-Rufino, J.D.; Espinosa-Lara, N.; Osugui, L.; Sanchez-Guijo, F. Targeting the Immune System With Mesenchymal Stromal Cell-Derived Extracellular Vesicles: What Is the Cargo’s Mechanism of Action? Front. Bioeng. Biotech. 2019, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, K.; Hirohata, S. The role of IL–10 in human B cell activation, proliferation, and differentiation. J. Immunol. 1995, 154, 4341–4350. [Google Scholar] [PubMed]
- Taga, K.; Tosato, G. IL–10 inhibits human T cell proliferation and IL–2 production. J. Immunol. 1992, 148, 1143–1148. [Google Scholar] [PubMed]
- Akdis, C.A.; Blaser, K. Mechanisms of interleukin–10–mediated immune suppression. Immunology 2001, 103, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.A.; Li, M.O. TGF–β: Guardian of T cell function. J. Immunol. 2013, 191, 3973–3979. [Google Scholar] [CrossRef]
- Lebman, D.A.; Edmiston, J.S. The role of TGF–beta in growth, differentiation, and maturation of B lymphocytes. Microbes Infect. 1999, 1, 1297–1304. [Google Scholar] [CrossRef]
- Liang, H.; Kidder, K.; Liu, Y. Extracellular microRNAs initiate immunostimulation via activating toll–like receptor signaling pathways. ExRNA 2019, 1, 9. [Google Scholar] [CrossRef] [Green Version]
- Bayraktar, R.; Bertilaccio, M.T.S.; Calin, G.A. The Interaction Between Two Worlds: MicroRNAs and Toll–Like Receptors. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Zou, L.; Yan, D.; Chen, H.; Xu, G.; Jian, W.; Cui, P.; Chao, W. Extracellular MicroRNAs Induce Potent Innate Immune Responses via TLR7/MyD88–Dependent Mechanisms. J. Immunol. 2017, 199, 2106–2117. [Google Scholar] [CrossRef]
- Wang, T.; Marken, J.; Chen, J.; Tran, V.B.; Li, Q.-Z.; Li, M.; Cerosaletti, K.; Elkon, K.B.; Zeng, X.; Giltiay, N.V. High TLR7 Expression Drives the Expansion of CD19+CD24hiCD38hi Transitional B Cells and Autoantibody Production in SLE Patients. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Suthers, A.N.; Sarantopoulos, S. TLR7/TLR9– and B Cell Receptor–Signaling Crosstalk: Promotion of Potentially Dangerous B Cells. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.-L.; Sun, P.; Li, Y.; Liu, S.-S.; Lu, Y. Exosomes as critical mediators of cell-to-cell communication in cancer pathogenesis and their potential clinical application. Transl. Cancer Res. 2019, 8, 298–311. [Google Scholar] [CrossRef]
- Su, S.A.; Xie, Y.; Fu, Z.; Wang, Y.; Wang, J.A.; Xiang, M. Emerging role of exosome–mediated intercellular communication in vascular remodeling. Oncotarget 2017, 8, 25700–25712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, B.H.; Ketova, T.; Hoshino, D.; Zijlstra, A.; Weaver, A.M. Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 2015, 6, 7164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, B.; Garcia, M.; Murakami, J.L.; Chen, C.-C. Exosome–mediated microenvironment dysregulation in leukemia. Biophys. Acta Mol. Cell Res. 2016, 1863, 464–470. [Google Scholar] [CrossRef]
- Phatak, S.; Chaurasia, S.; Mishra, S.K.; Gupta, R.; Agrawal, V.; Aggarwal, A.; Misra, R. Urinary B cell activating factor (BAFF) and a proliferation–inducing ligand (APRIL): Potential biomarkers of active lupus nephritis. Clin. Exp. Immunol. 2017, 187, 376–382. [Google Scholar] [CrossRef] [Green Version]
- Schwarting, A.; Relle, M.; Meineck, M.; Föhr, B.; Triantafyllias, K.; Weinmann, A.; Roth, W.; Weinmann-Menke, J. Renal tubular epithelial cell-derived BAFF expression mediates kidney damage and correlates with activity of proliferative lupus nephritis in mouse and men. Lupus 2018, 27, 243–256. [Google Scholar] [CrossRef]
- Liao, W.; Du, Y.; Zhang, C.; Pan, F.; Yao, Y.; Zhang, T.; Peng, Q. Exosomes: The next generation of endogenous nanomaterials for advanced drug delivery and therapy. Acta Biomater. 2019, 86, 1–14. [Google Scholar] [CrossRef]
- Sarvar, D.P.; Shamsasenjan, K.; Akbarzadehlaleh, P. Mesenchymal stem cell-derived exosomes: New opportunity in cell-free therapy. Adv. Pharm. Bull. 2016, 6, 293. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, L.; Lin, Z.; Tao, L.; Chen, M. More efficient induction of antitumor T cell immunity by exosomes from CD40L gene–modified lung tumor cells. Mol. Med. Rep. 2014, 9, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Petitprez, F.; de Reyniès, A.; Keung, E.Z.; Chen, T.W.-W.; Sun, C.-M.; Calderaro, J.; Jeng, Y.-M.; Hsiao, L.-P.; Lacroix, L.; Bougoüin, A.; et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020, 577, 556–560. [Google Scholar] [CrossRef]
