Differential Regulation of Gonadotropins as Revealed by Transcriptomes of Distinct LH and FSH Cells of Fish Pituitary
Abstract
:1. Introduction
2. Results and Discussion
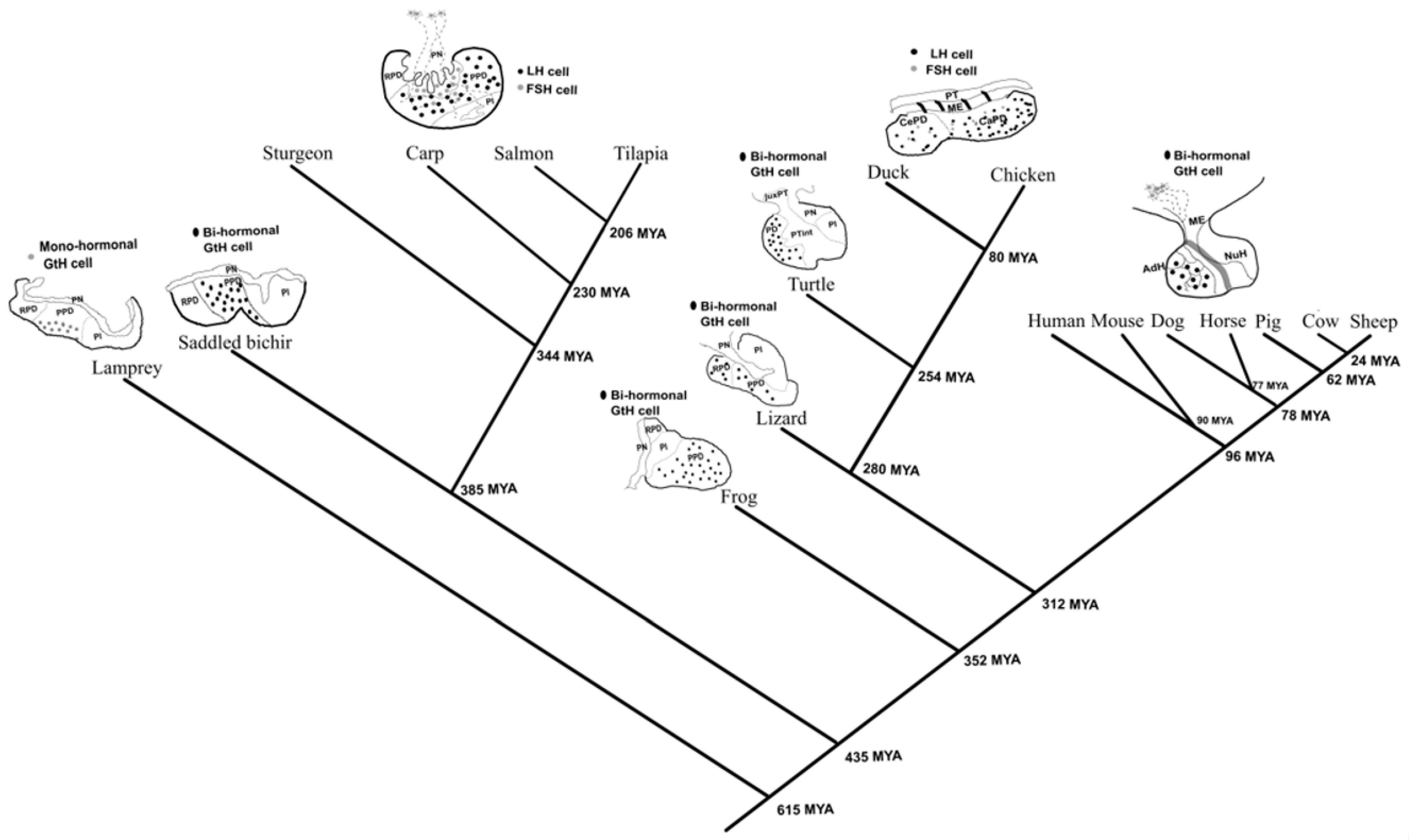
2.1. The Emergence of the Mono-Hormonal System from Bi-Hormonal Gonadotrophs
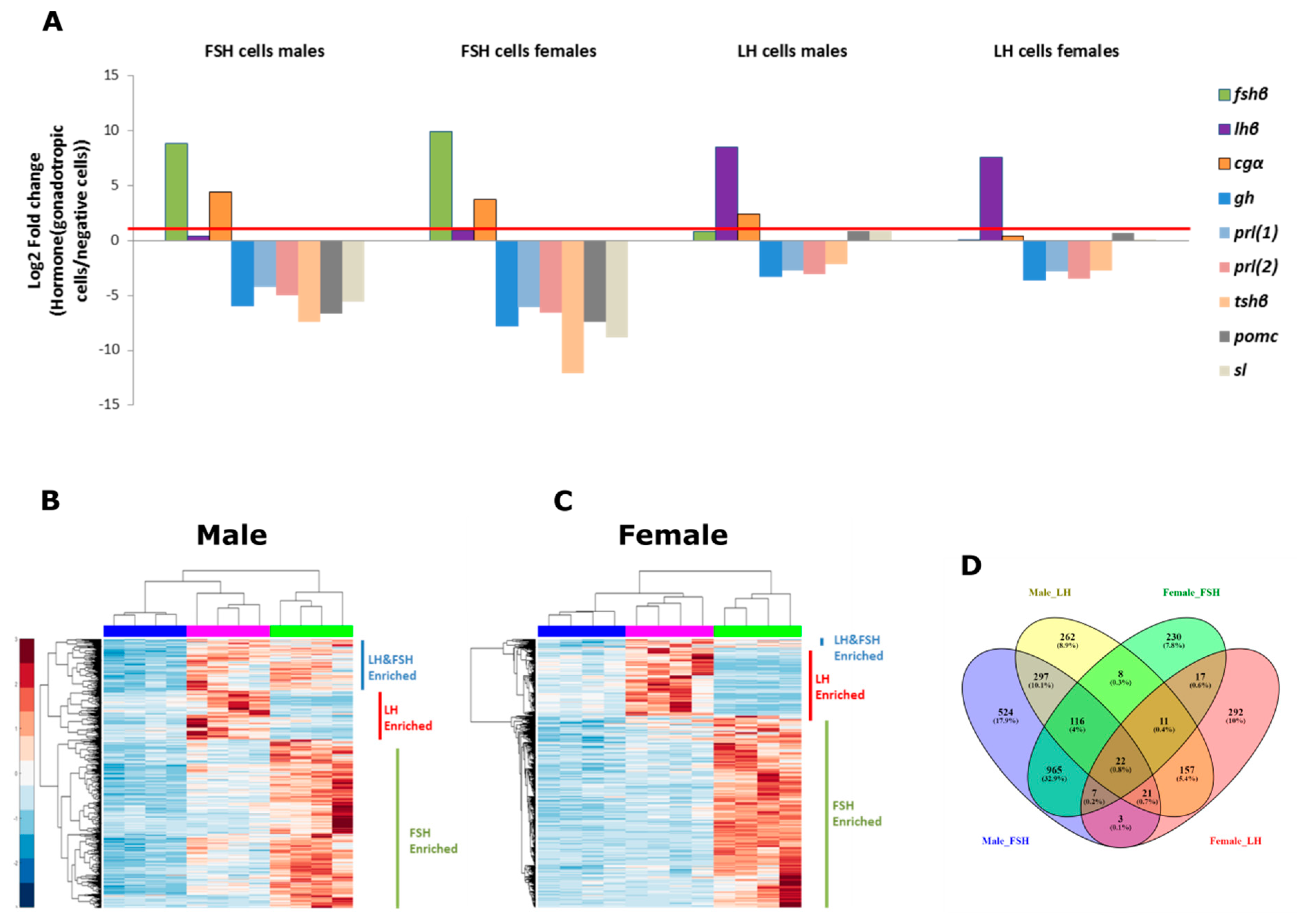
2.2. The Isolation of LH and FSH Cells
2.3. Identifying Enriched Genes in Both FSH and LH Cells
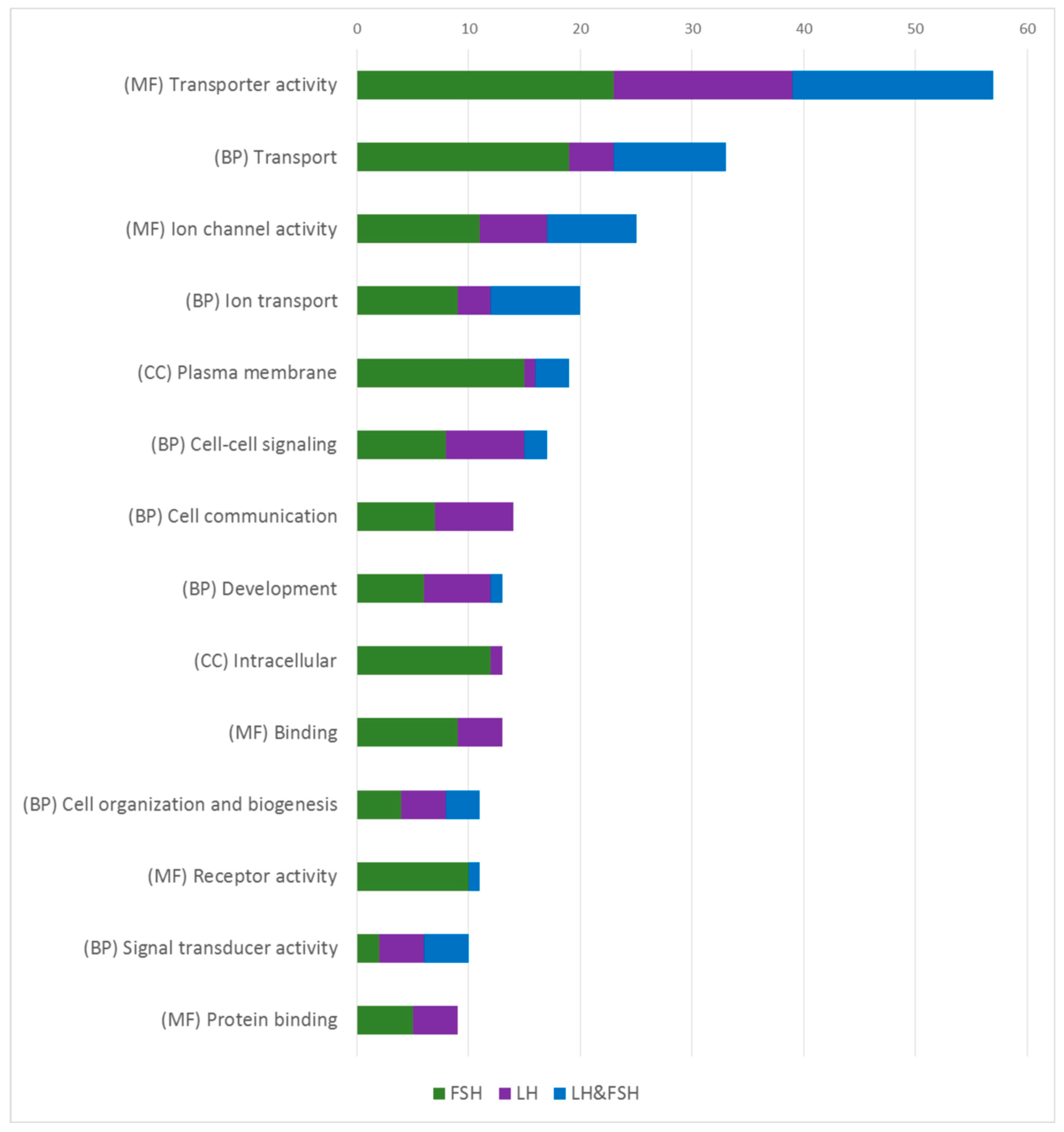
2.4. Functional Annotation of Enriched Genes in LH and FSH
2.5. Highly Enriched Genes in LH and FSH Cells
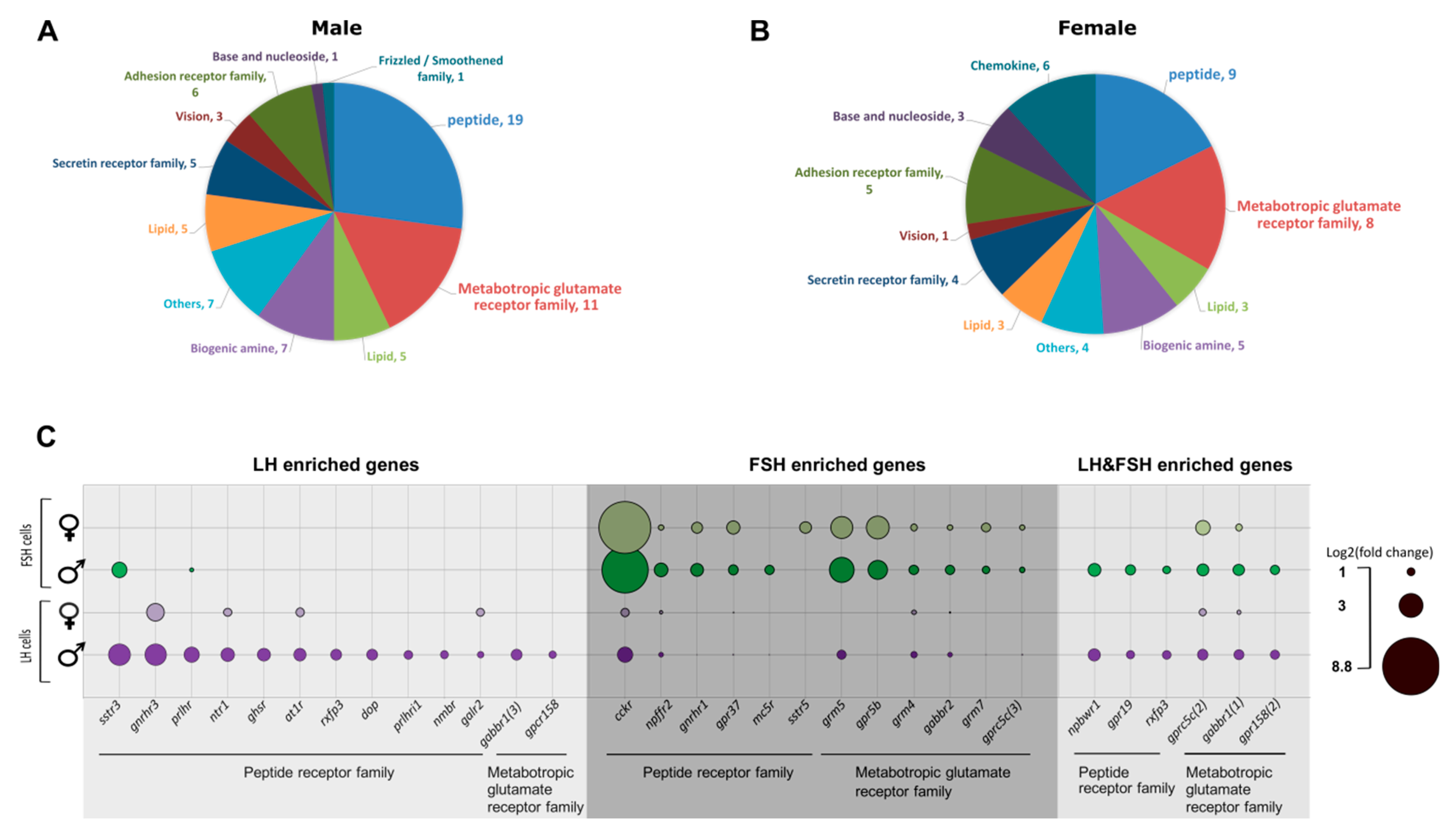
2.6. The Unique GPCR Signatures of Each Gonadotropic Cell
2.6.1. Hormones Directly Regulating LH Cells
2.6.2. Hormones Directly Regulating FSH Cells
2.7. Enriched Genes Conserved between Mammalian and Fish Gonadotrophs
3. Materials and Methods
3.1. Fish Care and Maintenance
3.2. Confocal Imaging
3.3. Isolation of LH and FSH Cells from Double Labeled Pituitaries
3.4. RNA-Seq Library Preparation of FACS Cells
3.5. RNA-Seq Library Processing, Mapping, and Annotation
3.6. Identifying Uniquely Expressed Genes in LH and FSH Cells
3.7. Functional Analysis of Enriched Genes in FSH and LH Cells
3.8. Real-Time PCR Validations
3.9. Comparing Tilapia and Rat Transcriptome Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Gonadotropin-releasing hormone | GnRH |
| Luteinizing-hormone | LH |
| Follicle-stimulating-hormone | FSH |
| Gonadotropin hormones | GtHs |
| Hypothalamus-pituitary-gonad | HPG |
| Rostral pars distalis | RPD |
| Proximal pars distalis | PPD |
| Pars intermedia | PI |
| Pars nervosa | PN |
| LHβ subunit | lhβ |
| FSHβ subunit | shβ |
| Growth hormone | gh |
| Prolactin | prl |
| Thyroid-stimulating hormone | tshβ |
| Somatolactin | sl |
| Pro-opiomelanocortin | pomc |
| Glycoprotein α-subunit | gcα |
| Normalized read | NR |
| Enriched genes | EG |
| Gene ontology | GO |
| Neurokinin B | NKB |
| Estrogen receptor 2 | esr2 |
| Progesterone receptor | pgr |
| 17α-20β-Dihydroprogesterone | DHP |
| Estradiol | E2 |
| Somatostatin receptor | sstr |
| Somatostatin | SST |
| Adropin receptor | Gpr19 |
| Neuropeptide W/neuropeptide B | NPWB |
| Relaxin-3 receptor | rxfp3 |
| Secretagogue receptor | ghsr |
| Prolactin-releasing peptide receptor | prlhr, gpr10 |
| Prolactin-releasing peptide | PrRP |
| Angiotensin II receptor type1 | at1r |
| Angiotensin-converting-enzyme 2 | ace2 |
| Neurotensin receptor | ntr1 |
| Delta opioid receptor | dop |
| Galanin receptor 2 | galr2 |
| Spexin | SPX |
| Neuromedin receptor B | nmbr |
| Neuromedin B | NMB |
| Cholecystokinin | CCK |
| Apelin | apln |
| Calmodulin regulator protein or PEP-19 | pcp4 |
| Phosphodiesterase 6H | pde6h |
| Calmodulin-dependent kinase II | CaMKII |
| G-protein coupled receptor | GPCR, gpr |
| Neuropeptide FF receptor 2 | npffr2 |
| Melanocortin receptor 5 | mc5r |
| Prosaposin receptor | gpcr37 |
| Gamma-Aminobutyric acid | GABA |
| GABA type B receptor subunit1 | gabbr1 |
| GABA type B receptor subunit | gabbr2 |
| Gap-junction delta 2 protein | gjd2b |
| Connexin 36 | Cx36 |
| Potassium voltage-gated channel, subfamily H | eag-related |
| member 6a | kcnh6a |
| Potassium voltage-gated channel subfamily A member 4 | kcna4 |
| Calcium-activated potassium channel subunit beta-2 | kcnmb2 |
| Androgen receptor | ar |
| G protein signaling | rgs |
| Adenylate cyclase-activating polypeptide type 1 receptor isoform 1 | adcyap1r1 |
| Latrophilin1 | adgrl1 |
| Dopamine receptor | drd |
References
- Levavi-Sivan, B.; Bogerd, J.; Mananos, E.L.; Gomez, A.; Lareyre, J.J. Perspectives on fish gonadotropins and their receptors. Gen. Comp. Endocrinol. 2010, 165, 412–437. [Google Scholar] [CrossRef]
- Marshall, G.R.; Bint Akhtar, F.; Weinbauer, G.F.; Nieschlag, E. Gonadotrophin-releasing hormone (GnRH) overcomes GnRH antagonist-induced suppression of LH secretion in primates. J. Endocrinol. 1986, 110, 145–150. [Google Scholar] [CrossRef]
- Stamatiades, G.A.; Kaiser, U.B. Gonadotropin regulation by pulsatile GnRH: Signaling and gene expression. Mol. Cell Endocrinol. 2018, 463, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Dalkin, A.C.; Haisenleder, D.J.; Ortolano, G.A.; Ellis, T.R.; Marshall, J.C. The Frequency of Gonadotropin-Releasing-Hormone Stimulation Differentially Regulates Gonadotropin Subunit Messenger Ribonucleic Acid Expression. Endocrinology 1989, 125, 917–923. [Google Scholar] [CrossRef] [PubMed]
- Thackray, V.G.; Mellon, P.L.; Coss, D. Hormones in synergy: Regulation of the pituitary gonadotropin genes. Mol. Cell Endocrinol. 2010, 314, 192–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denef, C. Paracrinicity: The story of 30 years of cellular pituitary crosstalk. J. Neuroendocrinol. 2008, 20, 1–70. [Google Scholar] [CrossRef]
- Zohar, Y.; Munoz-Cueto, J.A.; Elizur, A.; Kah, O. Neuroendocrinology of reproduction in teleost fish. Gen. Comp. Endocrinol. 2010, 165, 438–455. [Google Scholar] [CrossRef]
- Dufour, S.; Quérat, B.; Tostivint, H.; Pasqualini, C.; Vaudry, H.; Rousseau, K. Origin and Evolution of the Neuroendocrine Control of Reproduction in Vertebrates, With Special Focus on Genome and Gene Duplications. Physiol. Rev. 2020, 100, 869–943. [Google Scholar] [CrossRef]
