Fatty Acid Desaturases: Uncovering Their Involvement in Grapevine Defence against Downy Mildew
Abstract
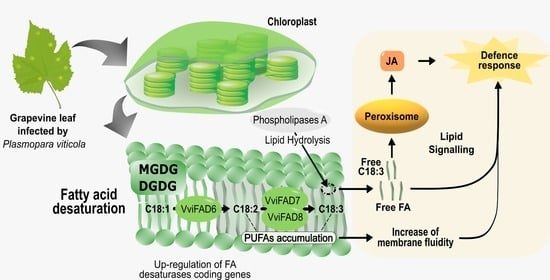
:1. Introduction
2. Results
2.1. Characterisation of Grapevine Fatty Acid Desaturases
2.1.1. Grapevine Fatty Acid Desaturase Gene Family Identification
2.1.2. Phylogenetic Analysis of Grapevine Fatty Acid Desaturases
2.1.3. Identification of Cis-Elements of Grapevine FA Desaturases Genes
2.1.4. Protein Structure and Domain Analysis
2.1.5. Grapevine FA Desaturases Subcellular Targeting Prediction
2.2. Fatty Acid Desaturases Gene Expression Correlates with Changes in the Fatty Acid Profiles upon Pathogen Inoculation
3. Discussion
3.1. Characterisation of Fatty Acid Desaturases in Grapevine
3.2. Fatty Acid Desaturases in Grapevine Defence
3.3. Fatty Acid Modulation in Grapevine-Plasmopara Viticola Interaction
4. Materials and Methods
4.1. Plant Material
4.2. Lipid Analysis
4.3. Characterisation of the Grapevine Fatty Acid Desaturases
4.3.1. Identification and Retrieval of Grapevine FA Desaturases Sequences
4.3.2. Cis-Element Analysis for Grapevine FA Desaturases GENE Promoter
4.3.3. Domain Structure Analysis, Sequence Properties, Subcellular Location Prediction and Chromosomal Location
4.3.4. Phylogenetic Analysis
4.4. Expression Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gessler, C.; Pertot, I.; Perazzolli, M. Plasmopara viticola: A review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 2011, 50, 3–44. [Google Scholar] [CrossRef]
- Wang, X. Lipid signaling. Curr. Opin. Plant Biol. 2004, 7, 329–336. [Google Scholar] [CrossRef]
- Kachroo, A.; Kachroo, P. Fatty Acid–Derived Signals in Plant Defense. Annu. Rev. Phytopathol. 2009, 47, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Ohlrogge, J.; Browse, J. Lipid biosynthesis. Plant Cell 1995, 7, 957–970. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.C.; Sinha, R.P.; Häder, D.-P. Role of Lipids and Fatty Acids in Stress Tolerance in Cyanobacteria. Acta Protozool. 2002, 41, 297–308. [Google Scholar]
- Alonso, D.L.; García-Maroto, F.; Rodríguez-Ruiz, J.; Garrido, J.; Vilches, M. Evolution of the membrane-bound fatty acid desaturases. Biochem. Syst. Ecol. 2003, 31, 1111–1124. [Google Scholar] [CrossRef]
- Yukawa, Y.; Shoji, K.; Takaiwa, F.; Masuda, K.; Yamada, K. Structure and Expression of Two Seed-Specific cDNA Clones Encoding Stearoyl-Acyl Carrier Protein Desaturase from Sesame, Sesamum indicum L. Plant Cell Physiol. 1996, 37, 201–205. [Google Scholar] [CrossRef]
- McCartney, A.W.; Dyer, J.M.; Dhanoa, P.K.; Kim, P.K.; Andrews, D.W.; McNew, J.A.; Mullen, R.T. Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J. 2003, 37, 156–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, N.S.; Wierzbicki, A.; Aegerter, M.; Caster, C.S.; Perez-Grau, L.; Kinney, A.J.; Hitz, W.D.; Booth, J.R., Jr.; Schweiger, B.; Stecca, K.L.; et al. Cloning of Higher Plant [omega]-3 Fatty Acid Desaturases. Plant Physiol. 1993, 103, 467–476. [Google Scholar] [CrossRef] [Green Version]
- Los, D.A.; Murata, N. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta (BBA) Lipids Lipid Metab. 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Somerville, C.; Browse, J. Plant Lipids: Metabolism, Mutants, and Membranes. Science 1991, 252, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Yang, Q.; Lu, Y.; Wang, J.; Zhang, Q.; Pan, L.; Chen, M.; He, Y.; Yu, S. Genome-Wide Analysis of Fatty Acid Desaturases in Soybean (Glycine max). Plant Mol. Biol. Rep. 2011, 29, 769–783. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; He, Q.; Daud, M.K.; Chen, J.; Zhu, S. Characterization of 19 Genes Encoding Membrane-Bound Fatty Acid Desaturases and their Expression Profiles in Gossypium raimondii Under Low Temperature. PLoS ONE 2015, 10, e0123281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, X.; Zhang, Z.; Chen, N.; Zhang, X.; Wang, M.; Chen, M.; Wang, T.; Pan, L.; Chen, J.; Yang, Z.; et al. Isolation and functional analysis of fatty acid desaturase genes from peanut (Arachis hypogaea L.). PLoS ONE 2017, 12, e0189759. [Google Scholar] [CrossRef] [Green Version]
