Expression and Role of Ubiquitin-Specific Peptidases in Osteoblasts
Abstract
:1. Ubiquitination
2. USPs: Conserved Structural and Functional Domains
3. A Glance at the Skeleton
4. Regulation of Bone Remodeling
The Ubiquitin-Proteasome System
5. USPs and Osteoblasts
5.1. Regulation of Signal Transduction Pathways
5.2. Mesenchymal Commitment and Differentiation
5.2.1. USP34 and USP7
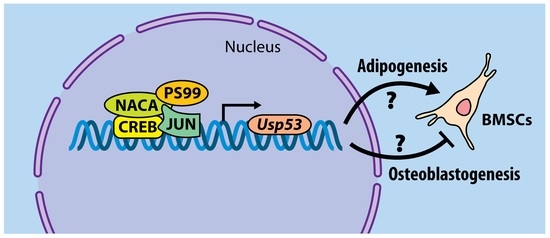
5.2.2. USP53
6. Conclusions
7. Materials and Methods
7.1. RNA-seq
7.2. Immunofluorescence
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [Green Version]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Akutsu, M.; Dikic, I.; Bremm, A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016, 129, 875–880. [Google Scholar] [CrossRef] [Green Version]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [Green Version]
- Ciechanover, A.; Kwon, Y.T. Degradation of misfolded proteins in neurodegenerative diseases: Therapeutic targets and strategies. Exp. Mol. Med. 2015, 47, e147. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal. Transduct. Target. Ther. 2020, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.F.; Gan, Z.Y.; Komander, D.; Dewson, G. Ubiquitin signalling in neurodegeneration: Mechanisms and therapeutic opportunities. Cell Death Differ. 2021, 28, 570–590. [Google Scholar] [CrossRef]
- Herhaus, L.; Dikic, I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015, 16, 1071–1083. [Google Scholar] [CrossRef] [Green Version]
- Ohtake, F.; Saeki, Y.; Sakamoto, K.; Ohtake, K.; Nishikawa, H.; Tsuchiya, H.; Ohta, T.; Tanaka, K.; Kanno, J. Ubiquitin acetylation inhibits polyubiquitin chain elongation. EMBO Rep. 2015, 16, 192–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatrin, C.; Gabrielsen, M.; Buetow, L.; Nakasone, M.A.; Ahmed, S.F.; Sumpton, D.; Sibbet, G.J.; Smith, B.O.; Huang, D.T. Structural insights into ADP-ribosylation of ubiquitin by Deltex family E3 ubiquitin ligases. Sci. Adv. 2020, 6, eabc0418. [Google Scholar] [CrossRef] [PubMed]
- Vivelo, C.A.; Ayyappan, V.; Leung, A.K.L. Poly(ADP-ribose)-dependent ubiquitination and its clinical implications. Biochem. Pharmacol. 2019, 167, 3–12. [Google Scholar] [CrossRef]
- Pérez Berrocal, D.A.; Witting, K.F.; Ovaa, H.; Mulder, M.P.C. Hybrid Chains: A Collaboration of Ubiquitin and Ubiquitin-Like Modifiers Introducing Cross-Functionality to the Ubiquitin Code. Front. Chem. 2020, 7, 931. [Google Scholar] [CrossRef]
- French, M.E.; Koehler, C.F.; Hunter, T. Emerging functions of branched ubiquitin chains. Cell Discov. 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Haakonsen, D.L.; Rape, M. Branching Out: Improved Signaling by Heterotypic Ubiquitin Chains. Trends Cell Biol. 2019, 29, 704–716. [Google Scholar] [CrossRef]
- Clague, M.J.; Urbé, S. Ubiquitin: Same molecule, different degradation pathways. Cell 2010, 143, 682–685. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Luo, Z.-Q. Post-translational regulation of ubiquitin signaling. J. Cell Biol. 2019, 218, 1776–1786. [Google Scholar] [CrossRef] [PubMed]
- Wauer, T.; Swatek, K.N.; Wagstaff, J.L.; Gladkova, C.; Pruneda, J.N.; Michel, M.A.; Gersch, M.; Johnson, C.M.; Freund, S.M.; Komander, D. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015, 34, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Basar, M.A.; Beck, D.B.; Werner, A. Deubiquitylases in developmental ubiquitin signaling and congenital diseases. Cell Death Differ. 2021, 28, 538–556. [Google Scholar] [CrossRef]
