Identification of Intercellular Crosstalk between Decidual Cells and Niche Cells in Mice
Abstract
:1. Introduction
2. Results
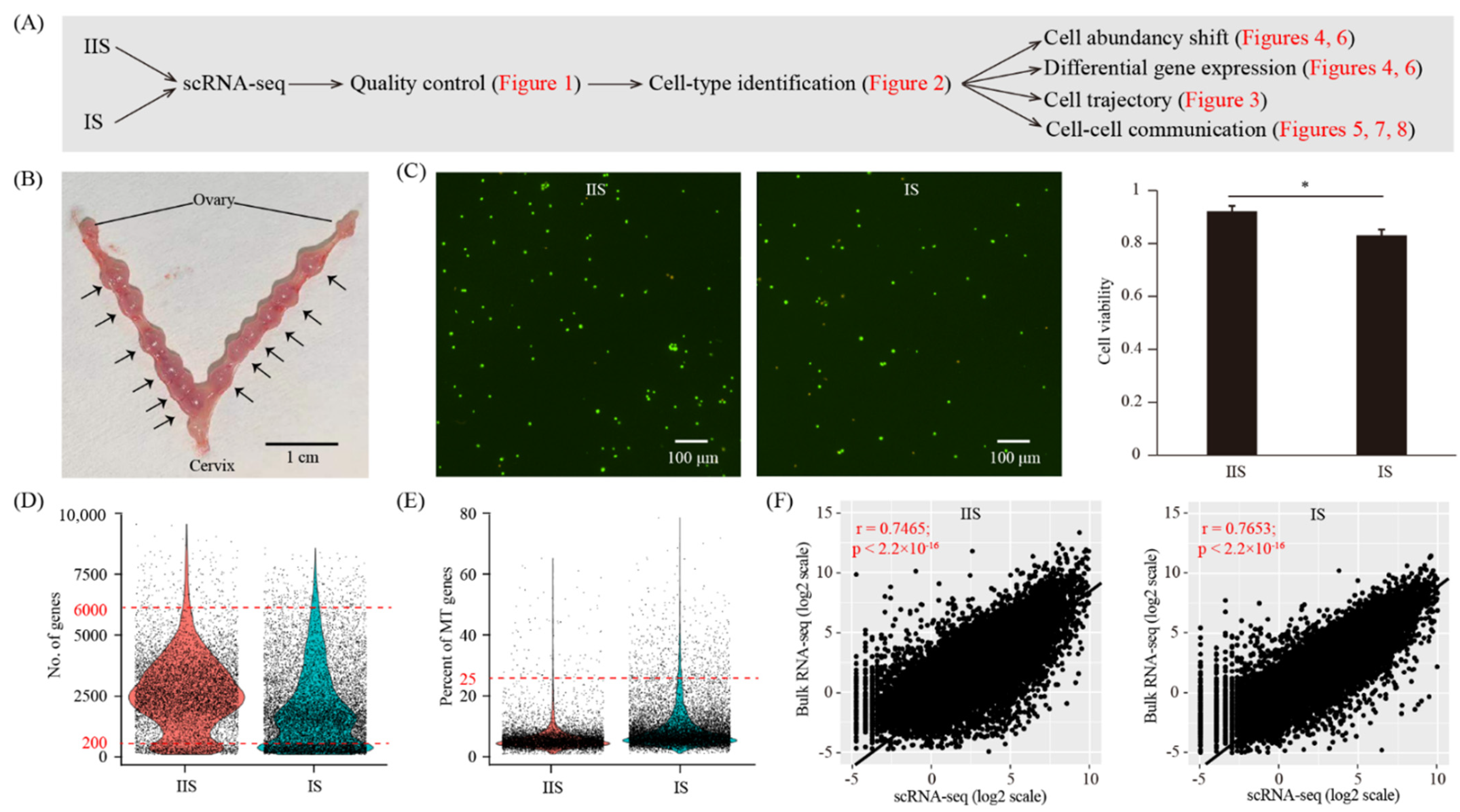
2.1. A Single-Cell Atlas of Mouse Uterus on Gestational Day Eight
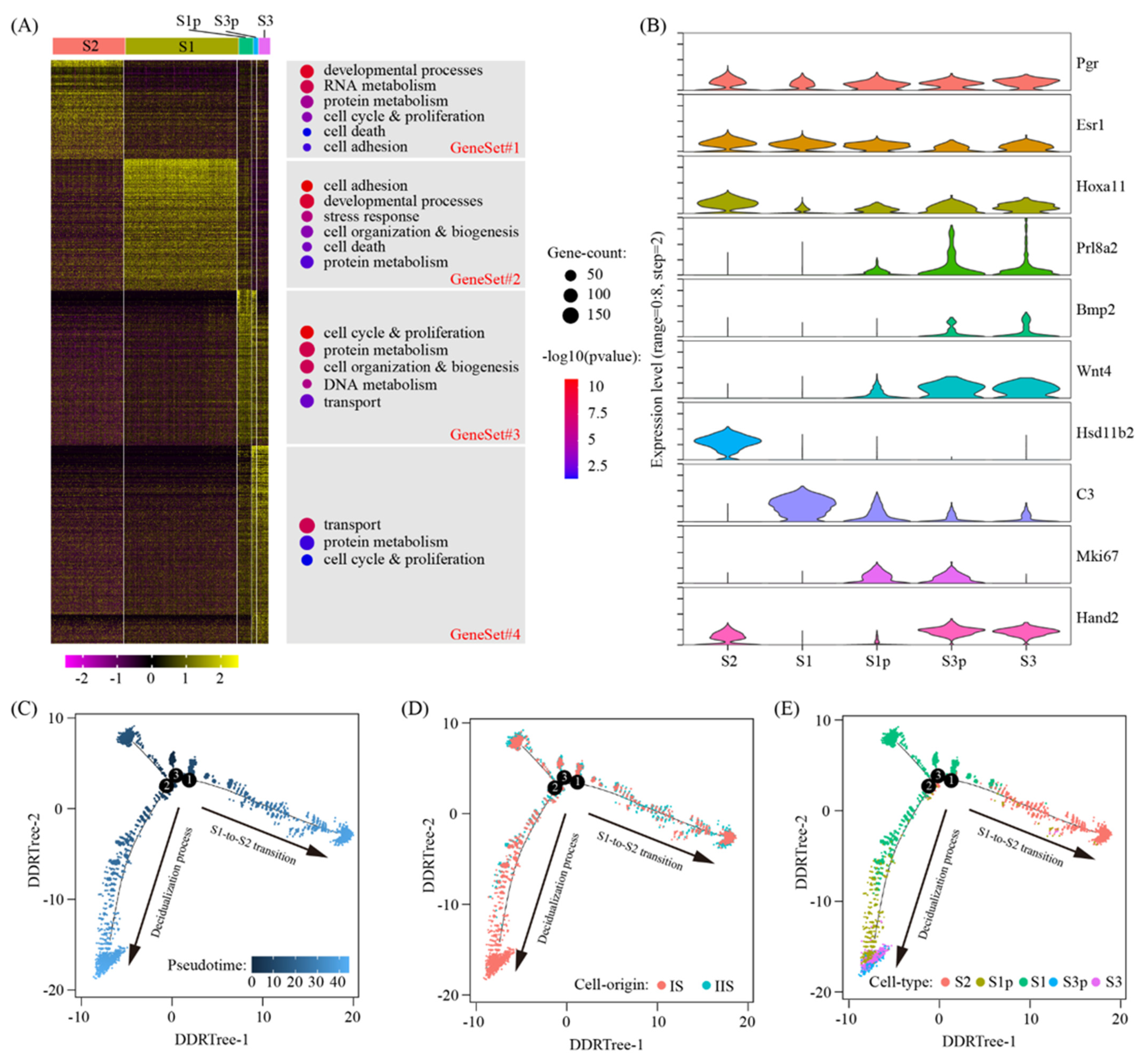
2.2. Reconstruction of Decidual Cell Trajectory Across Stromal Cells
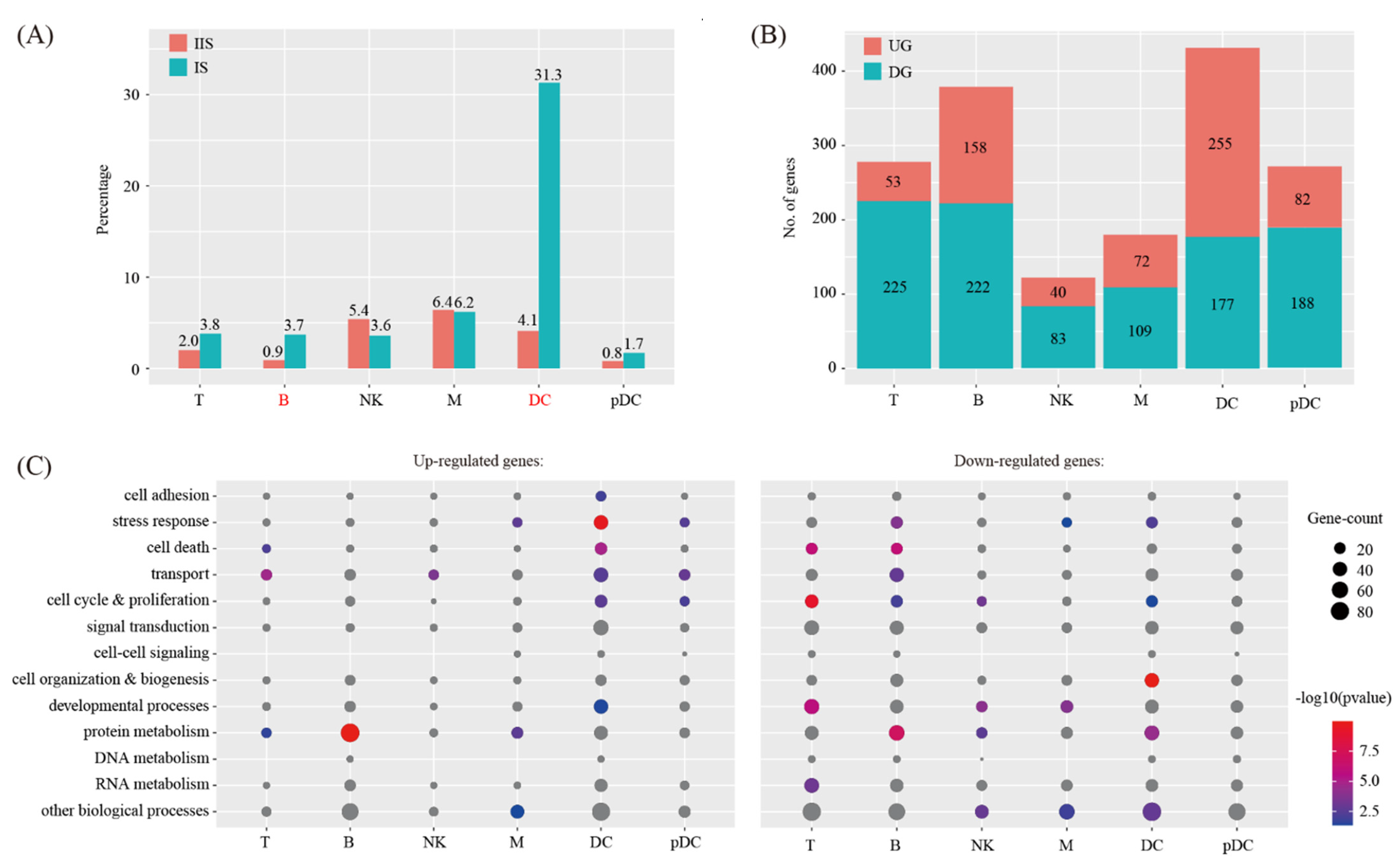
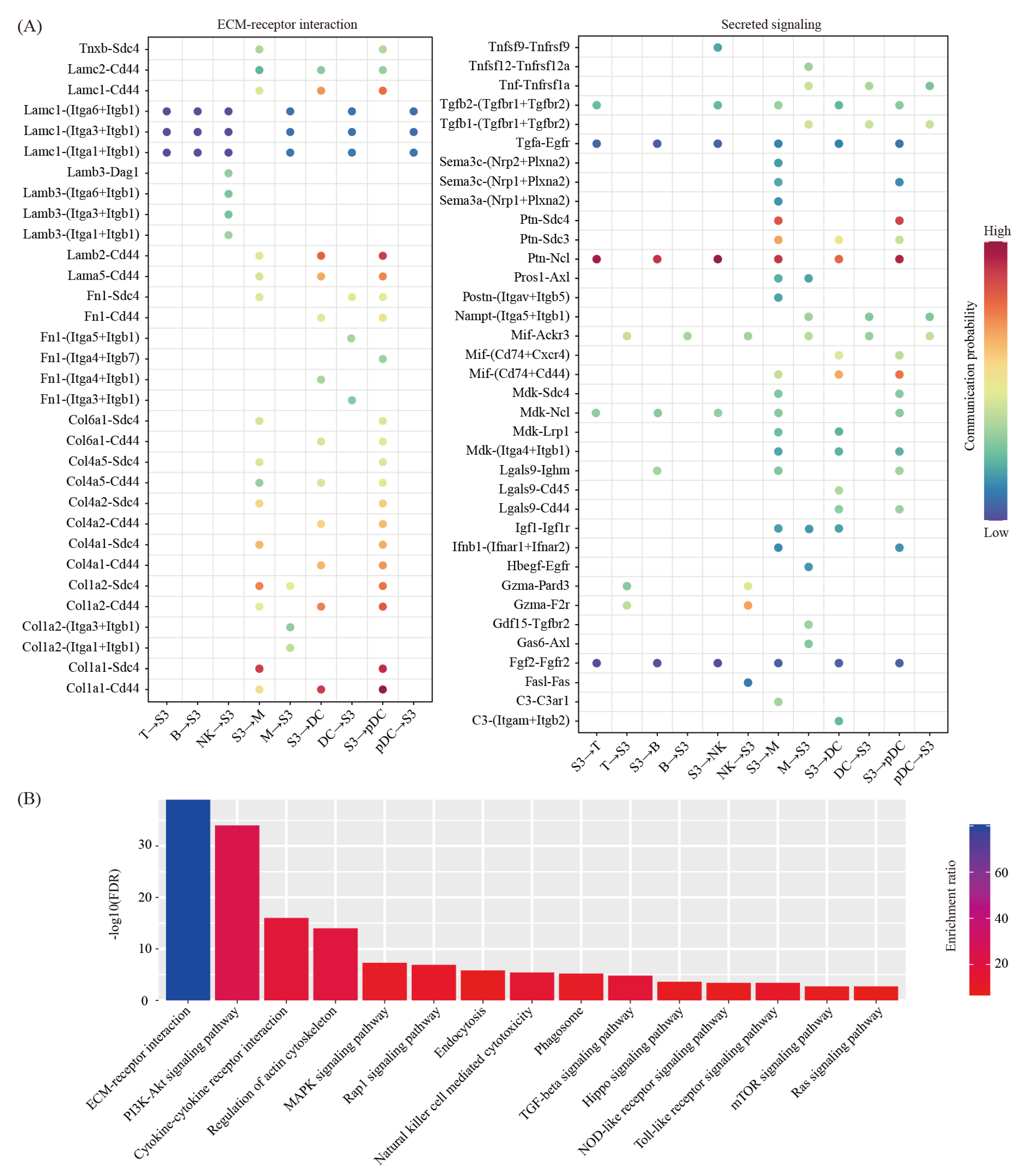
2.3. Cell-Cell Communication between Decidual Cells and Immune Cells
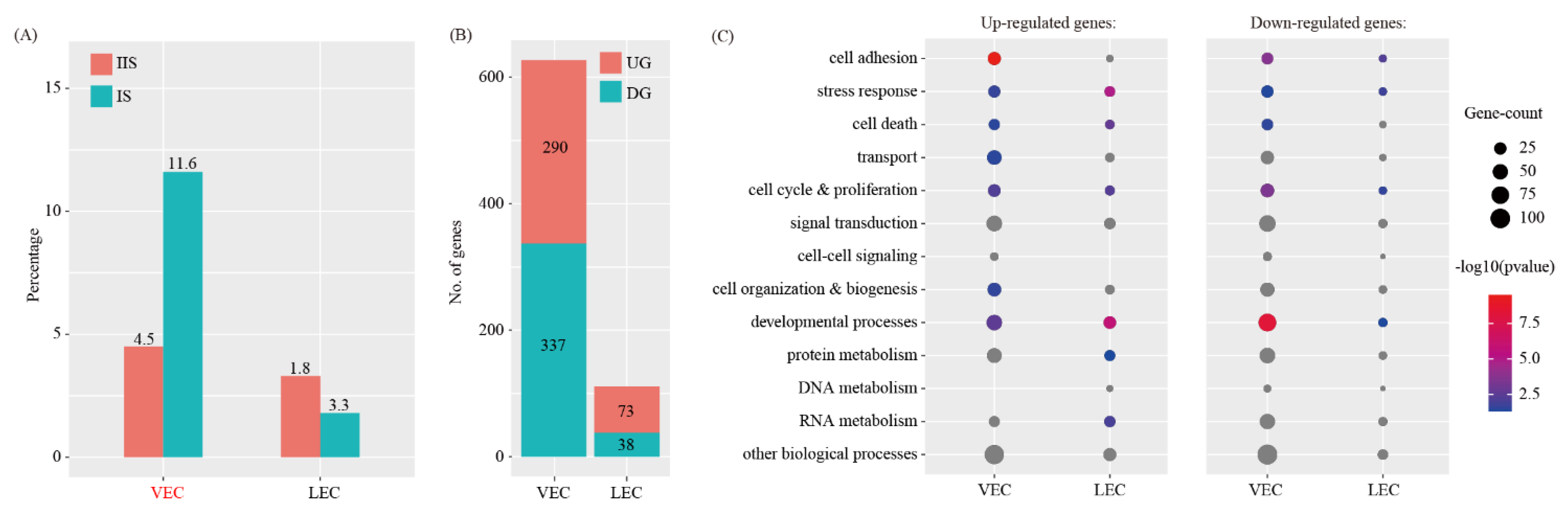
2.4. Cell-Cell Communication between Decidual Cells and Endothelial Cells
2.5. Cell-Cell Communication between Decidual Cells and Trophoblast Cells
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Bulk RNA-Seq Analysis
4.3. Single-Cell Dissociation of Mouse Uterus
4.4. Single-Cell RNA-Seq Library Preparation and Sequencing
4.5. Single Cell RNA-Seq Data Analysis
4.6. Pseudotime Analysis
4.7. Gene Ontology Analysis
4.8. Pathway Analysis
4.9. Cell-Cell Communication Analysis
4.10. Validation by Quantitative RT-PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, H.; Dey, S.K. Roadmap to embryo implantation: Clues from mouse models. Nat. Rev. Genet. 2006, 7, 185–199. [Google Scholar] [CrossRef]
- Ramathal, C.Y.; Bagchi, I.C.; Taylor, R.N.; Bagchi, M.K. Endometrial decidualization: Of mice and men. Semin. Reprod. Med. 2010, 28, 17–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christian, M.; Mak, I.; White, J.O.; Brosens, J.J. Mechanisms of decidualization. Reprod. Biomed. Online 2002, 4 (Suppl. 3), 24–30. [Google Scholar] [CrossRef]
