D-Amino Acid-Containing Lipopeptides Derived from the Lead Peptide BP100 with Activity against Plant Pathogens
Abstract
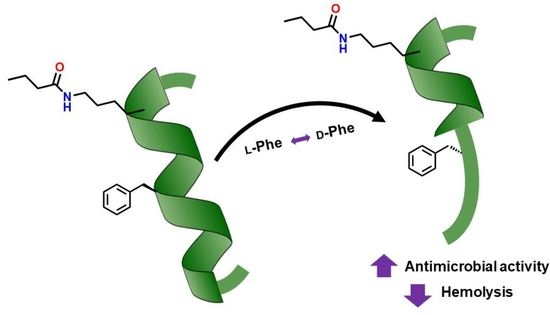
:1. Introduction
2. Results
2.1. Design and Solid-Phase Synthesis of the Lipopeptides
2.2. Antimicrobial Activity
2.3. Toxicity
2.4. Structural Characterization by NMR Spectroscopy
3. Discussion
4. Materials and Methods
4.1. General Methods
4.2. Synthesis of Lipopeptides
4.3. Bacterial and Fungal Strains and Growth Conditions
4.4. Antimicrobial Activity
4.5. Hemolytic Activity
4.6. Effect of Peptide Infiltration on Tobacco Leaves
4.7. Structural Characterization by NMR Spectroscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Department of Economic and Social Affairs, United Nations. World Population Prospects 2019. Available online: https://population.un.org/wpp/ (accessed on 21 May 2021).
- Oerke, E. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keymanesh, K.; Soltani, S.; Sardari, S. Application of antimicrobial peptides in agriculture and food industry. World J. Microbiol. Biotechnol. 2009, 25, 933–944. [Google Scholar] [CrossRef]
- Montesinos, E.; Badosa, E.; Cabrefiga, J.; Planas, M.; Feliu, L.; Bardaji, E. Antimicrobial Peptides for Plant Disease Control—From Discovery to Application. In Small Wonders: Peptides for Disease Control; Rajasekaran, K., Cary, J.W., Jaynes, J.M., Montesinos, E., Eds.; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 2012; pp. 235–262. [Google Scholar]
- De Souza Candido, E.; de Silva Cardoso, M.H.; Sousa, D.A.; Viana, J.C.; de Oliveira-Júnior, N.G.; Miranda, V.; Franco, O.L. The use of versatile plant antimicrobial peptides in agribusiness and human health. Peptides 2014, 55, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Ageitos, J.M.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T.G. Antimicrobial peptides (AMPs): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef]
- Hollmann, A.; Martinez, M.; Maturana, P.; Semorile, L.C.; Maffia, P.C. Antimicrobial Peptides: Interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem. 2018, 6, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Feijó Corrêa, J.A.; Gonçalves Evangelista, A.; de Melo Nazareth, T.; Bittencourt Luciano, F. Fundamentals on the molecular mechanism of action of antimicrobial peptides. Materialia 2019, 8, 100494. [Google Scholar] [CrossRef]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A new era of antibiotics: The clinical potential of antimicrobial peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial peptides as therapeutic agents: Opportunities and challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef]
- Gomes, B.; Augusto, M.T.; Felício, M.R.; Hollmann, A.; Franco, O.L.; Gonçalves, S.; Santos, N.C. Designing improved active peptides for therapeutic approaches against infectious diseases. Biotechnol. Adv. 2018, 36, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Ciumac, D.; Gong, H.; Hu, X.; Lu, J.R. Membrane targeting cationic antimicrobial peptides. J. Colloid Interface Sci. 2019, 537, 163–185. [Google Scholar] [CrossRef] [PubMed]