- Nijman, S.M.B.; Luna-Vargas, M.P.A.; Velds, A.; Brummelkamp, T.R.; Dirac, A.M.G.; Sixma, T.K.; Bernards, R. A Genomic and Functional Inventory of Deubiquitinating Enzymes. Cell 2005, 123, 773–786. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, K.D. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997, 11, 1245–1256. [Google Scholar] [CrossRef]
- Cai, X.; Wang, Z.; Hou, Y.; Liu, C.; Hendy, A.; Xing, J.; Chen, X.-L. Systematic characterization of the ubiquitin-specific proteases in Magnaporthe oryzae. Phytopathol. Res. 2020, 2, 8. [Google Scholar] [CrossRef]
- Tsou, W.-L.; Sheedlo, M.J.; Morrow, M.E.; Blount, J.R.; McGregor, K.M.; Das, C.; Todi, S.V. Systematic analysis of the physiological importance of deubiquitinating enzymes. PLoS ONE 2012, 7, e43112. [Google Scholar]
- Overstreet, E.; Fitch, E.; Fischer, J.A. Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells. Development 2004, 131, 5355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlasschaert, C.; Cook, D.; Xia, X.; Gray, D.A. The Evolution and Functional Diversification of the Deubiquitinating Enzyme Superfamily. Genome Biol. Evol. 2017, 9, 558–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Scheel, H.; Hofmann, K.; Komander, D. Dissection of USP catalytic domains reveals five common insertion points. Mol. BioSyst. 2009, 5, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Ronau, J.A.; Beckmann, J.F.; Hochstrasser, M. Substrate specificity of the ubiquitin and Ubl proteases. Cell Res. 2016, 26, 441–456. [Google Scholar] [CrossRef] [Green Version]
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Quesada, V.C.; Díaz-Perales, A.; Gutiérrez-Fernández, A.; Garabaya, C.; Cal, S.; López-Otín, C. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem. Biophys. Res. Commun. 2004, 314, 54–62. [Google Scholar] [CrossRef]
- Huang, Y.; Pan, X.W.; Li, L.; Chen, L.; Liu, X.; Lu, J.L.; Zhu, X.M.; Huang, H.; Yang, Q.W.; Ye, J.Q.; et al. Overexpression of USP39 predicts poor prognosis and promotes tumorigenesis of prostate cancer via promoting EGFR mRNA maturation and transcription elongation. Oncotarget 2016, 7, 22016–22030. [Google Scholar] [CrossRef] [Green Version]
- van Leuken, R.J.; Luna-Vargas, M.P.; Sixma, T.K.; Wolthuis, R.M.F.; Medema, R.H. Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of Aurora B. Cell Cycle 2008, 7, 2710–2719. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Ji, J.; Zhang, X.; Huang, B.; Chen, A.; Zhang, D.; Li, X.; Wang, X.; Wang, J. RNA splicing factor USP39 promotes glioma progression by inducing TAZ mRNA maturation. Oncogene 2019, 38, 6414–6428. [Google Scholar] [CrossRef] [PubMed]
- Behl, T.; Chadha, S.; Sachdeva, M.; Kumar, A.; Hafeez, A.; Mehta, V.; Bungau, S. Ubiquitination in rheumatoid arthritis. Life Sci. 2020, 261, 118459. [Google Scholar] [CrossRef]
- Fiore, A.; Liang, Y.; Lin, Y.H.; Tung, J.; Wang, H.; Langlais, D.; Nijnik, A. Deubiquitinase MYSM1 in the Hematopoietic System and beyond: A Current Review. Int. J. Mol. Sci. 2020, 21, 3007. [Google Scholar] [CrossRef]
- Gupta, I.; Singh, K.; Varshney, N.K.; Khan, S. Delineating Crosstalk Mechanisms of the Ubiquitin Proteasome System That Regulate Apoptosis. Front. Cell Dev. Biol. 2018, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Herhaus, L.; Sapkota, G.P. The emerging roles of deubiquitylating enzymes (DUBs) in the TGFβ and BMP pathways. Cell. Signal. 2014, 26, 2186–2192. [Google Scholar] [CrossRef] [Green Version]
- Kwon, S.K.; Saindane, M.; Baek, K.H. p53 stability is regulated by diverse deubiquitinating enzymes. Biochim. Biophys. Acta. Rev. Cancer 2017, 1868, 404–411. [Google Scholar] [CrossRef]