- Gellersen, B.; Brosens, J.J. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr. Rev. 2014, 35, 851–905. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, M.; Zhang, W.Q.; Huang, M.Y.; Zhu, C.; He, J.P.; Liu, J.L. Comparative analysis of mouse decidualization models at the molecular level. Genes 2020, 11, 935. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Wang, T.S. Systematic analysis of the molecular mechanism underlying decidualization using a text mining approach. PLoS ONE 2015, 10, e0134585. [Google Scholar] [CrossRef]
- Cheng, C.W.; Bielby, H.; Licence, D.; Smith, S.K.; Print, C.G.; Charnock-Jones, D.S. Quantitative cellular and molecular analysis of the effect of progesterone withdrawal in a murine model of decidualization. Biol. Reprod. 2007, 76, 871–883. [Google Scholar] [CrossRef] [Green Version]
- Kashiwagi, A.; DiGirolamo, C.M.; Kanda, Y.; Niikura, Y.; Esmon, C.T.; Hansen, T.R.; Shioda, T.; Pru, J.K. The postimplantation embryo differentially regulates endometrial gene expression and decidualization. Endocrinology 2007, 148, 4173–4184. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.L.; Zhang, W.Q.; Zhao, M.; Huang, M.Y. Integration of transcriptomic and metabolomic data reveals enhanced steroid hormone biosynthesis in mouse uterus during decidualization. Proteomics 2017, 17, 1700059. [Google Scholar] [CrossRef]
- McConaha, M.E.; Eckstrum, K.; An, J.; Steinle, J.J.; Bany, B.M. Microarray assessment of the influence of the conceptus on gene expression in the mouse uterus during decidualization. Reproduction 2011, 141, 511–527. [Google Scholar] [CrossRef] [Green Version]
