Key Regulators of Sucrose Metabolism Identified through Comprehensive Comparative Transcriptome Analysis in Peanuts
Abstract
:1. Introduction
2. Results
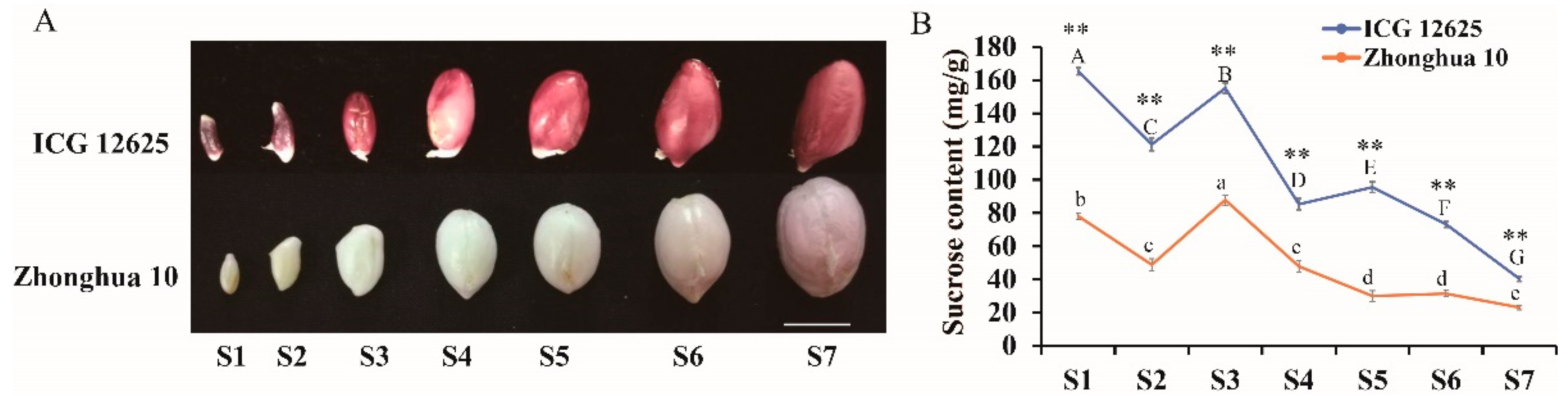
2.1. Significant Difference of Sucrose Content between ICG 12625 and Zhonghua 10 during Seed Development
2.2. Transcriptome Profile of Seed Development in ICG 12625 and Zhonghua 10
2.3. Stage-Specific Transcriptome Profile during Seed Development in ICG 12625 and Zhonghua 10
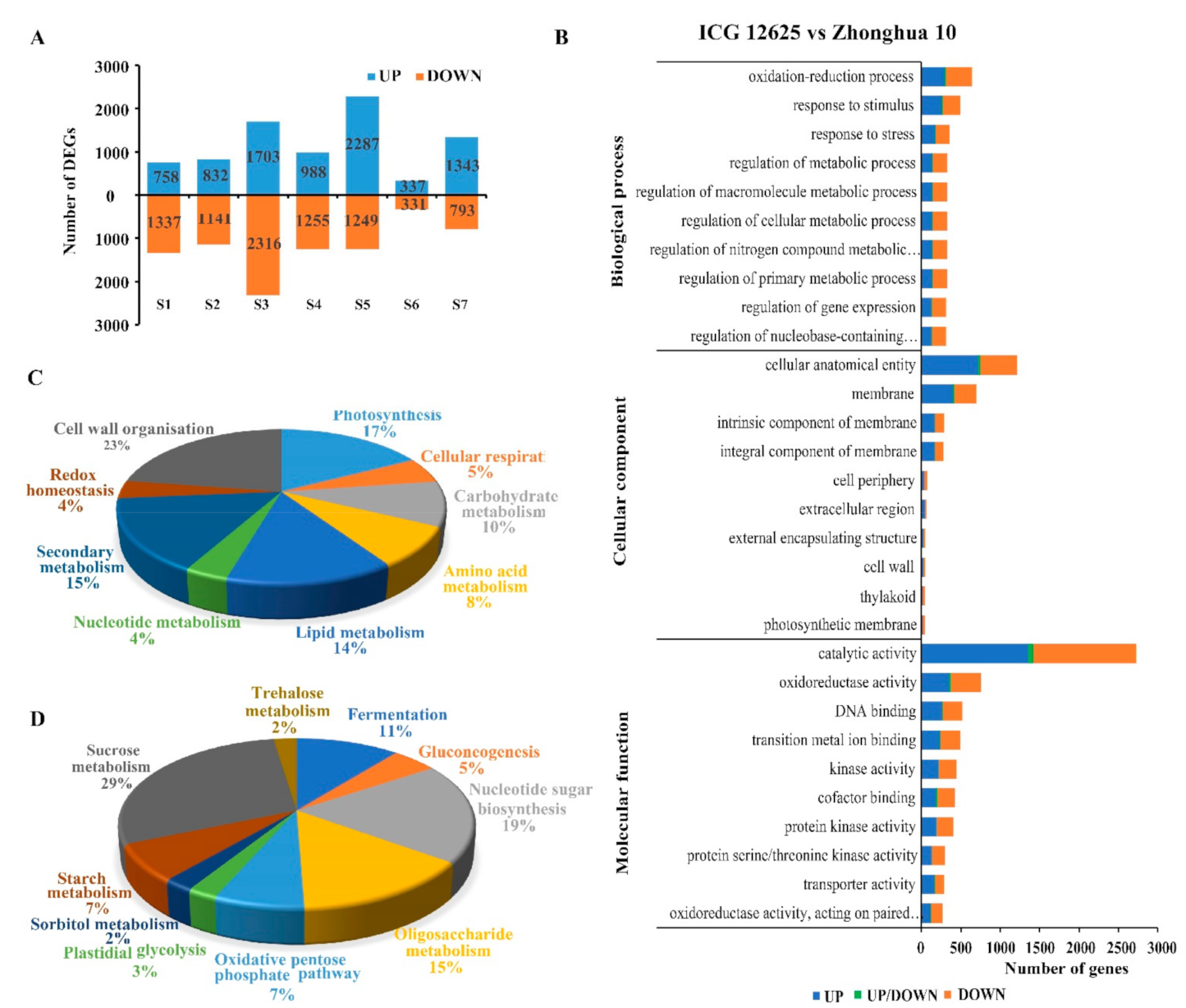
2.4. Differential Gene Expression between ICG 12,625 and Zhonghua 10
2.5. DEGs Related to Seed Sucrose Metabolism between ICG 12625 and Zhonghua 10
2.6. SWEET and Key Enzyme Genes Involved in the Regulatory Network of Sucrose Metabolism
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Sampling
4.2. Determination of Sucrose Content, Oil Content, and Protein Content
4.3. RNA-Seq and Data Processing
4.4. Identification of Stage-Special Expression Genes
4.5. Phylogenetic Analysis of SWEET Genes
4.6. Gene Ontology Enrichment Analysis and Metabolism Pathway Analysis
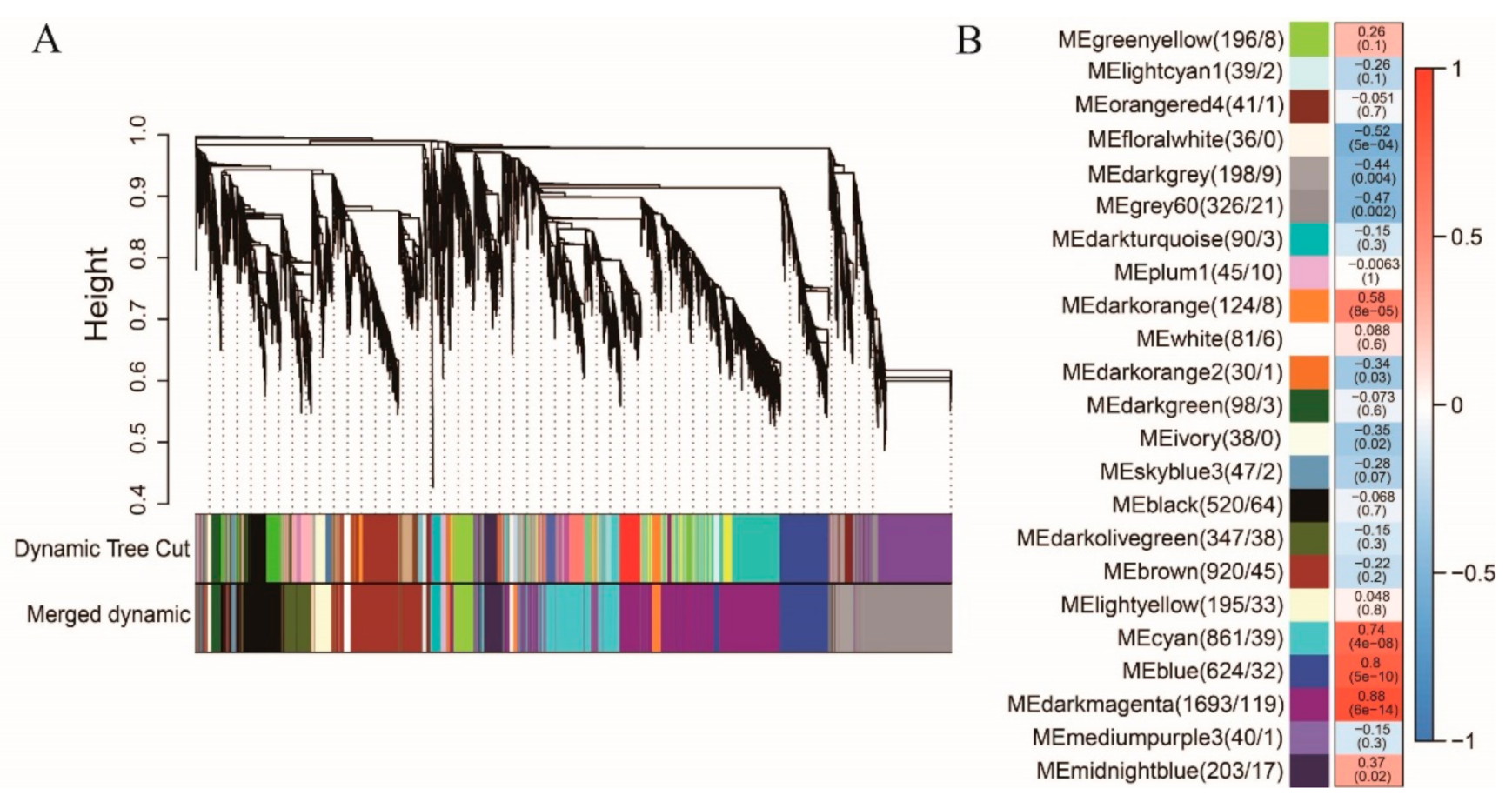
4.7. Gene Co-Expression Analysis by WGCNA
4.8. Regulation between TF and Genes Related to Sucrose Content
4.9. qRT-PCR Validation for DEGs
4.10. Expression Patterns of 10 Genes in RIL Population
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, J.; Liu, N.; Li, W.; Wu, B.; Chen, H.; Huang, L.; Chen, W.; Luo, H.; Zhou, X.; Jiang, H. Identification of two major loci and linked marker for oil content in peanut (Arachis hypogaea L.). Euphytica 2021, 217, 1–11. [Google Scholar] [CrossRef]
- Pandey, M.; Monyo, E.; Ozias-Akins, P.; Liang, X.; Guimarães, P.; Nigam, S.N.; Upadhyaya, H.D.; Janila, P.; Zhang, X.; Guo, B.; et al. Advances in Arachis genomics for peanut improvement. Biotechnol. Adv. 2012, 30, 639–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arya, S.S.; Salve, A.; Chauhan, S. Peanuts as functional food: A review. J. Food Sci. Technol. 2016, 53, 31–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pattee, H.E.; Isleib, T.G.; Giesbrecht, F.G. Variation in Intensity of Sweet and Bitter Sensory Attributes Across Peanut Genotypes1. Peanut Sci. 1998, 25, 63–69. [Google Scholar] [CrossRef]
- McDaniel, K.A.; White, B.L.; Dean, L.L.; Sanders, T.H.; Davis, J.P. Compositional and Mechanical Properties of Peanuts Roasted to Equivalent Colors using Different Time/Temperature Combinations. J. Food Sci. 2012, 77, C1293–C1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pattee, H.E.; Young, C.T.; Giesbrecht, F.G. Seed size and storage effects on carbohydrates of peanuts. J. Agric. Food Chem. 1981, 29, 800–802. [Google Scholar] [CrossRef]