- Zhiguo, E.; Chen, C.; Yang, J.; Tong, H.; Li, T.; Wang, L.; Chen, H. Genome-wide analysis of fatty acid desaturase genes in rice (Oryza sativa L.). Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Lightner, J.; Wu, J.; Browse, J. A Mutant of Arabidopsis with Increased Levels of Stearic Acid. Plant Physiol. 1994, 106, 1443–1451. [Google Scholar] [CrossRef] [Green Version]
- Fukuchi-Mizutani, M.; Tasaka, Y.; Tanaka, Y.; Ashikari, T.; Kusumi, T.; Murata, N. Characterization of A9 Acyl-lipid Desaturase Homologues from Arabidopsis thaliana. Plant Cell Physiol. 1998, 39, 247–253. [Google Scholar] [CrossRef] [Green Version]
- Heilmann, I.; Mekhedov, S.; King, B.; Browse, J.; Shanklin, J. Identification of the Arabidopsis Palmitoyl-Monogalactosyldiacylglycerol Δ7-Desaturase Gene FAD5, and Effects of Plastidial Retargeting of Arabidopsis Desaturases on the fad5 Mutant Phenotype. Plant Physiol. 2004, 136, 4237–4245. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Ajjawi, I.; Manoli, A.; Sawin, A.; Xu, C.; Froehlich, J.E.; Last, R.L.; Benning, C. Fatty Acid Desaturase4 of Arabidopsis Encodes a Protein Distinct from Characterized Fatty Acid Desaturases. Plant J. 2009, 60, 832–839. [Google Scholar] [CrossRef]
- Falcone, D.L.; Gibson, S.; Lemieux, B.; Somerville, C. Identification of a Gene that Complements an Arabidopsis Mutant Deficient in Chloroplast [omega] 6 Desaturase Activity. Plant Physiol. 1994, 106, 1453–1459. [Google Scholar] [CrossRef] [Green Version]
- Heppard, E.P.; Kinney, A.J.; Stecca, K.L.; Miao, G.H. Developmental and Growth Temperature Regulation of Two Different Microsomal [omega]-6 Desaturase Genes in Soybeans. Plant Physiol. 1996, 110, 311–319. [Google Scholar] [CrossRef] [Green Version]
- Arondel, V.; Lemieux, B.; Hwang, I.; Gibson, S.; Goodman, H.M.; Somerville, C.R. Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science 1992, 258, 1353–1355. [Google Scholar] [CrossRef]
- McConn, M.; Hugly, S.; Browse, J.; Somerville, C. A Mutation at the fad8 Locus of Arabidopsis Identifies a Second Chloroplast [omega]-3 Desaturase. Plant Physiol. 1994, 106, 1609–1614. [Google Scholar] [CrossRef] [Green Version]
- Sperling, P.; Zähringer, U.; Heinz, E. A Sphingolipid Desaturase from Higher Plants. J. Biol. Chem. 1998, 273, 28590–28596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ternes, P.; Franke, S.; Zähringer, U.; Sperling, P.; Heinz, E. Identification and Characterization of a Sphingolipid Δ4-Desaturase Family. J. Biol. Chem. 2002, 277, 25512–25518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueiredo, A.; Monteiro, F.; Sebastiana, M. First clues on a jasmonic acid role in grapevine resistance against the biotrophic fungus Plasmopara viticola. Eur. J. Plant Pathol. 2015, 142, 645–652. [Google Scholar] [CrossRef]
- Guerreiro, A.; Figueiredo, J.; Silva, M.S.; Figueiredo, A. Linking Jasmonic Acid to Grapevine Resistance against the Biotrophic Oomycete Plasmopara viticola. Front. Plant Sci. 2016, 7, 565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laureano, G.; Figueiredo, J.; Cavaco, A.R.; Duarte, B.; Caçador, I.; Malhó, R.; Silva, M.S.; Matos, A.R.; Figueiredo, A. The interplay between membrane lipids and phospholipase A family members in grapevine resistance against Plasmopara viticola. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.; Martins, J.; Sebastiana, M.; Guerreiro, A.; Silva, A.; Matos, A.R.; Monteiro, F.; Pais, M.S.; Roepstorff, P.; Coelho, A.V. Specific adjustments in grapevine leaf proteome discriminating resistant and susceptible grapevine genotypes to Plasmopara viticola. J. Proteom. 2017, 152, 48–57. [Google Scholar] [CrossRef]