- Svensson, V.; Vento-Tormo, R.; Teichmann, S.A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 2018, 13, 599–604. [Google Scholar] [CrossRef]
- Hamilton, K.J.; Arao, Y.; Korach, K.S. Estrogen hormone physiology: Reproductive findings from estrogen receptor mutant mice. Reprod. Biol. 2014, 14, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.P.; Li, R.; DeMayo, F.J. Progesterone receptor regulation of uterine adaptation for pregnancy. Trends Endocrinol. Metab. 2018, 29, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Kalucka, J.; de Rooij, L.; Goveia, J.; Rohlenova, K.; Dumas, S.J.; Meta, E.; Conchinha, N.V.; Taverna, F.; Teuwen, L.A.; Veys, K.; et al. Single-cell transcriptome atlas of murine endothelial cells. Cell 2020, 180, 764–779.e20. [Google Scholar] [CrossRef] [PubMed]
- Hilton, H.G.; Rubinstein, N.D.; Janki, P.; Ireland, A.T.; Bernstein, N.; Fong, N.L.; Wright, K.M.; Smith, M.; Finkle, D.; Martin-McNulty, B.; et al. Single-cell transcriptomics of the naked mole-rat reveals unexpected features of mammalian immunity. PLoS Biol. 2019, 17, e3000528. [Google Scholar] [CrossRef] [PubMed]
- Jin, S. Bipotent stem cells support the cyclical regeneration of endometrial epithelium of the murine uterus. Proc. Natl. Acad. Sci. USA 2019, 116, 6848–6857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gendron, R.L.; Paradis, H.; Hsieh-Li, H.M.; Lee, D.W.; Potter, S.S.; Markoff, E. Abnormal uterine stromal and glandular function associated with maternal reproductive defects in Hoxa-11 null mice. Biol. Reprod. 1997, 56, 1097–1105. [Google Scholar] [CrossRef]