- Biran, J.; Golan, M.; Mizrahi, N.; Ogawa, S.; Parhar, I.S.; Levavi-Sivan, B. Direct Regulation of Gonadotropin Release by Neurokinin B in Tilapia (Oreochromis niloticus). Endocrinology 2014, 155, 4831–4842. [Google Scholar] [CrossRef] [Green Version]
- Biran, J.; Ben-Dor, S.; Levavi-Sivan, B. Molecular identification and functional characterization of the kisspeptin/kisspeptin receptor system in lower vertebrates. Biol. Reprod. 2008, 79, 776–786. [Google Scholar] [CrossRef]
- Felip, A.; Zanuy, S.; Pineda, R.; Pinilla, L.; Carrillo, M.; Tena-Sempere, M.; Gómez, A. Evidence for two distinct KiSS genes in non-placental vertebrates that encode kisspeptins with different gonadotropin-releasing activities in fish and mammals. Mol. Cell. Endocrinol. 2009, 312, 61–71. [Google Scholar] [CrossRef] [Green Version]
- Hayakawa, Y.; Morita, T.; Kitamura, W.; Kanda, S.; Banba, A.; Nagaya, H.; Hotta, K.; Sohn, Y.C.; Yoshizaki, G.; Kobayashi, M. Biological activities of single-chain goldfish follicle-stimulating hormone and luteinizing hormone. Aquaculture 2008, 274, 408–415. [Google Scholar] [CrossRef]
- Trevisan, C.M.; Montagna, E.; de Oliveira, R.; Christofolini, D.M.; Barbosa, C.P.; Crandall, K.A.; Bianco, B. Kisspeptin/GPR54 System: What Do We Know About Its Role in Human Reproduction? Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 49, 1259–1276. [Google Scholar] [CrossRef] [PubMed]
- Biran, J.; Palevitch, O.; Ben-Dor, S.; Levavi Sivan, B. Neurokinin B and neurokinin B receptor: A novel system involved in controlling fish reproduction. Proc. Natl. Acad. Sci. USA 2012, 109, 10269–10274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, S.; Sivalingam, M.; Biran, J.; Golan, M.; Anthonysamy, R.S.; Levavi-Sivan, B.; Parhar, I.S. Distribution of LPXRFa, a gonadotropin-inhibitory hormone ortholog peptide, and LPXRFa receptor in the brain and pituitary of the tilapia. J. Comp. Neurol. 2016, 524, 2753–2775. [Google Scholar] [CrossRef]
- Cohen, Y.; Hausken, K.; Bonfil, Y.; Gutnick, M.; Levavi-Sivan, B. Spexin and a Novel Cichlid-Specific Spexin Paralog Both Inhibit FSH and LH Through a Specific Galanin Receptor (Galr2b) in Tilapia. Front. Endocrinol. 2020, 11, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golan, M.; Martin, A.O.; Mollard, P.; Levavi-Sivan, B. Anatomical and functional gonadotrope networks in the teleost pituitary. Sci. Rep. 2016, 6, 23777. [Google Scholar] [CrossRef] [Green Version]
- Golan, M.; Biran, J.; Levavi-Sivan, B. A novel model for development, organization and function of gonadotropes in fish pituitary. Front. Endocrinol. 2014, 5, 182. [Google Scholar] [CrossRef] [Green Version]
- Kah, O. A 45-years journey within the reproductive brain of fish. Gen. Comp. Endocrinol. 2020, 288, 113370. [Google Scholar] [CrossRef]
- Hausken, K.N.; Marquis, T.J.; Sower, S.A. Expression of two glycoprotein hormone receptors in larval, parasitic phase, and adult sea lampreys. Gen. Comp. Endocrinol. 2018, 264, 39–47. [Google Scholar] [CrossRef]
- Kawauchi, H.; Sower, S.A. The dawn and evolution of hormones in the adenohypophysis. Gen. Comp. Endocrinol. 2006, 148, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Marquis, T.J.; Nozaki, M.; Fagerberg, W.; Sower, S.A. Comprehensive histological and immunological studies reveal a novel glycoprotein hormone and thyrostimulin expressing proto-glycotrope in the sea lamprey pituitary. Cell Tissue Res. 2017, 367, 311–338. [Google Scholar] [CrossRef] [PubMed]
- Buechi, H.B.; Bridgham, J.T. Evolution of specificity in cartilaginous fish glycoprotein hormones and receptors. Gen. Comp. Endocrinol 2017, 246, 309–320. [Google Scholar] [CrossRef]
- Proudman, J.A.; Vandesande, F.; Berghman, L.R. Immunohistochemical evidence that follicle-stimulating hormone and luteinizing hormone reside in separate cells in the chicken pituitary. Biol. Reprod. 1999, 60, 1324–1328. [Google Scholar] [CrossRef] [PubMed]
- Hedges, S.B.; Marin, J.; Suleski, M.; Paymer, M.; Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015, 32, 835–845. [Google Scholar] [CrossRef]
- Fletcher, P.A.; Smiljanic, K.; Maso-Prévide, R.; Iben, J.R.; Li, T.; Rokic, M.B.; Sherman, A.; Coon, S.L.; Stojilkovic, S.S. Cell Type- and Sex-Dependent Transcriptome Profiles of Rat Anterior Pituitary Cells. Front. Endocrinol. 2019, 10, 623. [Google Scholar] [CrossRef]
- Cheung, L.Y.M.; George, A.S.; McGee, S.R.; Daly, A.Z.; Brinkmeier, M.L.; Ellsworth, B.S.; Camper, S.A. Single-Cell RNA Sequencing Reveals Novel Markers of Male Pituitary Stem Cells and Hormone-Producing Cell Types. Endocrinology 2018, 159, 3910–3924. [Google Scholar] [CrossRef] [Green Version]
- El Etreby, M.F.; El Bab, M.R.F. Localization of gonadotrophic hormones in the dog pituitary gland. Cell Tissue Res. 1977, 183, 167–175. [Google Scholar] [CrossRef]
- Dacheux, F. Ultrastructural localization of gonadotrophic hormones in the porcine pituitary using the immunoperoxidase technique. Cell Tissue Res. 1978, 191, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Newman, G.R.; Jasani, B.; Williams, E.D. Multiple hormone storage by cells of the human pituitary. J. Histochem. Cytochem. Off. J. Histochem. Soc. 1989, 37, 1183–1192. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.G.; Clarke, I.J. The Positive Feedback Action of Estrogen Mobilizes LH-Containing, but not FSH-Containing Secretory Granules in Ovine Gonadotropes. Endocrinology 1997, 138, 1347–1350. [Google Scholar] [CrossRef]