- Bilska-Kos, A.; Mytych, J.; Suski, S.; Magoń, J.; Ochodzki, P.; Zebrowski, J. Sucrose phosphate synthase (SPS), sucrose synthase (SUS) and their products in the leaves of Miscanthus × giganteus and Zea mays at low temperature. Planta 2020, 252, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, J.; Huang, Z.; Mi, L.; Xu, K.; Wu, J.; Fan, Y.; Ma, S.; Jiang, D. Effects of Low Temperature at Booting Stage on Sucrose Metabolism and Endogenous Hormone Contents in Winter Wheat Spikelet. Front. Plant Sci. 2019, 10, 498. [Google Scholar] [CrossRef]
- Khan, H.; Siddique, K.; Colmer, T.D. Vegetative and reproductive growth of salt-stressed chickpea are carbon-limited: Sucrose infusion at the reproductive stage improves salt tolerance. J. Exp. Bot. 2017, 68, 2001–2011. [Google Scholar] [CrossRef]
- Davis, J.P.; Dean, L.L. Peanut Composition, Flavor and Nutrition. Peanuts 2016, 289–345. [Google Scholar] [CrossRef]
- Gallardo, K.; Thompson, R.; Burstin, J. Reserve accumulation in legume seeds. Comptes Rendus Biol. 2008, 331, 755–762. [Google Scholar] [CrossRef]
- Aguirre, M.; Kiegle, E.; Leo, G.; Ezquer, I. Carbohydrate reserves and seed development: An overview. Plant Reprod. 2018, 31, 263–290. [Google Scholar] [CrossRef]
- Aslam, M.M.; Deng, L.; Wang, X.; Wang, Y.; Pan, L.; Liu, H.; Niu, L.; Lu, Z.; Cui, G.; Zeng, W.; et al. Expression patterns of genes involved in sugar metabolism and accumulation during peach fruit development and ripening. Sci. Hortic. 2019, 257, 108633. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Zheng, H.; Peng, Q.; Jiang, Q.; Wang, H.; Fang, T.; Liao, L.; Wang, L.; He, H.; Han, Y. Assessment of Sugar Components and Genes Involved in the Regulation of Sucrose Accumulation in Peach Fruit. J. Agric. Food Chem. 2016, 64, 6723–6729. [Google Scholar] [CrossRef]
- Lü, J.; Tao, X.; Yao, G.; Zhang, S.; Zhang, H. Transcriptome Analysis of Low- and High-Sucrose Pear Cultivars Identifies Key Regulators of Sucrose Biosynthesis in Fruits. Plant Cell Physiol. 2020, 61, 1493–1506. [Google Scholar] [CrossRef]
- Ma, Q.-J.; Sun, M.-H.; Lu, J.; Liu, Y.-J.; Hu, D.-G.; Hao, Y.-J. Transcription Factor AREB2 Is Involved in Soluble Sugar Accumulation by Activating Sugar Transporter and Amylase Genes. Plant Physiol. 2017, 174, 2348–2362. [Google Scholar] [CrossRef]
- Zhen, Q.; Fang, T.; Peng, Q.; Liao, L.; Zhao, L.; Owiti, A.; Han, Y. Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic. Res. 2018, 5, 14. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-G.; Zhao, T.-T.; Wang, W.-Z.; Feng, C.-L.; Feng, X.-Y.; Xiong, G.-R.; Shen, L.-B.; Zhang, S.-Z.; Wang, W.-Q.; Zhang, Z.-X. Culm transcriptome sequencing of Badila (Saccharum officinarum L.) and analysis of major genes involved in sucrose accumulation. Plant Physiol. Biochem. 2019, 144, 455–465. [Google Scholar] [CrossRef]