- Suryawanshi, H.; Morozov, P.; Straus, A.; Sahasrabudhe, N.; Max, K.E.A.; Garzia, A.; Kustagi, M.; Tuschl, T.; Williams, Z. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 2018, 4, eaau4788. [Google Scholar] [CrossRef] [Green Version]
- Mucenski, M.L.; Mahoney, R.; Adam, M.; Potter, A.S.; Potter, S.S. Single cell rna-seq study of wild type and Hox9,10,11 mutant developing uterus. Sci. Rep. 2019, 9, 4557. [Google Scholar] [CrossRef] [Green Version]
- Fu, D.J.; De Micheli, A.J.; Bidarimath, M.; Ellenson, L.H.; Cosgrove, B.D.; Flesken-Nikitin, A.; Nikitin, A.Y. Cells expressing Pax8 are the main source of homeostatic regeneration of adult mouse endometrial epithelium and give rise to serous endometrial carcinoma. Dis. Model. Mech. 2020, 13, dmm047035. [Google Scholar] [CrossRef]
- Li, Q.; Kannan, A.; DeMayo, F.J.; Lydon, J.P.; Cooke, P.S.; Yamagishi, H.; Srivastava, D.; Bagchi, M.K.; Bagchi, I.C. The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science 2011, 331, 912–916. [Google Scholar] [CrossRef] [Green Version]
- Alam, S.M.; Konno, T.; Soares, M.J. Identification of target genes for a prolactin family paralog in mouse decidua. Reproduction 2015, 149, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Franco, H.L.; Dai, D.; Lee, K.Y.; Rubel, C.A.; Roop, D.; Boerboom, D.; Jeong, J.W.; Lydon, J.P.; Bagchi, I.C.; Bagchi, M.K.; et al. Wnt4 is a key regulator of normal postnatal uterine development and progesterone signaling during embryo implantation and decidualization in the mouse. FASEB J. 2011, 25, 1176–1187. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Kannan, A.; Wang, W.; Demayo, F.J.; Taylor, R.N.; Bagchi, M.K.; Bagchi, I.C. Bone morphogenetic protein 2 functions via a conserved signaling pathway involving Wnt4 to regulate uterine decidualization in the mouse and the human. J. Biol. Chem. 2007, 282, 31725–31732. [Google Scholar] [CrossRef] [Green Version]