- Hazam, P.K.; Goyal, R.; Ramakirshnan, V. Peptide based antimicrobials: Design strategies and therapeutic potential. Prog. Biophys. Mol. Biol. 2019, 142, 10–22. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, J.; Tong, Z.; Jia, Y.; Yang, B.; Wang, Z. The revitalization of antimicrobial peptides in the resistance area. Pharmacol. Res. 2021, 163, 105276. [Google Scholar] [CrossRef] [PubMed]
- Jerala, R. Synthetic lipopeptides: A novel class of anti-infectives. Expert Opin. Investig. Drugs 2007, 16, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Ahmed, S.; Eswari, J.S. Therapeutic cyclic lipopeptides mining from microbes: Latest strides and hurdles. World J. Microbiol. Biotechnol. 2015, 31, 1177–1193. [Google Scholar] [CrossRef]
- Zhao, H.; Shao, D.; Jiang, C.; Shi, J.; Li, Q.; Huang, Q.; Rajoka, M.S.R.; Yang, H.; Jin, M. Biological activity of lipopeptides from Bacillus. Appl. Microbiol. Biotechnol. 2017, 101, 5951–5960. [Google Scholar] [CrossRef] [PubMed]
- Malina, A.; Shai, Y. Conjugation of fatty acids with different lengths modulates the antibacterial and antifungal activity of a cationic biologically inactive peptide. Biochem. J. 2005, 390, 695–702. [Google Scholar] [CrossRef]
- Mandal, S.M.; Barbosa, A.E.; Franco, O.L. Lipopeptides in microbial infection control: Scope and reality for industry. Biotechnol. Adv. 2013, 31, 338–345. [Google Scholar] [CrossRef]
- Meena, K.R.; Kanwar, S.S. Lipopeptides as the antifungal and antibacterial agents: Applications in food safety and therapeutics. BioMed Res. Int. 2015, 473050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malmsten, M. Interactions of antimicrobial peptides with bacterial membranes and membrane components. Curr. Top. Med. Chem. 2016, 16, 16–24. [Google Scholar] [CrossRef]
- Vilà, S.; Badosa, E.; Montesinos, E.; Planas, M.; Feliu, L. Synthetic cyclolipopeptides selective agaisnt microbial, plant and animal cell targets by incorporation of D-amino acids or histidine. PLoS ONE 2016, 11, e0151639. [Google Scholar] [CrossRef] [Green Version]
- Koh, J.-J.; Lin, S.; Beuerman, R.W.; Liu, S. Recent advances in synthetic lipopeptides as anti-microbial agents: Designs and synthetic approaches. Amino Acids 2017, 49, 1653–1677. [Google Scholar] [CrossRef]
- Avitabile, C.; D’Andrea, L.D.; D’Aversa, E.; Milani, R.; Gambari, R.; Romanelli, A. Effect of acylation on the antimicrobial activity of temporin B analogues. ChemMedChem 2018, 13, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Sim, J.-Y.; Kim, S.; Lee, J.; Lim, H.; Kim, H.H.; Park, Z.-Y.; Kim, J.I. A significantly enhanced antibacterial spectrum of D-enantiomeric lipopeptide bactenecin. Biochem. Biophys. Res. Commun. 2019, 514, 497–502. [Google Scholar] [CrossRef]
- Albada, B. Tuning Activity of Antimicrobial Peptides by Lipidation. In Health Consequences of Microbial Interactions with Hydrocarbons, Oils, and Lipids: Handbook of Hydrocarbon and Lipid Microbiology; Goldfine, H., Ed.; Springer: Cham, Switzerland, 2020; pp. 317–334. [Google Scholar] [CrossRef]