- McKinley, B.; Rooney, W.; Wilkerson, C.; Mullet, J. Dynamics of biomass partitioning, stem gene expression, cell wall biosynthesis, and sucrose accumulation during development of Sorghum bicolor. Plant J. 2016, 88, 662–680. [Google Scholar] [CrossRef]
- Li, Y.; Wang, W.; Feng, Y.; Tu, M.; Wittich, P.E.; Bate, N.J.; Messing, J. Transcriptome and metabolome reveal distinct carbon allocation patterns during internode sugar accumulation in different sorghum genotypes. Plant Biotechnol. J. 2018, 17, 472–487. [Google Scholar] [CrossRef] [Green Version]
- Ruan, Y.-L. Sucrose Metabolism: Gateway to Diverse Carbon Use and Sugar Signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nat. Cell Biol. 2010, 468, 527–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.-Q.; Lin, I.W.; Qu, X.-Q.; Sosso, D.; McFarlane, H.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A Cascade of Sequentially Expressed Sucrose Transporters in the Seed Coat and Endosperm Provides Nutrition for the Arabidopsis Embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Yokosho, K.; Guo, R.; Whelan, J.; Ruan, Y.-L.; Ma, J.F.; Shou, H. The Soybean Sugar Transporter GmSWEET15 Mediates Sucrose Export from Endosperm to Early Embryo. Plant Physiol. 2019, 180, 2133–2141. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Guo, W.; Li, J.; Yue, P.; Bu, H.; Jiang, J.; Liu, W.; Xu, Y.; Yuan, H.; Li, T.; et al. Histone Acetylation at the Promoter for the Transcription Factor PuWRKY31 Affects Sucrose Accumulation in Pear Fruit. Plant Physiol. 2020, 182, 2035–2046. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Mao, W.; Jia, M.; Xing, S.; Ali, U.; Zhao, Y.; Chen, Y.; Cao, M.; Dai, Z.; Zhang, K.; et al. FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J. Exp. Bot. 2018, 69, 4805–4820. [Google Scholar] [CrossRef] [Green Version]
- Thalor, S.K.; Berberich, T.; Lee, S.S.; Yang, S.H.; Zhu, X.; Imai, R.; Takahashi, Y.; Kusano, T. Deregulation of Sucrose-Controlled Translation of a bZIP-Type Transcription Factor Results in Sucrose Accumulation in Leaves. PLoS ONE 2012, 7, e33111. [Google Scholar] [CrossRef]
- Pattee, H.E.; Isleib, T.G.; Giesbrecht, F.G.; McFeeters, R.F. Investigations into Genotypic Variations of Peanut Carbohydrates. J. Agric. Food Chem. 2000, 48, 750–756. [Google Scholar] [CrossRef]
- Isleib, T.G.; Pattee, H.E.; Giesbrecht, F.G. Oil, Sugar, and Starch Characteristics in Peanut Breeding Lines Selected for Low and High Oil Content and Their Combining Ability. J. Agric. Food Chem. 2004, 52, 3165–3168. [Google Scholar] [CrossRef]