- Rahmanian, M.S.; Thompson, D.L., Jr.; Melrose, P.A. Immunocytochemical localization of luteinizing hormone and follicle-stimulating hormone in the equine pituitary. J. Anim. Sci. 1998, 76, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Kadokawa, H.; Pandey, K.; Nahar, A.; Nakamura, U.; Rudolf, F.O. Gonadotropin-releasing hormone (GnRH) receptors of cattle aggregate on the surface of gonadotrophs and are increased by elevated GnRH concentrations. Anim. Reprod. Sci. 2014, 150, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Sower, S.A.; Decatur, W.A.; Hausken, K.N.; Marquis, T.J.; Barton, S.L.; Gargan, J.; Freamat, M.; Wilmot, M.; Hollander, L.; Hall, J.A.; et al. Emergence of an Ancestral Glycoprotein Hormone in the Pituitary of the Sea Lamprey, a Basal Vertebrate. Endocrinology 2015, 156, 3026–3037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miki, M.; Shimotani, T.; Nozaki, M. Distribution of immunoreactivities for adenohypophysial hormones in the pituitary gland of the polypteriform fish, Polypterus endlicheri. Zool. Sci 2005, 22, 1123–1130. [Google Scholar] [CrossRef]
- Hollander-Cohen, L.; Golan, M.; Aizen, J.; Shpilman, M.; Levavi-Sivan, B. Characterization of carp gonadotropins: Structure, annual profile, and carp and zebrafish pituitary topographic organization. Gen. Comp. Endocrinol. 2018, 264, 28–38. [Google Scholar] [CrossRef]
- Nozaki, M.; Naito, N.; Swanson, P.; Dickhoff, W.W.; Nakai, Y.; Suzuki, K.; Kawauchi, H. Salmonid pituitary gonadotrophs. II. Ontogeny of GTH I and GTH II cells in the rainbow trout (Salmo gairdneri irideus). Gen. Comp. Endocrinol. 1990, 77, 358–367. [Google Scholar] [CrossRef]
- Yom-Din, S.; Hollander-Cohen, L.; Aizen, J.; Boehm, B.; Shpilman, M.; Golan, M.; Hurvitz, A.; Degani, G.; Levavi-Sivan, B. Gonadotropins in the Russian Sturgeon: Their Role in Steroid Secretion and the Effect of Hormonal Treatment on Their Secretion. PLoS ONE 2016, 11, e0162344. [Google Scholar] [CrossRef] [Green Version]
- Mikami, S.; Miyasaka, S.; Taniguchi, K. Light and electron microscopic immunocytochemistry of the pituitary gland of the tortoise. Arch. Histol. Jpn. 1985, 48, 373–388. [Google Scholar] [CrossRef] [Green Version]
- Chiba, A.; Sakai, H.; Sato, M.; Honma, R.; Murata, K.; Sugimori, F. Pituitary-gonadal axis and secondary sex characters in the spontaneously masculinized pintail, Anas acuta (Anatidae, Aves), with special regard to the gonadotrophs. Gen. Comp. Endocrinol. 2004, 137, 50–61. [Google Scholar] [CrossRef]
- Malashetty, V.B.; Sonar, A.; Patil, S.B. Anatomy and Histophysiological Changes in Pituitary of Calotes versicolor During Breeding and Nonbreeding Seasons. Int. J. Morphol. 2009, 27, 1223–1234. [Google Scholar] [CrossRef] [Green Version]
- Mizutani, F.; Iwasawa, H.; Tanaka, S. A morphometric analysis of the subcellular distribution of LH beta and FSH beta in secretory granules in the pituitary gonadotrophs of the frog (Rana japonica). Cell Tissue Res. 1994, 277, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Kanda, S. Evolution of the regulatory mechanisms for the hypothalamic-pituitary-gonadal axis in vertebrates-hypothesis from a comparative view. Gen. Comp. Endocrinol. 2019, 284, 113075. [Google Scholar] [CrossRef]
- Chang, J.P.; Pemberton, J.G. Comparative aspects of GnRH-Stimulated signal transduction in the vertebrate pituitary—Contributions from teleost model systems. Mol. Cell Endocrinol. 2018, 463, 142–167. [Google Scholar] [CrossRef] [PubMed]
- Golan, M.; Levavi-Sivan, B. Social dominance in tilapia is associated with gonadotroph hyperplasia. Gen. Comp. Endocrinol. 2013, 192, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Soengas, J.L.; Cerdá-Reverter, J.M.; Delgado, M.J. Central regulation of food intake in fish: An evolutionary perspective. J. Mol. Endocrinol. 2018, 60, R171–R199. [Google Scholar] [CrossRef]
- Yaron, Z.; Gur, G.; Melamed, P.; Rosenfeld, H.; Elizur, A.; Levavi-Sivan, B. Regulation of fish gonadotropins. Int. Rev. Cytol. 2003, 225, 131–185. [Google Scholar]
- Blanco, A.M. Hypothalamic- and pituitary-derived growth and reproductive hormones and the control of energy balance in fish. Gen. Comp. Endocrinol. 2020, 287, 113322. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.K.; Southey, B.R.; Romanova, E.V.; Maruska, K.P.; Sweedler, J.V.; Fernald, R.D. Identification of prohormones and pituitary neuropeptides in the African cichlid, Astatotilapia burtoni. BMC Genom. 2016, 17, 660. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, A.; Mizusawa, K. Posttranslational modifications of proopiomelanocortin in vertebrates and their biological significance. Front. Endocrinol. 2013, 4, 143. [Google Scholar] [CrossRef] [Green Version]
- Ocampo-Daza, D.; Larhammar, D. Evolution of the growth hormone, prolactin, prolactin 2 and somatolactin family. Gen. Comp. Endocrinol. 2018, 264, 94–112. [Google Scholar] [CrossRef] [PubMed]