- Santos, R.B.; Nascimento, R.; Coelho, A.V.; Figueiredo, A. Grapevine–Downy Mildew Rendezvous: Proteome Analysis of the First Hours of an Incompatible Interaction. Plants 2020, 9, 1498. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Grimplet, J.; Adam-Blondon, A.-F.; Bert, P.-F.; Bitz, O.; Cantu, D.; Davies, C.; Delrot, S.; Pezzotti, M.; Rombauts, S.; Cramer, G.R. The grapevine gene nomenclature system. BMC Genom. 2014, 15, 1077. [Google Scholar] [CrossRef] [Green Version]
- Sato, N.; Moriyama, T. Genomic and Biochemical Analysis of Lipid Biosynthesis in the Unicellular Rhodophyte Cyanidioschyzon merolae: Lack of a Plastidic Desaturation Pathway Results in the Coupled Pathway of Galactolipid Synthesis. Eukaryot. Cell 2007, 6, 1006–1017. [Google Scholar] [CrossRef] [Green Version]
- Berestovoy, M.A.; Pavlenko, O.S.; Goldenkova-Pavlova, I.V. Plant Fatty Acid Desaturases: Role in the Life of Plants and Biotechnological Potential. Biol. Bull. Rev. 2020, 10, 127–139. [Google Scholar] [CrossRef]
- Kachroo, A.; Shanklin, J.; Whittle, E.; Lapchyk, L.; Hildebrand, D.; Kachroo, P. The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol. Biol. 2006, 63, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Shanklin, J.; Cahoon, E.B. Desaturation and Related Modifications of Fatty Acids. Annu. Rev. Plant Biol. 1998, 49, 611–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández, M.L.; Sicardo, M.D.; Martínez-Rivas, J.M. Differential Contribution of Endoplasmic Reticulum and Chloroplast ω-3 Fatty Acid Desaturase Genes to the Linolenic Acid Content of Olive (Olea europaea) Fruit. Plant Cell Physiol. 2015, 57, 138–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, M.A.; Dauk, M.; Ramadan, H.; Yang, H.; Seamons, L.E.; Haslam, R.P.; Beaudoin, F.; Ramirez-Erosa, I.; Forseille, L. Involvement of Arabidopsis ACYL-COENZYME A DESATURASE-LIKE2 (At2g31360) in the Biosynthesis of the Very-Long-Chain Monounsaturated Fatty Acid Components of Membrane Lipids. Plant Physiol. 2012, 161, 81–96. [Google Scholar] [CrossRef] [Green Version]
- Kunst, L.; Browse, J.; Somerville, C. A Mutant of Arabidopsis Deficient in Desaturation of Palmitic Acid in Leaf Lipids. Plant Physiol. 1989, 90, 943–947. [Google Scholar] [CrossRef] [Green Version]
- Jamieson, G.; Reid, E. The occurrence of hexadeca-7,10,13-trienoic acid in the leaf lipids of angiosperms. Phytochemistry 1971, 10, 1837–1843. [Google Scholar] [CrossRef]
- Chen, M.; Thelen, J.J. Acyl-Lipid Desaturase2 is Required for Chilling and Freezing Tolerance in Arabidopsis. Plant Cell 2013, 25, 1430–1444. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Ma, D.; Zhang, Z.; Wang, S.; Du, S.; Deng, X.; Yin, L. Plant lipid remodeling in response to abiotic stresses. Environ. Exp. Bot. 2019, 165, 174–184. [Google Scholar] [CrossRef]
- Bjornson, M.; Dandekar, A.; Dehesh, K. Determinants of timing and amplitude in the plant general stress response. J. Integr. Plant Biol. 2015, 58, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Song, N.; Hu, Z.; Li, Y.; Li, C.; Peng, F.; Yao, Y.; Peng, H.; Ni, Z.; Xie, C.; Sun, Q. Overexpression of a wheat stearoyl-ACP desaturase (SACPD) gene TaSSI2 in Arabidopsis ssi2 mutant compromise its resistance to powdery mildew. Gene 2013, 524, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.-Q.; Mu, J.-J.; Su, P.-S.; Wu, H.-Y.; Yu, G.; Wang, G.-P.; Wang, L.; Ma, X.; Li, A.-F.; Wang, H.-W.; et al. Multi-functional roles of TaSSI2 involved in Fusarium head blight and powdery mildew resistance and drought tolerance. J. Integr. Agric. 2018, 17, 368–380. [Google Scholar] [CrossRef]
- Kachroo, P.; Shanklin, J.; Shah, J.; Whittle, E.J.; Klessig, D.F. A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 2001, 98, 9448–9453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madi, L.; Wang, X.; Kobiler, I.; Lichter, A.; Prusky, D. Stress on avocado fruits regulates Δ9-stearoyl ACP desaturase expression, fatty acid composition, antifungal diene level and resistance to Colletotrichum gloeosporioides attack. Physiol. Mol. Plant Pathol. 2003, 62, 277–283. [Google Scholar] [CrossRef]