- Chen, X.; Li, H.; Pandey, M.; Yang, Q.; Wang, X.; Garg, V.; Li, H.; Chi, X.; Doddamani, D.; Hong, Y.; et al. Draft genome of the peanut A-genome progenitor (Arachis duranensis) provides insights into geocarpy, oil biosynthesis, and allergens. Proc. Natl. Acad. Sci. USA 2016, 113, 6785–6790. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.-C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef] [Green Version]
- Bertioli, S.C.M.L.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Burow, M.D.; Simpson, C.E.; Faries, M.W.; Starr, J.L.; Paterson, A.H. Molecular biogeographic study of recently described B- and A-genome Arachis species, also providing new insights into the origins of cultivated peanut. Genome 2009, 52, 107–119. [Google Scholar] [CrossRef]
- Chen, X.; Lu, Q.; Liu, H.; Zhang, J.; Hong, Y.; Lan, H.; Li, H.; Wang, J.; Liu, H.; Li, S.; et al. Sequencing of Cultivated Peanut, Arachis hypogaea, Yields Insights into Genome Evolution and Oil Improvement. Mol. Plant 2019, 12, 920–934. [Google Scholar] [CrossRef] [Green Version]
- Yin, D.; Ji, C.; Ma, X.; Li, H.; Zhang, W.; Li, S.; Liu, F.; Zhao, K.; Li, F.; Li, K.; et al. Genome of an allotetraploid wild peanut Arachis monticola: A de novo assembly. GigaScience 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Pandey, M.K.; Pandey, A.K.; Kumar, R.; Nwosu, C.V.; Guo, B.; Wright, G.C.; Bhat, R.S.; Chen, X.; Bera, S.K.; Yuan, M.; et al. Translational genomics for achieving higher genetic gains in groundnut. Theor. Appl. Genet. 2020, 133, 1679–1702. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Liu, X.; Pandey, M.K.; Ren, X.; Chen, H.; Xue, X.; Liu, N.; Huai, D.; Chen, Y.; Zhou, X.; et al. Genome-wide expression quantitative trait locus analysis in a recombinant inbred line population for trait dissection in peanut. Plant Biotechnol. J. 2020, 18, 779–790. [Google Scholar] [CrossRef] [Green Version]
- Saldivar, X.; Wang, Y.-J.; Chen, P.; Hou, A. Changes in chemical composition during soybean seed development. Food Chem. 2011, 124, 1369–1375. [Google Scholar] [CrossRef]
- Hajduch, M.; Matusova, R.; Houston, N.L.; Thelen, J.J. Comparative proteomics of seed maturation in oilseeds reveals differences in intermediary metabolism. Proteomics 2011, 11, 1619–1629. [Google Scholar] [CrossRef]
- Hill, L.M.; Morley-Smith, E.R.; Rawsthorne, S. Metabolism of Sugars in the Endosperm of Developing Seeds of Oilseed Rape. Plant Physiol. 2003, 131, 228–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borisjuk, L.; Rolletschek, H.; Radchuk, R.; Weschke, W.; Wobus, U.; Weber, H. Seed Development and Differentiation: A Role for Metabolic Regulation. Plant Biol. 2004, 6, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Pickett, T.A. Composition of developing peanut seed. Plant Physiol. 1950, 25, 210–224. [Google Scholar] [CrossRef] [PubMed]