- Chu-Kung, A.F.; Nguyen, R.; Bozzelli, K.N.; Tirrell, M. Chain length dependence of antimicrobial peptide-fatty acid conjugate activity. J. Colloid Interface Sci. 2010, 345, 160–167. [Google Scholar] [CrossRef]
- Güell, I.; Cabrefiga, J.; Badosa, E.; Ferre, R.; Talleda, M.; Bardají, E.; Planas, M.; Feliu, L.; Montesinos, E. Improvement of the efficacy of linear undecapeptides against plant-pathogenic bacteria by incorporation of D-amino acids. Appl. Environ. Microbiol. 2011, 77, 2667–2675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; He, L.; Li, G.; Zhai, N.; Jiang, H.; Chen, Y. Role of helicity of α-helical antimicrobial peptides to improve specificity. Protein Cell 2014, 5, 631–642. [Google Scholar] [CrossRef] [Green Version]
- Ong, Z.Y.; Wiradharma, N.; Yang, Y.Y. Strategies employed in the design and optimization of synthetic antimicrobial peptide amphiphiles with enhance therapeutic potentials. Adv. Drug Deliv. Rev. 2014, 78, 28–45. [Google Scholar] [CrossRef]
- Li, H.; Anuwongcharoen, N.; Malik, A.A.; Prachayasittikul, V.; Wikberg, J.E.S.; Nantasenamat, C. Roles of D-amino acids on the bioactivity of host defense peptides. Int. J. Mol. Sci. 2016, 17, 1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Zhao, Q.; Li, S.; Yan, Z.; Li, J.; Li, Y.; Mou, L.; Zhang, B.; Yang, W.; Miao, X.; et al. Novel antimicrobial peptide CPF-C1 analogs with superior stabilities and activities against multidrug-resistant bacteria. Chem. Biol. Drug Des. 2017, 90, 690–702. [Google Scholar] [CrossRef]
- Jia, F.; Wang, J.; Peng, J.; Zhao, P.; Kong, Z.; Wang, K.; Yan, W.; Wang, R. D-Amino acid substitution enhances the stability of antimicrobial peptide polybia-CP. Acta Biochim. Biophys. Sin. 2017, 49, 916–925. [Google Scholar] [CrossRef] [Green Version]
- Konai, M.M.; Adhikary, U.; Haldar, J. Design and solution-phase synthesis of membrane-targeting lipopeptides with selective antibacterial activity. Chemistry 2017, 23, 12853–12860. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Li, Y.; Li, J.; Yan, Z.; Wang, D.; Guo, X.; Zhang, J.; Zhang, B.; Mou, L.; Yang, W.; et al. Potent effects of amino acid scanned antimicrobial peptide Feleucin-K3 analogues against both multidrug-resistant strains and biofilms of Pseudomonas aeruginosa. Amino Acids 2018, 50, 1471–1483. [Google Scholar] [CrossRef]
- Topman, S.; Tamir-Ariel, D.; Bochnic-Tamir, H.; Stern Bauer, T.; Shafir, S.; Burdman, S.; Hayouka, Z. Random peptide mixtures as new crop protection agents. Microb. Biotechnol. 2018, 11, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Casciaro, B.; Cappiello, F.; Loffredo, M.R.; Ghirga, F.; Mangoni, M.L. The potential of frog skin peptides for anti-infective therapies: The case of Esculentin-1a(1–21)NH2. Curr. Med. Chem. 2020, 27, 1405–1419. [Google Scholar] [CrossRef]
- Avrahami, D.; Shai, Y. Bestowing antifungal and antibacterial activities by lipophilic acid conjugation to D,L-amino acid-containing antimicrobial peptides: A plausible mode of action. Biochemistry 2003, 42, 14946–14956. [Google Scholar] [CrossRef] [PubMed]