- Walker, S.; Busatto, S.; Pham, A.; Tian, M.; Suh, A.; Carson, K.; Quintero, A.; Lafrence, M.; Malik, H.; Santana, M.X.; et al. Extracellular vesicle–based drug delivery Syst. for cancer treatment. Theranostics 2019, 9, 8001–8017. [Google Scholar] [CrossRef] [PubMed]
- Talaei-Khozani, T.; Borhani-Haghighi, M.; Ayatollahi, M.; Vojdani, Z. An in vitro model for hepatocyte–like cell differentiation from Wharton’s jelly derived–mesenchymal stem cells by cell-base aggregates. Gastroenterol. Hepatol. Bed Bench 2015, 8, 188. [Google Scholar] [PubMed]
- Johkura, K.; Cui, L.; Asanuma, K.; Okouchi, Y.; Ogiwara, N.; Sasaki, K. Cytochemical and ultrastructural characterization of growing colonies of human embryonic stem cells. J. Anat. 2004, 205, 247–255. [Google Scholar] [CrossRef]
- Graham, L.; Orenstein, J.M. Processing tissue and cells for transmission electron microscopy in diagnostic pathology and research. Nat. Protoc. 2007, 2, 2439. [Google Scholar] [CrossRef] [PubMed]
- Ciuffreda, M.C.; Malpasso, G.; Musarò, P.; Turco, V.; Gnecchi, M. Protocols for in vitro differentiation of human mesenchymal stem cells into osteogenic, chondrogenic and adipogenic lineages. In Mesenchymal Stem Cells; Humana Press: New York, NY, USA, 2016; pp. 149–158. [Google Scholar]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Prot. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar]
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef]
- Uphoff, C.C.; Drexler, H.G. Detecting mycoplasma contamination in cell cultures by polymerase chain reaction. Meth. Mol. Biol. 2011, 731, 93–103. [Google Scholar]
- Soares Martins, T.; Catita, J.; Martins Rosa, I.; da Cruz e Silva, O.; Henriques, A.G. Exosome isolation from distinct biofluids using precipitation and column–based approaches. PLoS ONE 2018, 13, e0198820. [Google Scholar] [CrossRef] [Green Version]
- Kowal, E.J.; Ter-Ovanesyan, D.; Regev, A.; Church, G.M. Extracellular vesicle isolation and analysis by western blotting. In Extracellular Vesicles; Humana Press: New York, NY, USA, 2017; pp. 143–152. [Google Scholar]
- Jung, M.K.; Mun, J.Y. Sample Preparation and Imaging of Exosomes by Transmission Electron Microscopy. J. Vis. Exp. 2018. [Google Scholar] [CrossRef]
- Fuss, I.J.; Kanof, M.E.; Smith, P.D.; Zola, H. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr. Prot. Immunol. 2009. [Google Scholar] [CrossRef]
- Zhang, B.; Yeo, R.W.Y.; Lai, R.C.; Sim, E.W.K.; Chin, K.C.; Lim, S.K. Mesenchymal stromal cell exosome–enhanced regulatory T-cell production through an antigen–presenting cell-mediated pathway. Cytotherapy 2018, 20, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Gao, K.; Huang, C.; Li, J.; Luo, H.; Li, L.; Huang, Y.; Jin, J. Exosomes derived from septic mouse serum modulate immune responses via exosome–associated cytokines. Front. Immunol. 2019, 10, 1560. [Google Scholar]
- Krieg, A.M.; Yi, A.-K.; Matson, S.; Waldschmidt, T.J.; Bishop, G.A.; Teasdale, R.; Koretzky, G.A.; Klinman, D.M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 1995, 374, 546–549. [Google Scholar] [CrossRef]
- Trickett, A.; Kwan, Y.L. T cell stimulation and expansion using anti–CD3/CD28 beads. J. Immunol. Meth. 2003, 275, 251–255. [Google Scholar] [CrossRef]
- Quah, B.J.; Parish, C.R. The use of carboxyfluorescein diacetate succinimidyl ester (CFSE) to monitor lymphocyte proliferation. J. Vis. Exp. 2010. [Google Scholar] [CrossRef] [Green Version]
- Yeh, C.-J.G.; Hsi, B.-L.; Faulk, W.P. Propidium iodide as a nuclear marker in immunofluorescence. II. Use with cellular identification and viability studies. J. Immunol. Meth. 1981, 43, 269–275. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zidan, A.A.; Al-Hawwas, M.; Perkins, G.B.; Mourad, G.M.; Stapledon, C.J.M.; Bobrovskaya, L.; Zhou, X.-F.; Hurtado, P.R. Characterization of Urine Stem Cell-Derived Extracellular Vesicles Reveals B Cell Stimulating Cargo. Int. J. Mol. Sci. 2021, 22, 459. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22010459
Zidan AA, Al-Hawwas M, Perkins GB, Mourad GM, Stapledon CJM, Bobrovskaya L, Zhou X-F, Hurtado PR. Characterization of Urine Stem Cell-Derived Extracellular Vesicles Reveals B Cell Stimulating Cargo. International Journal of Molecular Sciences. 2021; 22(1):459. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22010459
Chicago/Turabian StyleZidan, Asmaa A., Mohammed Al-Hawwas, Griffith B. Perkins, Ghada M. Mourad, Catherine J. M. Stapledon, Larisa Bobrovskaya, Xin-Fu Zhou, and Plinio R. Hurtado. 2021. "Characterization of Urine Stem Cell-Derived Extracellular Vesicles Reveals B Cell Stimulating Cargo" International Journal of Molecular Sciences 22, no. 1: 459. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22010459