- Jeena, G.S.; Kumar, S.; Shukla, R.K. Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants. Plant Mol. Biol. 2019, 100, 351–365. [Google Scholar] [CrossRef]
- Feng, L.; Frommer, W.B. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem. Sci. 2015, 40, 480–486. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2011, 335, 207–211. [Google Scholar] [CrossRef]
- Lin, I.W.; Sosso, D.; Chen, L.-Q.; Gase, K.; Kim, S.-G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.-H.; Qu, X.-Q.; et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nat. Cell Biol. 2014, 508, 546–549. [Google Scholar] [CrossRef]
- Guan, Y.-F.; Huang, X.-Y.; Zhu, J.; Gao, J.-F.; Zhang, H.-X.; Yang, Z.-N. RUPTURED POLLEN GRAIN1, a Member of the MtN3/saliva Gene Family, Is Crucial for Exine Pattern Formation and Cell Integrity of Microspores in Arabidopsis. Plant Physiol. 2008, 147, 852–863. [Google Scholar] [CrossRef] [Green Version]
- Seo, P.J.; Park, J.-M.; Kang, S.K.; Kim, S.-G.; Park, C.-M. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 2010, 233, 189–200. [Google Scholar] [CrossRef]
- Durand, M.; Porcheron, B.; Hennion, N.; Maurousset, L.; Lemoine, R.; Pourtau, N. Water Deficit Enhances C Export to the Roots in Arabidopsis thaliana Plants with Contribution of Sucrose Transporters in Both Shoot and Roots. Plant Physiol. 2016, 170, 1460–1479. [Google Scholar] [CrossRef] [Green Version]
- Wobus, U.; Weber, H. Sugars as Signal Molecules in Plant Seed Development. Biol. Chem. 1999, 380, 937–944. [Google Scholar] [CrossRef]
- Morley-Smith, E.R.; Pike, M.J.; Findlay, K.; Köckenberger, W.; Hill, L.M.; Smith, A.M.; Rawsthorne, S. The Transport of Sugars to Developing Embryos Is Not via the Bulk Endosperm in Oilseed Rape Seeds. Plant Physiol. 2008, 147, 2121–2130. [Google Scholar] [CrossRef] [Green Version]
- Ru, L.; He, Y.; Zhu, Z.; Patrick, J.W.; Ruan, Y.-L. Integrating Sugar Metabolism with Transport: Elevation of Endogenous Cell Wall Invertase Activity Up-Regulates SlHT2 and SlSWEET12c Expression for Early Fruit Development in Tomato. Front. Genet. 2020, 11, 592596. [Google Scholar] [CrossRef]
- Kumar, R.; Mukherjee, S.; Ayele, B.T. Molecular aspects of sucrose transport and its metabolism to starch during seed development in wheat: A comprehensive review. Biotechnol. Adv. 2018, 36, 954–967. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, Y.; Haque, M.E.; Xu, W.; Li, F.; Liu, A. Transcriptomic analyses reveal complex and interconnected sucrose signaling cascades in developing seeds of castor bean. J. Plant Physiol. 2018, 221, 1–10. [Google Scholar] [CrossRef]