- Fukamachi, S.; Sugimoto, M.; Mitani, H.; Shima, A. Somatolactin selectively regulates proliferation and morphogenesis of neural-crest derived pigment cells in medaka. Proc. Natl. Acad. Sci. USA 2004, 101, 10661–10666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stojilkovic, S.S.; Bjelobaba, I.; Zemkova, H. Ion Channels of Pituitary Gonadotrophs and Their Roles in Signaling and Secretion. Front. Endocrinol. 2017, 8, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waring, D.W.; Turgeon, J.L. Ca2+-activated K+ channels in gonadotropin-releasing hormone-stimulated mouse gonadotrophs. Endocrinology 2009, 150, 2264–2272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levavi-Sivan, B.; Bloch, C.L.; Gutnick, M.J.; Fleidervish, I.A. Electrotonic coupling in the anterior pituitary of a teleost fish. Endocrinology 2005, 146, 1048–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlegel, W.; Winiger, B.P.; Mollard, P.; Vacher, P.; Wuarin, F.; Zahnd, G.R.; Wollheim, C.B.; Dufy, B. Oscillations of cytosolic Ca2+ in pituitary cells due to action potentials. Nature 1987, 329, 719–721. [Google Scholar] [CrossRef] [PubMed]
- Mollard, P.; Schlegel, W. Why are endocrine pituitary cells excitable? Trends Endocrinol. Metab. Tem 1996, 7, 361–365. [Google Scholar] [CrossRef]
- Hodne, K.; Strandabo, R.A.; von Krogh, K.; Nourizadeh-Lillabadi, R.; Sand, O.; Weltzien, F.A.; Haug, T.M. Electrophysiological differences between fshb- and lhb-expressing gonadotropes in primary culture. Endocrinology 2013, 154, 3319–3330. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, P.A.; Sherman, A.; Stojilkovic, S.S. Common and diverse elements of ion channels and receptors underlying electrical activity in endocrine pituitary cells. Mol. Cell. Endocrinol. 2018, 463, 23–36. [Google Scholar] [CrossRef]
- Karigo, T.; Aikawa, M.; Kondo, C.; Abe, H.; Kanda, S.; Oka, Y. Whole Brain-Pituitary In Vitro Preparation of the Transgenic Medaka (Oryzias latipes) as a Tool for Analyzing the Differential Regulatory Mechanisms of LH and FSH Release. Endocrinology 2014, 155, 536–547. [Google Scholar] [CrossRef] [Green Version]
- Levavi-Sivan, B.; Biran, J.; Fireman, E. Sex steroids are involved in the regulation of gonadotropin-releasing hormone and dopamine D2 receptors in female tilapia pituitary. Biol. Reprod. 2006, 75, 642–650. [Google Scholar] [CrossRef] [Green Version]
- Prizant, H.; Gleicher, N.; Sen, A. Androgen actions in the ovary: Balance is key. J. Endocrinol. 2014, 222, R141–R151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molés, G.; Hausken, K.; Carrillo, M.; Zanuy, S.; Levavi-Sivan, B.; Gómez, A. Generation and use of recombinant gonadotropins in fish. Gen. Comp. Endocrinol. 2020, 299, 113555. [Google Scholar] [CrossRef]
- Michalakis, K.; Mintziori, G.; Kaprara, A.; Tarlatzis, B.C.; Goulis, D.G. The complex interaction between obesity, metabolic syndrome and reproductive axis: A narrative review. Metab. Clin. Exp. 2013, 62, 457–478. [Google Scholar] [CrossRef] [PubMed]
- Shahjahan, M.; Kitahashi, T.; Parhar, I.S. Central pathways integrating metabolism and reproduction in teleosts. Front. Endocrinol. 2014, 5, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, J.E. Energy balance and reproduction. Physiol. Behav. 2004, 81, 289–317. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Fernandez, R.; Martini, A.C.; Navarro, V.M.; Castellano, J.M.; Dieguez, C.; Aguilar, E.; Pinilla, L.; Tena-Sempere, M. Novel signals for the integration of energy balance and reproduction. Mol. Cell. Endocrinol. 2006, 254–255, 127–132. [Google Scholar] [CrossRef]
- Castellano, J.M.; Roa, J.; Luque, R.M.; Dieguez, C.; Aguilar, E.; Pinilla, L.; Tena-Sempere, M. KiSS-1/kisspeptins and the metabolic control of reproduction: Physiologic roles and putative physiopathological implications. Peptides 2009, 30, 139–145. [Google Scholar] [CrossRef]
- Hasebe, M.; Kanda, S.; Oka, Y. Female-Specific Glucose Sensitivity of GnRH1 Neurons Leads to Sexually Dimorphic Inhibition of Reproduction in Medaka. Endocrinology 2016, 157, 4318–4329. [Google Scholar] [CrossRef]
- Rehfeld, J.F. Cholecystokinin-From Local Gut Hormone to Ubiquitous Messenger. Front. Endocrinol. 2017, 8, 47. [Google Scholar] [CrossRef]
- Roche, J.; Ramé, C.; Reverchon, M.; Mellouk, N.; Rak, A.; Froment, P.; Dupont, J. Apelin (APLN) regulates progesterone secretion and oocyte maturation in bovine ovarian cells. Reproduction 2017, 153, 589–603. [Google Scholar] [CrossRef] [Green Version]
- Kurowska, P.; Barbe, A.; Różycka, M.; Chmielińska, J.; Dupont, J.; Rak, A. Apelin in Reproductive Physiology and Pathology of Different Species: A Critical Review. Int. J. Endocrinol. 2018, 2018, 9170480. [Google Scholar] [CrossRef] [PubMed]
- Sandal, S.; Tekin, S.; Seker, F.B.; Beytur, A.; Vardi, N.; Colak, C.; Tapan, T.; Yildiz, S.; Yilmaz, B. The effects of intracerebroventricular infusion of apelin-13 on reproductive function in male rats. Neurosci. Lett. 2015, 602, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Taheri, S.; Murphy, K.; Cohen, M.; Sujkovic, E.; Kennedy, A.; Dhillo, W.; Dakin, C.; Sajedi, A.; Ghatei, M.; Bloom, S. The effects of centrally administered apelin-13 on food intake, water intake and pituitary hormone release in rats. Biochem. Biophys. Res. Commun. 2002, 291, 1208–1212. [Google Scholar] [CrossRef]
- Tuziak, S.M.; Rise, M.L.; Volkoff, H. An investigation of appetite-related peptide transcript expression in Atlantic cod (Gadus morhua) brain following a Camelina sativa meal-supplemented feeding trial. Gene 2014, 550, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Volkoff, H. In vitro assessment of interactions between appetite-regulating peptides in brain of goldfish (Carassius auratus). Peptides 2014, 61, 61–68. [Google Scholar] [CrossRef]