- Makovitzki, A.; Shai, Y. pH-dependent antifungal lipopeptides and their plausible mode of action. Biochemistry 2005, 44, 9775–9784. [Google Scholar] [CrossRef]
- Rosenfeld, Y.; Lev, N.; Shai, Y. Effect of the hydrophobicity to net positive charge ratio on antibacterial and anti-endotoxin activities of structurally similar antimicrobial peptides. Biochemistry 2010, 49, 853–861. [Google Scholar] [CrossRef]
- Lee, J.; Kim, S.; Sim, J.Y.; Lee, D.; Kim, H.H.; Hwang, J.S.; Lee, D.G.; Park, Z.Y.; Kim, J.I. A potent antibacterial activity of new short D-enantiomeric lipopeptide against multi drug resistant bacteria. Biochim. Biophys. Acta Biomembr. 2019, 1861, 34–42. [Google Scholar] [CrossRef]
- Oliveras, A.; Baro, A.; Montesinos, L.; Badosa, E.; Montesinos, E.; Feliu, L.; Planas, M. Antimicrobial activity of linear lipopeptides derived from BP100 towards plant pathogens. PLoS ONE 2018, 13, e0201571. [Google Scholar] [CrossRef] [PubMed]
- Tengel, T.; Sethson, I.; Francis, M.S. Conformational analysis by CD and NMR spectroscopy of a peptide encompassing the amphipathic domain of YopD from Yersinia. Eur. J. Biochem. 2002, 15, 3659–3668. [Google Scholar] [CrossRef] [PubMed]
- Di Grazia, A.; Cappiello, F.; Cohen, H.; Casciaro, B.; Luca, V.; Pini, A.; Di, Y.P.; Shai, Y.; Mangoni, M.L. D-Amino acids incorporation in the frog skin-derived peptide esculentin-1a(1–21)NH2 is beneficial for its multiple functions. Amino Acids 2015, 47, 2505–2519. [Google Scholar] [CrossRef]
- Makovitzki, A.; Avrahamai, D.; Shai, Y. Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. USA 2006, 103, 15997–16002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangoni, M.L.; Shai, Y. Short native antimicrobial peptides and engineered ultrashort lipopeptides: Similarities and differences in cell specificities and modes of action. Cell. Mol. Life Sci. 2011, 68, 2267–2280. [Google Scholar] [CrossRef]
- Laverty, G.; McLaughlin, M.; Shaw, C.; Gorman, S.P.; Gilmore, B.F. Antimicrobial activity of short, synthetic cationic lipopeptides. Chem. Biol. Drug Des. 2010, 75, 563–569. [Google Scholar] [CrossRef]
- Greber, K.E.; Dawgul, M.; Kamysz, W.; Sawicki, W. Cationic net charge and counter ion type as antimicrobial activity determinant factors of short lipopeptides. Front. Microbiol. 2017, 8, 123. [Google Scholar] [CrossRef] [Green Version]
- Blondelle, S.E.; Lohner, K. Combinatorial libraries: A tool to design antimicrobial and antifungal peptide analogues having lytic specificities for structure-activity relationship studies. Biopolymers 2000, 55, 74–87. [Google Scholar] [CrossRef]
- Oh, D.; Shin, S.Y.; Lee, S.; Kang, J.H.; Kim, S.D.; Ryu, P.D.; Hahm, K.S.; Kim, Y. Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A(1–8)-magainin 2(1–12) and its analogues, on their antibiotic activities and structures. Biochemistry 2000, 39, 11855–11864. [Google Scholar] [CrossRef] [PubMed]