- Wind, J.; Smeekens, S.; Hanson, J. Sucrose: Metabolite and signaling molecule. Phytochemistry 2010, 71, 1610–1614. [Google Scholar] [CrossRef]
- Stein, O.; Granot, D. An Overview of Sucrose Synthases in Plants. Front. Plant Sci. 2019, 10, 95. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Zhang, P.; Zhang, Y.; Wang, S.; Qu, L.; Liu, X.; Luo, J. Identification of a peroxisomal-targeted aldolase involved in chlorophyll biosynthesis and sugar metabolism in rice. Plant Sci. 2016, 250, 205–215. [Google Scholar] [CrossRef]
- Kunz, H.-H.; Zamani-Nour, S.; Häusler, R.E.; Ludewig, K.; Schroeder, J.I.; Malinova, I.; Fettke, J.; Flügge, U.-I.; Gierth, M. Loss of Cytosolic Phosphoglucose Isomerase Affects Carbohydrate Metabolism in Leaves and Is Essential for Fertility of Arabidopsis. Plant Physiol. 2014, 166, 753–765. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Liu, Y.; Liu, M.; Zheng, Q.; Li, B.; Li, Z.; Li, H. Sugar accumulation is associated with leaf senescence induced by long-term high light in wheat. Plant Sci. 2019, 287, 110169. [Google Scholar] [CrossRef]
- Juárez-Colunga, S.; López-González, C.; Morales-Elías, N.C.; Massange-Sánchez, J.A.; Trachsel, S.; Tiessen, A. Genome-wide analysis of the invertase gene family from maize. Plant Mol. Biol. 2018, 97, 385–406. [Google Scholar] [CrossRef]
- Lee, D.-W.; Lee, S.-K.; Rahman, M.; Kim, Y.-J.; Zhang, D.; Jeon, J.-S. The Role of Rice Vacuolar Invertase2 in Seed Size Control. Mol. Cells 2019, 42, 711–720. [Google Scholar] [CrossRef]
- Welham, T.; Pike, J.; Horst, I.; Flemetakis, E.; Katinakis, P.; Kaneko, T.; Sato, S.; Tabata, S.; Perry, J.; Parniske, M.; et al. A cytosolic invertase is required for normal growth and cell development in the model legume, Lotus japonicus. J. Exp. Bot. 2009, 60, 3353–3365. [Google Scholar] [CrossRef]
- Rende, U.; Wang, W.; Gandla, M.L.; Jönsson, L.; Niittylä, T. Cytosolic invertase contributes to the supply of substrate for cellulose biosynthesis in developing wood. New Phytol. 2017, 214, 796–807. [Google Scholar] [CrossRef]
- Winter, H.; Huber, S.C. Regulation of Sucrose Metabolism in Higher Plants: Localization and regulation of Activity of Key Enzymes. Crit. Rev. Plant Sci. 2000, 19, 31–67. [Google Scholar] [CrossRef]
- Koch, K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [CrossRef]
- Baroja-Fernández, E.; Muñoz, F.; Montero, M.; Etxeberria, E.; Sesma, M.T.; Ovecka, M.; Bahaji, A.; Ezquer, I.; Li, J.; Prat, S.; et al. Enhancing Sucrose Synthase Activity in Transgenic Potato (Solanum tuberosum L.) Tubers Results in Increased Levels of Starch, ADPglucose and UDPglucose and Total Yield. Plant Cell Physiol. 2009, 50, 1651–1662. [Google Scholar] [CrossRef]