- Hao, J.; Liu, Q.; Zhang, X.; Wu, Y.; Zhu, J.; Qi, J.; Tang, N.; Wang, S.; Wang, H.; Chen, D.; et al. The evidence of apelin has the bidirectional effects on feeding regulation in Siberian sturgeon (Acipenser baerii). Peptides 2017, 94, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Pauli, A.; Norris, M.L.; Valen, E.; Chew, G.L.; Gagnon, J.A.; Zimmerman, S.; Mitchell, A.; Ma, J.; Dubrulle, J.; Reyon, D.; et al. Toddler: An embryonic signal that promotes cell movement via Apelin receptors. Science 2014, 343, 1248636. [Google Scholar] [CrossRef] [PubMed]
- Rajeswari, J.J.; Unniappan, S. Phoenixin-20 Stimulates mRNAs Encoding Hypothalamo-Pituitary-Gonadal Hormones, is Pro-Vitellogenic, and Promotes Oocyte Maturation in Zebrafish. Sci. Rep. 2020, 10, 6264. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Chen, H.-P.; Zhai, Y.; Jiang, D.-n.; Liu, J.-Y.; Tian, C.-x.; Wu, T.-l.; Zhu, C.-H.; Deng, S.-P.; Li, G.-L. Phoenixin: Expression at different ovarian development stages and effects on genes ralated to reproduction in spotted scat, Scatophagus argus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 228, 17–25. [Google Scholar] [CrossRef]
- Avitan, A.; Zelinger, E.; Levavi-Sivan, B. Homologous desensitization and visualization of the tilapia GnRH type 3 receptor. Gen. Comp. Endocrinol. 2007, 153, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Parhar, I.S.; Ogawa, S.; Sakuma, Y. Three GnRH receptor types in laser-captured single cells of the cichlid pituitary display cellular and functional heterogeneity. Proc. Natl. Acad. Sci. USA. 2005, 102, 2204–2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunningham, M.J.; Clifton, D.K.; Steiner, R.A. Leptin’s actions on the reproductive axis: Perspectives and mechanisms. Biol. Reprod. 1999, 60, 216–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, J.W.; Elmquist, J.K.; Elias, C.F. Hypothalamic pathways linking energy balance and reproduction. Am. J. Physiol. Endocrinol. Metab. 2008, 294, E827–E832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melamed, P.; Eliahu, N.; Levavi-Sivan, B.; Ofir, M.; Farchi-Pisanty, O.; Rentier-Delrue, F.; Smal, J.; Yaron, Z.; Naor, Z. Hypothalamic and thyroidal regulation of growth hormone in tilapia. Gen. Comp. Endocrinol. 1995, 97, 13–30. [Google Scholar] [CrossRef]
- Gahete, M.D.; Cordoba-Chacón, J.; Duran-Prado, M.; Malagón, M.M.; Martinez-Fuentes, A.J.; Gracia-Navarro, F.; Luque, R.M.; Castaño, J.P. Somatostatin and its receptors from fish to mammals. Ann. N. Y. Acad. Sci. 2010, 1200, 43–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillon, D.; Caraty, A.; Fabre-Nys, C.; Lomet, D.; Cateau, M.; Bruneau, G. Regulation by estradiol of hypothalamic somatostatin gene expression: Possible involvement of somatostatin in the control of luteinizing hormone secretion in the ewe. Biol. Reprod. 2004, 71, 38–44. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, A.; Tsuchida, H.; Ieda, N.; Ikegami, K.; Inoue, N.; Uenoyama, Y.; Tsukamura, H. Somatostatin-Somatostatin Receptor 2 Signaling Mediates LH Pulse Suppression in Lactating Rats. Endocrinology 2019, 160, 473–483. [Google Scholar] [CrossRef] [Green Version]
- Jasaszwili, M.; Billert, M.; Strowski, M.Z.; Nowak, K.W.; Skrzypski, M. Adropin as A Fat-Burning Hormone with Multiple Functions-Review of a Decade of Research. Molecules 2020, 25, 549. [Google Scholar] [CrossRef] [Green Version]
- Ghoshal, S.; Stevens, J.R.; Billon, C.; Girardet, C.; Sitaula, S.; Leon, A.S.; Rao, D.C.; Skinner, J.S.; Rankinen, T.; Bouchard, C.; et al. Adropin: An endocrine link between the biological clock and cholesterol homeostasis. Mol. Metab. 2018, 8, 51–64. [Google Scholar] [CrossRef]
- Chottova-Dvorakova, M. Distribution and Function of Neuropeptides W/B Signaling System. Front. Physiol. 2018, 9, 981. [Google Scholar] [CrossRef]
- Ganella, D.; Ma, S.; Gundlach, A. Relaxin-3/RXFP3 Signaling and Neuroendocrine Function—A Perspective on Extrinsic Hypothalamic Control. Front. Endocrinol. 2013, 4, 128. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Yang, P.; Zhang, Q.; Jiang, Q. Tilapia adropin: The localization and regulation of growth hormone gene expression in pituitary cells. Peptides 2017, 97, 1–7. [Google Scholar] [CrossRef]
- McGowan, B.M.; Stanley, S.A.; Donovan, J.; Thompson, E.L.; Patterson, M.; Semjonous, N.M.; Gardiner, J.V.; Murphy, K.G.; Ghatei, M.A.; Bloom, S.R. Relaxin-3 stimulates the hypothalamic-pituitary-gonadal axis. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E278–E286. [Google Scholar] [CrossRef]
- Kanasaki, H.; Tumurbaatar, T.; Tumurgan, Z.; Oride, A.; Okada, H.; Kyo, S. Effect of relaxin-3 on Kiss-1, gonadotropin-releasing hormone, and gonadotropin subunit gene expression. Reprod. Med. Biol. 2019, 18, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.C.; Burnett, D.; Rappaport, R.; Parry, L.J.; Fletcher, E.K. Relaxin-3 and RXFP3 expression, and steroidogenic actions in the ovary of teleost fish. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2009, 153, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Tena-Sempere, M. Ghrelin and reproduction: Ghrelin as novel regulator of the gonadotropic axis. Vitam Horm. 2008, 77, 285–300. [Google Scholar] [CrossRef]