- Keenihan, S.N.; Robertson, S.A. Diversity in phenotype and steroid hormone dependence in dendritic cells and macrophages in the mouse uterus. Biol. Reprod. 2004, 70, 1562–1572. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Gaugler, B.; Mohty, M.; Malard, F. Plasmacytoid dendritic cell biology and its role in immune-mediated diseases. Clin. Transl. Immunol. 2020, 9, e1139. [Google Scholar] [CrossRef]
- Qiu, X.; Hill, A.; Packer, J.; Lin, D.; Ma, Y.A.; Trapnell, C. Single-cell mrna quantification and differential analysis with census. Nat. Methods 2017, 14, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using cellchat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.M.; Smith, Z.D.; Grosswendt, S.; Kretzmer, H.; Norman, T.M.; Adamson, B.; Jost, M.; Quinn, J.J.; Yang, D.; Jones, M.G.; et al. Molecular recording of mammalian embryogenesis. Nature 2019, 570, 77–82. [Google Scholar] [CrossRef] [Green Version]
- Donnison, M.; Broadhurst, R.; Pfeffer, P.L. Elf5 and Ets2 maintain the mouse extraembryonic ectoderm in a dosage dependent synergistic manner. Dev. Biol. 2015, 397, 77–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmons, D.G.; Natale, D.R.; Begay, V.; Hughes, M.; Leutz, A.; Cross, J.C. Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development 2008, 135, 2083–2091. [Google Scholar] [CrossRef] [Green Version]