- McConn, M.; Browse, J. Polyunsaturated membranes are required for photosynthetic competence in a mutant of Arabidopsis. Plant J. 1998, 15, 521–530. [Google Scholar] [CrossRef]
- Lim, G.-H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty acid– and lipid-mediated signaling in plant defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef] [PubMed]
- Yaeno, T.; Matsuda, O.; Iba, K. Role of chloroplast trienoic fatty acids in plant disease defense responses. Plant J. 2004, 40, 931–941. [Google Scholar] [CrossRef]
- Vijayan, P.; Shockey, J.; Lévesque, C.A.; Cook, R.J.; Browse, J. A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 7209–7214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Q.-M.; Yu, K.; Xia, Y.; Shine, M.B.; Wang, C.; Navarre, D.; Kachroo, A.; Kachroo, P. Mono- and digalactosyldiacylglycerol lipids function nonredundantly to regulate systemic acquired resistance in plants. Cell Rep. 2014, 9, 1681–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueiredo, A.; Monteiro, F.; Fortes, A.M.; Bonow-Rex, M.; Zyprian, E.; Sousa, L.; Pais, M.S. Cultivar-specific kinetics of gene induction during downy mildew early infection in grapevine. Funct. Integr. Genom. 2012, 12, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Kortekamp, A.; Zyprian, E. Characterization ofPlasmopara-Resistance in grapevine usingin vitroplants. J. Plant Physiol. 2003, 160, 1393–1400. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Huala, E. The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001, 29, 102–105. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, Y.; De La Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2018, 47, D427–D432. [Google Scholar] [CrossRef]
- Vitulo, N.N.; Forcato, C.C.; Carpinelli, E.C.E.; Telatin, A.A.; Campagna, D.D.; D’Angelo, M.M.; Zimbello, R.R.; Corso, M.; Vannozzi, A.A.; Bonghi, C.C.; et al. A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 2014, 14, 99. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Small, I.; Peeters, N.; Legeai, F.; Lurin, C. Predotar: A tool for rapidly screening proteomes forN-terminal targeting sequences. Proteomics 2004, 4, 1581–1590. [Google Scholar] [CrossRef]
- Sperschneider, J.; Catanzariti, A.-M.; DeBoer, K.; Petre, B.; Gardiner, D.M.; Singh, K.B.; Dodds, P.N.; Taylor, J.M. Localizer: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017, 7, srep44598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelsson, O.; Brunak, S.; Von Heijne, G.; Nielsen, H.A. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteiro, F.; Sebastiana, M.; Pais, M.S.; Figueiredo, A. Reference Gene Selection and Validation for the Early Responses to Downy Mildew Infection in Susceptible and Resistant Vitis vinifera Cultivars. PLoS ONE 2013, 8, e72998. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laureano, G.; Cavaco, A.R.; Matos, A.R.; Figueiredo, A. Fatty Acid Desaturases: Uncovering Their Involvement in Grapevine Defence against Downy Mildew. Int. J. Mol. Sci. 2021, 22, 5473. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22115473
Laureano G, Cavaco AR, Matos AR, Figueiredo A. Fatty Acid Desaturases: Uncovering Their Involvement in Grapevine Defence against Downy Mildew. International Journal of Molecular Sciences. 2021; 22(11):5473. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22115473
Chicago/Turabian StyleLaureano, Gonçalo, Ana Rita Cavaco, Ana Rita Matos, and Andreia Figueiredo. 2021. "Fatty Acid Desaturases: Uncovering Their Involvement in Grapevine Defence against Downy Mildew" International Journal of Molecular Sciences 22, no. 11: 5473. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22115473