- Unniappan, S.; Peter, R.E. In vitro and in vivo effects of ghrelin on luteinizing hormone and growth hormone release in goldfish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004, 286, R1093–R1101. [Google Scholar] [CrossRef] [PubMed]
- Grey, C.L.; Chang, J.P. Ghrelin-induced growth hormone release from goldfish pituitary cells involves voltage-sensitive calcium channels. Gen. Comp. Endocrinol 2009, 160, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Sokolowska-Mikolajczyk, M.; Socha, M.; Szczerbik, P.; Epler, P. The effects of ghrelin on the in vitro spontaneous and sGnRH-A stimulated luteinizing hormone (LH) release from the pituitary cells of common carp (Cyprinus carpio L.). Comp. Biochem. Physiol. Part. A Mol. Integr. Physiol. 2009, 153, 386–390. [Google Scholar] [CrossRef]
- Seale, A.P.; Yamaguchi, Y.; Johnstone, W.M., 3rd; Borski, R.J.; Lerner, D.T.; Grau, E.G. Endocrine regulation of prolactin cell function and modulation of osmoreception in the Mozambique tilapia. Gen. Comp. Endocrinol. 2013, 192, 191–203. [Google Scholar] [CrossRef]
- Aruna, A.; Nagarajan, G.; Chang, C.F. The acute salinity changes activate the dual pathways of endocrine responses in the brain and pituitary of tilapia. Gen. Comp. Endocrinol. 2015, 211, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, Y.; Iijima, N.; Yano, T.; Kakihara, K.; Hayashi, S.; Hinuma, S.; Honjo, H.; Hayashi, S.; Tanaka, M.; Ibata, Y. Gonadal regulation of PrRP mRNA expression in the nucleus tractus solitarius and ventral and lateral reticular nuclei of the rat. Brain Res. Mol. Brain Res. 2001, 87, 42–47. [Google Scholar] [CrossRef]
- Ganong, W.F. Blood, pituitary, and brain renin-angiotensin systems and regulation of secretion of anterior pituitary gland. Front. Neuroendocrinol. 1993, 14, 233–249. [Google Scholar] [CrossRef]
- Wong, M.K.; Takei, Y.; Woo, N.Y. Differential status of the renin-angiotensin system of silver sea bream (Sparus sarba) in different salinities. Gen. Comp. Endocrinol. 2006, 149, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Bobe, J.; Montfort, J.; Nguyen, T.; Fostier, A. Identification of new participants in the rainbow trout (Oncorhynchus mykiss) oocyte maturation and ovulation processes using cDNA microarrays. Reprod. Biol. Endocrinol. RbE 2006, 4, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boules, M.; Li, Z.; Smith, K.; Fredrickson, P.; Richelson, E. Diverse roles of neurotensin agonists in the central nervous system. Front. Endocrinol. 2013, 4, 36. [Google Scholar] [CrossRef] [Green Version]
- Torruella-Suárez, M.L.; McElligott, Z.A. Neurotensin in reward processes. Neuropharmacology 2020, 167, 108005. [Google Scholar] [CrossRef]
- McCann, S.M.; Vijayan, E.; Koenig, J.; Krulich, L. The effects of neurotensin on anterior pituitary hormone secretion. Ann. N. Y. Acad. Sci. 1982, 400, 160–171. [Google Scholar] [CrossRef]
- Dobek, E.; Górski, K.; Romanowicz, K.; Misztal, T. Different types of opioid receptors involved in the suppression of LH secretion in lactating sheep. Anim. Reprod. Sci. 2013, 141, 62–67. [Google Scholar] [CrossRef]
- Drobnis, E.Z.; Nangia, A.K. Pain Medications and Male Reproduction. Adv. Exp. Med. Biol. 2017, 1034, 39–57. [Google Scholar] [CrossRef]
- Sokolowska-Mikolajczyk, M.; Socha, M.; Mikolajczyk, T.; Chyb, J.; Epler, P. The effects of naltrexone, an opioid receptor antagonist, on plasma LH levels in common carp (Cyprinus carpio L.). Comp. Biochem. Physiol. Toxicol. Pharmacol. Cbp 2002, 131, 417–424. [Google Scholar] [CrossRef]
- Sokolowska-Mikolajczyk, M.; Socha, M.; Mikolajczyk, T.; Chyb, J.; Epler, P. Seasonal short-term effects of naltrexone on LH secretion in male carp (Cyprinus carpio L.). Comp. Biochem. Physiol. Toxicol. Pharmacol. Cbp 2002, 131, 379–385. [Google Scholar] [CrossRef]
- Vijayalaxmi; Ganesh, C.B. Influence of leucine-enkephalin on pituitary-ovary axis of the cichlid fish Oreochromis mossambicus. Fish. Physiol. Biochem. 2017, 43, 1253–1264. [Google Scholar] [CrossRef]
- López, F.J.; Merchenthaler, I.; Ching, M.; Wisniewski, M.G.; Negro-Vilar, A. Galanin: A hypothalamic-hypophysiotropic hormone modulating reproductive functions. Proc. Natl. Acad. Sci. USA 1991, 88, 4508–4512. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.K.; Yun, S.; Son, G.H.; Hwang, J.I.; Park, C.R.; Kim, J.I.; Kim, K.; Vaudry, H.; Seong, J.Y. Coevolution of the spexin/galanin/kisspeptin family: Spexin activates galanin receptor type II and III. Endocrinology 2014, 155, 1864–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohki-Hamazaki, H. Neuromedin B. Prog. Neurobiol. 2000, 62, 297–312. [Google Scholar] [CrossRef]
- Ma, Z.; Su, J.; Guo, T.; Jin, M.; Li, X.; Lei, Z.; Hou, Y.; Li, X.; Jia, C.; Zhang, Z.; et al. Neuromedin B and Its Receptor: Gene Cloning, Tissue Distribution and Expression Levels of the Reproductive Axis in Pigs. PLoS ONE 2016, 11, e0151871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Culler, M.D.; Negro-Vilar, A. Pulsatile Follicle-Stimulating Hormone Secretion Is Independent of Luteinizing Hormone-Releasing Hormone (LHRH): Pulsatile Replacement of LHRH Bioactivity in LHRH-Immunoneutralized Rats. Endocrinology 1987, 120, 2011–2021. [Google Scholar] [CrossRef] [PubMed]