- Fugate, K.K.; Eide, J.D.; Martins, D.N.; Grusak, M.A.; Deckard, E.L.; Finger, F.L. Colocalization of sucrose synthase expression and sucrose storage in the sugarbeet taproot indicates a potential role for sucrose catabolism in sucrose accumulation. J. Plant Physiol. 2019, 240, 153016. [Google Scholar] [CrossRef]
- Xiao, G.; Qin, H.; Zhou, J.; Quan, R.; Lu, X.; Huang, R.; Zhang, H. OsERF2 controls rice root growth and hormone responses through tuning expression of key genes involved in hormone signaling and sucrose metabolism. Plant Mol. Biol. 2016, 90, 293–302. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Tang, Y.-M.; Wang, Y.; Sun, B.; Chen, T.; Lei, D.-Y.; Zhang, F.; Luo, Y.; Zhang, Y.; Wang, X.-R.; et al. Enhance sucrose accumulation in strawberry fruits by eliminating the translational repression of FabZIPs1.1. Sci. Hortic. 2020, 259, 108850. [Google Scholar] [CrossRef]
- Sun, C.; Palmqvist, S.; Olsson, H.; Borén, M.; Ahlandsberg, S.; Jansson, C. A Novel WRKY Transcription Factor, SUSIBA2, Participates in Sugar Signaling in Barley by Binding to the Sugar-Responsive Elements of the iso1 Promoter. Plant Cell 2003, 15, 2076–2092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pattee, H.E.; Johns, E.B.; Singleton, J.A.; Sanders, T.H. Composition Changes of Peanut Fruit Parts during Maturation1. Peanut Sci. 1974, 1, 57–62. [Google Scholar] [CrossRef]
- Dumont, E.; Fontaine, V.; Vuylsteker, C.; Sellier, H.; Bodèle, S.; Voedts, N.; Devaux, R.; Frise, M.; Avia, K.; Hilbert, J.-L.; et al. Association of sugar content QTL and PQL with physiological traits relevant to frost damage resistance in pea under field and controlled conditions. Theor. Appl. Genet. 2009, 118, 1561–1571. [Google Scholar] [CrossRef] [PubMed]
- Beljkaš, B.; Matić, J.; Milovanović, I.; Jovanov, P.; Misan, A.; Šarić, L. Rapid method for determination of protein content in cereals and oilseeds: Validation, measurement uncertainty and comparison with the Kjeldahl method. Accredit. Qual. Assur. 2010, 15, 555–561. [Google Scholar] [CrossRef]
- Sathe, S.K.; Venkatachalam, M.; Sharma, G.M.; Kshirsagar, H.H.; Teuber, S.S.; Roux, K.H. Solubilization and Electrophoretic Characterization of Select Edible Nut Seed Proteins. J. Agric. Food Chem. 2009, 57, 7846–7856. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Garg, R.; Singh, V.K.; Rajkumar, M.S.; Kumar, V.; Jain, M. Global transcriptome and coexpression network analyses reveal cultivar-specific molecular signatures associated with seed development and seed size/weight determination in chickpea. Plant J. 2017, 91, 1088–1107. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Wang, X. Application of the Gini Correlation Coefficient to Infer Regulatory Relationships in Transcriptome Analysis. Plant Physiol. 