- Makovitzki, A.; Viterbo, A.; Brotman, Y.; Chet, I.; Shai, Y. Inhibition of fungal and bacterial plant pathogens in vitro and in planta with ultrashort cationic lipopeptides. Appl. Environ. Microbiol. 2007, 73, 6629–6636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamora-Carreras, H.; Strandberg, E.; Muhlhauser, P.; Burch, J.; Wadhwani, P.; Jimenez, M.A.; Bruix, M.; Ulrich, A.S. Alanine scan and 2H NMR analysis of the membrane-active peptide BP100 point to a distinct carpet mechanism of action. Biochim. Biophys. Acta 2016, 1858, 1328–1338. [Google Scholar] [CrossRef]
- Kaiser, E.; Colescott, R.L.; Bossinger, C.D.; Cook, P. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal. Biochem. 1970, 34, 595–598. [Google Scholar] [CrossRef]
- Hazel, W.J.; Civerolo, E.L. Procedures for growth and inoculation of Xanthomonas fragariae, causal organism of angular leaf spot of strawberry. Plant Dis. 1980, 64, 178–181. [Google Scholar] [CrossRef]
- Badosa, E.; Ferre, R.; Planas, M.; Feliu, L.; Besalu, E.; Cabrefiga, J.; Bardaji, E.; Montesinos, E. A library of linear undecapeptides with bactericidal activity against phytopathogenic bacteria. Peptides 2007, 28, 2276–2285. [Google Scholar] [CrossRef] [PubMed]
- Badosa, E.; Moiset, G.; Montesinos, L.; Talleda, M.; Bardají, E.; Feliu, L.; Planas, M.; Montesinos, E. Derivatives of the antimicrobial peptide BP100 for expression in plant systems. PLoS ONE 2013, 8, e85515. [Google Scholar] [CrossRef] [Green Version]
- Berjanskii, M.V.; Wishart, D.S. A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 2005, 127, 14970–14971. [Google Scholar] [CrossRef]
- Hafsa, N.E.; Arndt, D.; Wishart, D.S. CSI 3.0: A web server for identifying secondary and super-secondary structure in proteins using NMR chemical shifts. Nucleic Acids Res. 2015, 43, W370–W377. [Google Scholar] [CrossRef] [Green Version]






| Peptide | Sequence 1 | Code | tR (min) 2 | Purity (%) 3 | HRMS (ESI) | ||
|---|---|---|---|---|---|---|---|
| Calcd. | Found | ||||||
| BP472 | C5H11CO-KKLfKKILKYL-NH2 | C5H11CO-D-F4 | 6.14 | >99 | C78H137N17O13 [M + 2H]2+ | 760.0285 | 760.0260 |
| BP473 | Ac-KKLfKK(COC3H7)ILKYL-NH2 | D-F4-K6(COC3H7) | 5.99 | >99 | C78H135N17O14 [M + 2H]2+ | 767.0182 | 767.0158 |
| BP474 | Ac-KKLfKKIK(COC3H7)KYL-NH2 | D-F4-K8(COC3H7) | 5.36 | >99 | C78H136N18O14 [M + 2H]2+ | 774.5236 | 774.5208 |
| BP475 | Ac-KKLfKKILKK(COC3H7)L-NH2 | D-F4-K10(COC3H7) | 5.69 | >99 | C75H138N18O13 [M + 2H]2+ | 749.5340 | 749.5343 |
| BP476 | Ac-KKLfKKILKYK(COC11H23)-NH2 | D-F4-K11(COC11H23) | 6.67 | >99 | C86H152N18O14 [M + 2H]2+ | 830.5862 | 830.5839 |
| BP484 | Ac-KKLk(COC5H11)KKILKYL-NH2 | D-K4(COC5H11) | 6.45 | >99 | C77H142N18O14 [M + 2H]2+ | 771.5471 | 771.5477 |
| BP485 | C3H7CO-KKLfKKILKYL-NH2 | C3H7CO-D-F4 | 6.79 | >99 | C76H133N17O13 [M + 2H]2+ | 746.0129 | 746.0097 |
| BP486 | Ac-KK(COC3H7)LfKKILKYL-NH2 | D-F4-K2(COC3H7) | 6.17 | >99 | C78H134N17O14 [M + H]+ | 1533.0291 | 1533.0266 |
| BP488 | Ac-KKLk(COC11H23)KKILKYL-NH2 | D-K4(COC11H23) | 6.80 | >99 | C83H153N18O14 [M + H]+ | 1626.1808 | 1626.1787 |
| BP489 | Ac-KKLfKKK(COC11H23)LKYL-NH2 | D-F4-K7(COC11H23) | 6.76 | >99 | C86H152N18O14 [M + 2H]2+ | 830.5862 | 830.5825 |