- Pau, K.Y.F.; Gliessman, P.M.; Oyama, T.; Spies, H.G. Disruption of GnRH Pulses by Anti-GnRH Serum and Phentolamine Obliterates Pulsatile LH but not FSH Secretion in Ovariectomized Rabbits. Neuroendocrinology 1991, 53, 382–391. [Google Scholar] [CrossRef]
- Padmanabhan, V.; McFadden, K.; Mauger, D.T.; Karsch, F.J.; Midgley, A.R., Jr. Neuroendocrine control of follicle-stimulating hormone (FSH) secretion. I. Direct evidence for separate episodic and basal components of FSH secretion. Endocrinology 1997, 138, 424–432. [Google Scholar] [CrossRef]
- Padmanabhan, V.; Cardoso, R.C. Neuroendocrine, autocrine, and paracrine control of follicle-stimulating hormone secretion. Mol. Cell. Endocrinol. 2020, 500, 110632. [Google Scholar] [CrossRef]
- Dickey, J.T.; Swanson, P. Effects of salmon gonadotropin-releasing hormone on follicle stimulating hormone secretion and subunit gene expression in coho salmon (Oncorhynchus kisutch). Gen. Comp. Endocrinol. 2000, 118, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, W.; Zhang, M.; Xu, B.; Shi, X.; Zhang, Q.; Guo, Y.; Xiao, J.; Chen, D.; Zheng, T.; et al. Analgesic activities of the mixed opioid and NPFF receptors agonist DN-9 in a mouse model of formalin-induced orofacial inflammatory pain. Peptides 2018, 110, 30–39. [Google Scholar] [CrossRef]
- Lin, Y.T.; Chen, J.C. Neuropeptide FF modulates neuroendocrine and energy homeostasis through hypothalamic signaling. Chin. J. Physiol. 2019, 62, 47–52. [Google Scholar] [CrossRef]
- Hu, K.L.; Chang, H.M.; Li, R.; Yu, Y.; Qiao, J. Regulation of LH secretion by RFRP-3—From the hypothalamus to the pituitary. Front. Neuroendocrinol. 2019, 52, 12–21. [Google Scholar] [CrossRef]
- Schioth, H.B.; Watanobe, H. Melanocortins and reproduction. Brain Res. Brain Res. Rev. 2002, 38, 340–350. [Google Scholar] [CrossRef]
- Wikberg, J.E.; Muceniece, R.; Mandrika, I.; Prusis, P.; Lindblom, J.; Post, C.; Skottner, A. New aspects on the melanocortins and their receptors. Pharmacol. Res. 2000, 42, 393–420. [Google Scholar] [CrossRef] [PubMed]
- Dores, R.M.; Oberer, N.; Hoglin, B.; Thomas, A.; Faught, E.; Vijayan, M.M. Evaluating interactions between the melanocortin-5 receptor, MRAP1, and ACTH(1-24): A phylogenetic study. Gen. Comp. Endocrinol. 2020, 294, 113476. [Google Scholar] [CrossRef]
- Morgan, C.; Cone, R.D. Melanocortin-5 receptor deficiency in mice blocks a novel pathway influencing pheromone-induced aggression. Behav. Genet. 2006, 36, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Metz, J.R.; Peters, J.J.; Flik, G. Molecular biology and physiology of the melanocortin system in fish: A review. Gen. Comp. Endocrinol. 2006, 148, 150–162. [Google Scholar] [CrossRef]
- Meyer, R.C.; Giddens, M.M.; Coleman, B.M.; Hall, R.A. The protective role of prosaposin and its receptors in the nervous system. Brain Res. 2014, 1585, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van den Pol, A.N. Weighing the role of hypothalamic feeding neurotransmitters. Neuron 2003, 40, 1059–1061. [Google Scholar] [CrossRef] [Green Version]
- Pinilla, L.; González, L.C.; Tena-Sempere, M.; Aguilar, E. Interactions between GABAergic and aminoacidergic pathways in the control of gonadotropin and GH secretion in pre-pubertal female rats. J. Endocrinol. Investig. 2002, 25, 96–100. [Google Scholar] [CrossRef]
- Margeta-Mitrovic, M.; Jan, Y.N.; Jan, L.Y. Function of GB1 and GB2 subunits in G protein coupling of GABA(B) receptors. Proc. Natl. Acad. Sci. USA 2001, 98, 14649–14654. [Google Scholar] [CrossRef] [Green Version]
- Göngrich, C.; García-González, D.; Le Magueresse, C.; Roth, L.C.; Watanabe, Y.; Burks, D.J.; Grinevich, V.; Monyer, H. Electrotonic Coupling in the Pituitary Supports the Hypothalamic-Pituitary-Gonadal Axis in a Sex Specific Manner. Front. Mol. Neurosci. 2016, 9, 65. [Google Scholar] [CrossRef]
- Latorre, R.; Castillo, K.; Carrasquel-Ursulaez, W.; Sepulveda, R.V.; Gonzalez-Nilo, F.; Gonzalez, C.; Alvarez, O. Molecular Determinants of BK Channel Functional Diversity and Functioning. Physiol Rev. 2017, 97, 39–87. [Google Scholar] [CrossRef] [PubMed]
- Contreras, G.F.; Neely, A.; Alvarez, O.; Gonzalez, C.; Latorre, R. Modulation of BK channel voltage gating by different auxiliary beta subunits. Proc. Natl. Acad. Sci. USA 2012, 109, 18991–18996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, K.A.; Edwards, M.C.; Tesic, D.; Caldwell, A.S.L.; Jimenez, M.; Smith, J.T.; Handelsman, D.J. The Role of Central Androgen Receptor Actions in Regulating the Hypothalamic-Pituitary-Ovarian Axis. Neuroendocrinology 2018, 106, 389–400. [Google Scholar] [CrossRef]
- Guzmán, J.M.; Luckenbach, J.A.; da Silva, D.A.M.; Hayman, E.S.; Ylitalo, G.M.; Goetz, F.W.; Swanson, P. Seasonal variation of pituitary gonadotropin subunit, brain-type aromatase and sex steroid receptor mRNAs, and plasma steroids during gametogenesis in wild sablefish. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2018, 219–220, 48–57. [Google Scholar] [CrossRef]
- Peñaranda, D.S.; Mazzeo, I.; Gallego, V.; Hildahl, J.; Nourizadeh-Lillabadi, R.; Pérez, L.; Weltzien, F.A.; Asturiano, J.F. The regulation of aromatase and androgen receptor expression during gonad development in male and female European eel. Reprod. Domest. Anim. 2014, 49, 512–521. [Google Scholar] [CrossRef]