2012, 160, 192–203. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Xin, M.; Feldmann, K.A.; Wang, X. Machine Learning–Based Differential Network Analysis: A Study of Stress-Responsive Transcriptomes in Arabidopsis. Plant Cell 2014, 26, 520–537. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Ma, Z.; Wang, A.; Zheng, T.; Huang, L.; Sun, W.; Zhang, Y.; Jin, W.; Zhan, J.; Cai, Y.; et al. Genome-Wide Investigation of the Auxin Response Factor Gene Family in Tartary Buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 2018, 19, 3526. [Google Scholar] [CrossRef] [Green Version]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, Article17. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Paul, S.; Andrew, M.; Owen, O.; Nitin, S.B.; Jonathan, T.W.; Daniel, R.; Nada, A.; Benno, S.; Trey, I. Cytoscape: A software environment for integrated models. Genome Res. 1971, 13, 426. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [Green Version]
- Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2019, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Yang, S.; Wang, F.; Guo, F.; Meng, J.-J.; Li, X.-G.; Dong, S.-T.; Wan, S.-B. Exogenous Calcium Alleviates Photoinhibition of PSII by Improving the Xanthophyll Cycle in Peanut (Arachis hypogaea) Leaves during Heat Stress under High Irradiance. PLoS ONE 2013, 8, e71214. [Google Scholar] [CrossRef] [Green Version]
- Brand, Y.; Hovav, R. Identification of Suitable Internal Control Genes for Quantitative Real-Time PCR Expression Analyses in Peanut (Arachis hypogaea). Peanut Sci. 2010, 37, 12–19. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Huang, L.; Liu, N.; Pandey, M.K.; Chen, Y.; Cheng, L.; Guo, J.; Yu, B.; Luo, H.; Zhou, X.; et al. Key Regulators of Sucrose Metabolism Identified through Comprehensive Comparative Transcriptome Analysis in Peanuts. Int. J. Mol. Sci. 2021, 22, 7266. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147266
Li W, Huang L, Liu N, Pandey MK, Chen Y, Cheng L, Guo J, Yu B, Luo H, Zhou X, et al. Key Regulators of Sucrose Metabolism Identified through Comprehensive Comparative Transcriptome Analysis in Peanuts. International Journal of Molecular Sciences. 2021; 22(14):7266. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147266
Chicago/Turabian StyleLi, Weitao, Li Huang, Nian Liu, Manish K. Pandey, Yuning Chen, Liangqiang Cheng, Jianbin Guo, Bolun Yu, Huaiyong Luo, Xiaojing Zhou, and et al. 2021. "Key Regulators of Sucrose Metabolism Identified through Comprehensive Comparative Transcriptome Analysis in Peanuts" International Journal of Molecular Sciences 22, no. 14: 7266. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147266