| BP490 | Ac-KKLfKKIK(COC11H23)KYL-NH2 | D-F4-K8(COC11H23) | 7.12 | >99 | C86H151N18O14 [M + H]+ | 1660.1652 | 1660.1635 |
| BP494 | Ac-KKLfKKK(COC5H11)LKYL-NH2 | D-F4-K7(COC5H11) | 5.42 | >99 | C80H139N18O14 [M + H]+ | 1576.0713 | 1576.0683 |
| BP495 | Ac-KKLfKKILKYK(COC5H11)-NH2 | D-F4-K11(COC5H11) | 5.44 | >99 | C80H139N18O14 [M + H]+ | 1576.0713 | 1576.0683 |
| BP496 | Ac-KKLfKKILKYK(COC3H7)-NH2 | D-F4-K11(COC3H7) | 5.11 | >99 | C78H135N18O14 [M + H]+ | 1548.0400 | 1548.0367 |
| BP497 | Ac-KKLfKKILK(COC11H23)YL-NH2 | D-F4-K9(COC11H23) | 7.21 | >99 | C86H150N17O14 [M + H]+ | 1645.1543 | 1645.1516 |
| BP498 | Ac-KKLfK(COC3H7)KILKYL-NH2 | D-F4-K5(COC3H7) | 6.12 | >99 | C78H135N17O14 [M + 2H]2+ | 767.0182 | 767.0147 |
| BP499 | Ac-KKLfKKILK(COC3H7)YL-NH2 | D-F4-K9(COC3H7) | 5.89 | >99 | C78H134N17O14 [M + H]+ | 1533.0291 | 1533.0269 |
| BP500 | Ac-KKK(COC11H23)fKKILKYL-NH2 | D-F4-K3(COC11H23) | 6.80 | >99 | C86H151N18O14 [M + H]+ | 1661.1684 | 1661.1667 |
| Peptide | Code | Hemolysis (%) 1 250 µM | Size of the Lesion (mm) 2 250 µM |
|---|---|---|---|
| BP472 | C5H11CO-D-F4 | 78 ± 5 | 4 ± 1 |
| BP473 | D-F4-K6(COC3H7) | 84 ± 11 | 5 ± 2 |
| BP474 | D-F4-K8(COC3H7) | 0 ± 0 | 4 ± 2 |
| BP475 | D-F4-K10(COC3H7) | 0 ± 0 | 10 ± 1 |
| BP476 | D-F4-K11(COC11H23) | 94 ± 10 | 5 ± 0.5 |
| BP484 | D-K4(COC5H11) | 10 ± 2 | 7 ± 1 |
| BP485 | C3H7CO-D-F4 | 24 ± 9 | 12 ± 1 |
| BP486 | D-F4-K2(COC3H7) | 14 ± 5 | 17 ± 2 |
| BP488 | D-K4(COC11H23) | 86 ± 14 | 13 ± 5 |
| BP489 | D-F4-K7(COC11H23) | 100 ± 6 | 11 ± 1 |
| BP490 | D-F4-K8(COC11H23) | 100 ± 6 | 11 ±3 |
| BP494 | D-F4-K7(COC5H11) | 0.2 ± 0.2 | 8 ± 1 |
| BP495 | D-F4-K11(COC5H11) | 0.6 ± 1 | 11 ± 2 |
| BP496 | D-F4-K11(COC3H7) | 1 ± 1 | 10 ± 3 |
| BP497 | D-F4-K9(COC11H23) | 100 ± 2 | 18 ± 0 |
| BP498 | D-F4-K5(COC3H7) | 21 ± 3 | 6 ± 1 |
| BP499 | D-F4-K9(COC3H7) | 11 ± 1 | 8 ± 2 |
| BP500 | D-F4-K3(COC11H23) | 71 ± 8 | 9 ± 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveras, À.; Moll, L.; Riesco-Llach, G.; Tolosa-Canudas, A.; Gil-Caballero, S.; Badosa, E.; Bonaterra, A.; Montesinos, E.; Planas, M.; Feliu, L. D-Amino Acid-Containing Lipopeptides Derived from the Lead Peptide BP100 with Activity against Plant Pathogens. Int. J. Mol. Sci. 2021, 22, 6631. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22126631
Oliveras À, Moll L, Riesco-Llach G, Tolosa-Canudas A, Gil-Caballero S, Badosa E, Bonaterra A, Montesinos E, Planas M, Feliu L. D-Amino Acid-Containing Lipopeptides Derived from the Lead Peptide BP100 with Activity against Plant Pathogens. International Journal of Molecular Sciences. 2021; 22(12):6631. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22126631
Chicago/Turabian StyleOliveras, Àngel, Luís Moll, Gerard Riesco-Llach, Arnau Tolosa-Canudas, Sergio Gil-Caballero, Esther Badosa, Anna Bonaterra, Emilio Montesinos, Marta Planas, and Lidia Feliu. 2021. "D-Amino Acid-Containing Lipopeptides Derived from the Lead Peptide BP100 with Activity against Plant Pathogens" International Journal of Molecular Sciences 22, no. 12: 6631. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22126631