- Burmeister, S.S.; Kailasanath, V.; Fernald, R.D. Social dominance regulates androgen and estrogen receptor gene expression. Horm. Behav. 2007, 51, 164–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köves, K.; Szabó, E.; Kántor, O.; Heinzlmann, A.; Szabó, F.; Csáki, Á. Current State of Understanding of the Role of PACAP in the Hypothalamo-Hypophyseal Gonadotropin Functions of Mammals. Front. Endocrinol. 2020, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.P.; Johnson, J.D.; Sawisky, G.R.; Grey, C.L.; Mitchell, G.; Booth, M.; Volk, M.M.; Parks, S.K.; Thompson, E.; Goss, G.G.; et al. Signal transduction in multifactorial neuroendocrine control of gonadotropin secretion and synthesis in teleosts-studies on the goldfish model. Gen. Comp. Endocrinol. 2009, 161, 42–52. [Google Scholar] [CrossRef]
- Gur, G.; Bonfil, D.; Safarian, H.; Naor, Z.; Yaron, Z. Pituitary adenylate cyclase activating polypeptide and neuropeptide Y regulation of gonadotropin subunit gene expression in tilapia: Role of PKC, PKA and ERK. Neuroendocrinology 2002, 75, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Hlubek, M.; Tian, D.; Stuenkel, E.L. Mechanism of alpha-latrotoxin action at nerve endings of neurohypophysis. Brain Res. 2003, 992, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Levavi-Sivan, B.; Yaron, Z. Involvement of cyclic adenosine monophosphate in the stimulation of gonadotropin secretion from the pituitary of the teleost fish, tilapia. Mol. Cell. Endocrinol. 1992, 85, 175–182. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M. NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, S.; Ishida, T.; Ohue, M.; Kakuta, M.; Akiyama, Y. GHOSTX: A Fast Sequence Homology Search Tool for Functional Annotation of Metagenomic Data. Methods Mol. Biol. 2017, 1611, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [Green Version]
- Hollander-Cohen, L.; Böhm, B.; Hausken, K.; Levavi-Sivan, B. Ontogeny of the specificity of gonadotropin receptors and gene expression in carp. Endocr. Connect. 2019, 8, 1433–1446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]


| Name | Protein Name | TOP 10 | Up Regulated Also In | |
|---|---|---|---|---|
| Gonadotrophs secretion | fshβ | follitropin subunit beta isoform X1 | FSH(M&F) | non |
| lhβ | gonadotropin subunit beta-2 | LH(M&F) | non | |
| esr2 | estrogen receptor beta isoform X2 | LH&FSH(M&F) | ||
| pgr | progesterone receptor | LH(F) | LH(M) | |
| Neuroactive ligand receptor interaction | cckr | cholecystokinin receptor | FSH(M&F) | LH (M&F) |
| apln | apelin | LH(M) | FSH(M) LH&FSH(F) | |
| Cell signaling | pdeh6h | retinal cone rhodopsin-sensitive cGMP 3\′,5\′-cyclic phosphodiesterase subunit gamma | FSH(M&F) | |
| pcp4 | calmodulin regulator protein PCP4 isoform X3 | LH(M) FSH(F) | FSH(M) LH(F) | |
| Cell morphology | xirp2 | xin actin-binding repeat-containing protein 2 isoform X1 | FSH(F) | FSH(M) LH(F) |
| thbs4b | thrombospondin-4-B | LH(M&F) | ||
| col5a2 | collagen alpha-2(V) chain | LH(F) | FSH(F) LH(M) | |
| cdh16 | cadherin-16 | FSH(F) | FSH(M) | |
| Post translation modification | ndst4 | bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 isoform X1 | LH(M) | FSH(M&F) LH(F) |
| dclk3 | serine/threonine-protein kinase DCLK3 isoform X1 | LH(M) | FSH(M) LH&FSH(F) | |
| pcdstpk | probable cyclin-dependent serine/threonine-protein kinase DDB_G0292550 | LH(M&F) | ||
| tyr | tyrosinase | FSH(M) | ||
| Protein trafficking | tnfaip2 | tumor necrosis factor alpha-induced protein 2 | FSH(M&F) | |
| tnfaip2 | tumor necrosis factor alpha-induced protein 2 | FSH(M&F) | ||
| tnfaip2 | tumor necrosis factor alpha-induced protein 2 | FSH(M) | FSH(F) | |
| Cell differentiation | fgfr4 | fibroblast growth factor receptor 4 | LH(M) | LH(F) |
| Transcription factore | zax | homeobox protein zampogna | FSH(M) | LH(M&F) FSH(F) |
| hnf4a | hepatocyte nuclear factor 4-alpha isoform X1 | LH(F) | FSH(F) LH&FSH(M) | |
| esrrb | LOW QUALITY PROTEIN: steroid hormone receptor ERR2 | FSH(F) | LH (F) LH&FSH(M) | |
| Chemokine receptore | xcr1 | chemokine XC receptor 1-like isoform X1 | LH(F) | |
| Other | ucp | uncharacterized protein LOC102076330 | FSH(M) | LH(M) LH&FSH(F) |
| ucp | uncharacterized protein LOC102077685 isoform X1 | LH(M&F) | FSH(M) | |
| f6i1 | fucolectin-6 isoform X1 | LH(F) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hollander-Cohen, L.; Golan, M.; Levavi-Sivan, B. Differential Regulation of Gonadotropins as Revealed by Transcriptomes of Distinct LH and FSH Cells of Fish Pituitary. Int. J. Mol. Sci. 2021, 22, 6478. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22126478
Hollander-Cohen L, Golan M, Levavi-Sivan B. Differential Regulation of Gonadotropins as Revealed by Transcriptomes of Distinct LH and FSH Cells of Fish Pituitary. International Journal of Molecular Sciences. 2021; 22(12):6478. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22126478
Chicago/Turabian StyleHollander-Cohen, Lian, Matan Golan, and Berta Levavi-Sivan. 2021. "Differential Regulation of Gonadotropins as Revealed by Transcriptomes of Distinct LH and FSH Cells of Fish Pituitary" International Journal of Molecular Sciences 22, no. 12: 6478. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22126478