- El-Hashash, A.H.; Warburton, D.; Kimber, S.J. Genes and signals regulating murine trophoblast cell development. Mech. Dev. 2010, 127, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; He, J.P.; Liu, J.L. Cell-cell communication at the embryo implantation site of mouse uterus revealed by single-cell analysis. Int. J. Mol. Sci. 2021, 22, 5177. [Google Scholar] [CrossRef]
- Van den Brink, S.C.; Sage, F.; Vertesy, A.; Spanjaard, B.; Peterson-Maduro, J.; Baron, C.S.; Robin, C.; van Oudenaarden, A. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 2017, 14, 935–936. [Google Scholar] [CrossRef]
- Krjutskov, K.; Katayama, S.; Saare, M.; Vera-Rodriguez, M.; Lubenets, D.; Samuel, K.; Laisk-Podar, T.; Teder, H.; Einarsdottir, E.; Salumets, A.; et al. Single-cell transcriptome analysis of endometrial tissue. Hum. Reprod. 2016, 31, 844–853. [Google Scholar] [CrossRef] [Green Version]
- Parasar, P.; Sacha, C.R.; Ng, N.; McGuirk, E.R.; Chinthala, S.; Ozcan, P.; Lindsey, J.; Salas, S.; Laufer, M.R.; Missmer, S.A.; et al. Differentiating mouse embryonic stem cells express markers of human endometrium. Reprod. Biol. Endocrinol. 2017, 15, 52. [Google Scholar] [CrossRef] [PubMed]
- Zenclussen, A.C.; Hammerling, G.J. Cellular regulation of the uterine microenvironment that enables embryo implantation. Front. Immunol. 2015, 6, 321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaks, V.; Birnberg, T.; Berkutzki, T.; Sela, S.; BenYashar, A.; Kalchenko, V.; Mor, G.; Keshet, E.; Dekel, N.; Neeman, M.; et al. Uterine DCs are crucial for decidua formation during embryo implantation in mice. J. Clin. Invest. 2008, 118, 3954–3965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, S.W.; Norwitz, G.A.; Pavlicev, M.; Tilburgs, T.; Simon, C.; Norwitz, E.R. Endometrial decidualization: The primary driver of pregnancy health. Int. J. Mol. Sci. 2020, 21, 4092. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. Tophat: Discovering splice junctions with rna-seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive integration of single-cell data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Zuo, R.J.; Peng, Y.; Fu, Y.S. The impact of multiparity on uterine gene expression and decidualization in mice. Reprod. Sci. 2016, 23, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Law, M.; Shaw, D.R. Mouse Genome Informatics (MGI) is the international resource for information on the laboratory mouse. Methods Mol. Biol. 2018, 1757, 141–161. [Google Scholar] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.-P.; Tian, Q.; Zhu, Q.-Y.; Liu, J.-L. Identification of Intercellular Crosstalk between Decidual Cells and Niche Cells in Mice. Int. J. Mol. Sci. 2021, 22, 7696. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147696
He J-P, Tian Q, Zhu Q-Y, Liu J-L. Identification of Intercellular Crosstalk between Decidual Cells and Niche Cells in Mice. International Journal of Molecular Sciences. 2021; 22(14):7696. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147696
Chicago/Turabian StyleHe, Jia-Peng, Qing Tian, Qiu-Yang Zhu, and Ji-Long Liu. 2021. "Identification of Intercellular Crosstalk between Decidual Cells and Niche Cells in Mice" International Journal of Molecular Sciences 22, no. 14: 7696. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147696