- Sévère, N.; Dieudonné, F.X.; Marie, P.J. E3 ubiquitin ligase-mediated regulation of bone formation and tumorigenesis. Cell Death Dis. 2013, 4, e463. [Google Scholar] [CrossRef] [Green Version]
- Sulkshane, P.; Ram, J.; Glickman, M.H. Ubiquitination of Intramitochondrial Proteins: Implications for Metabolic Adaptability. Biomolecules 2020, 10, 1559. [Google Scholar] [CrossRef]
- Parfitt, A.M. Targeted and nontargeted bone remodeling: Relationship to basic multicellular unit origination and progression. Bone 2002, 30, 5–7. [Google Scholar] [CrossRef]
- Bonewald, L.F. The Role of the Osteocyte in Bone and Nonbone Disease. Endocrinol. Metab. Clin. N. Am. 2017, 46, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Sims, N.A.; Martin, T.J. Osteoclasts Provide Coupling Signals to Osteoblast Lineage Cells Through Multiple Mechanisms. Annu. Rev. Physiol. 2020, 82, 507–529. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; McDonald, J.M. Disorders of bone remodeling. Annu. Rev. Pathol. 2011, 6, 121–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddiqui, J.A.; Partridge, N.C. Physiological Bone Remodeling: Systemic Regulation and Growth Factor Involvement. Physiology 2016, 31, 233–245. [Google Scholar] [CrossRef]
- Teti, A. Bone development: Overview of bone cells and signaling. Curr. Osteoporos. Rep. 2011, 9, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.T.; Gao, Y.H.; Inada, M.; et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef] [Green Version]
- Amarasekara, D.S.; Kim, S.; Rho, J. Regulation of Osteoblast Differentiation by Cytokine Networks. Int. J. Mol. Sci. 2021, 22, 2851. [Google Scholar] [CrossRef]
- Kim, J.M.; Yang, Y.S.; Park, K.H.; Ge, X.; Xu, R.; Li, N.; Song, M.; Chun, H.; Bok, S.; Charles, J.F.; et al. A RUNX2 stabilization pathway mediates physiologic and pathologic bone formation. Nat. Commun. 2020, 11, 2289. [Google Scholar] [CrossRef]
- Zhou, F.; Li, F.; Fang, P.; Dai, T.; Yang, B.; van Dam, H.; Jia, J.; Zheng, M.; Zhang, L. Ubiquitin-Specific Protease 4 Antagonizes Osteoblast Differentiation Through Dishevelled. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2016, 31, 1888–1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Lv, L.; Li, W.; Zhang, X.; Jiang, Y.; Ge, W.; Zhou, Y. Protein deubiquitinase USP7 is required for osteogenic differentiation of human adipose-derived stem cells. Stem Cell Res. Ther. 2017, 8, 186. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Wang, M.; Xue, H.; Liu, W.; Guo, Y.; Xu, R.; Shao, B.; Yuan, Q. Ubiquitin-Specific Protease 34 Inhibits Osteoclast Differentiation by Regulating NF-κB Signaling. J. Bone Miner. Res. 2020, 35, 1597–1608. [Google Scholar] [CrossRef]
- Yim, H.Y.; Park, C.; Lee, Y.D.; Arimoto, K.; Jeon, R.; Baek, S.H.; Zhang, D.E.; Kim, H.H.; Kim, K.I. Elevated Response to Type I IFN Enhances RANKL-Mediated Osteoclastogenesis in Usp18-Knockout Mice. J. Immunol. 2016, 196, 3887–3895. [Google Scholar] [CrossRef]
- Zangari, M.; Suva, L.J. The effects of proteasome inhibitors on bone remodeling in multiple myeloma. Bone 2016, 86, 131–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyajobi, B.O.; Garrett, I.R.; Gupta, A.; Flores, A.; Esparza, J.; Muñoz, S.; Zhao, M.; Mundy, G.R. Stimulation of new bone formation by the proteasome inhibitor, bortezomib: Implications for myeloma bone disease. Br. J. Haematol. 2007, 139, 434–438. [Google Scholar] [CrossRef]
- Uy, G.L.; Trivedi, R.; Peles, S.; Fisher, N.M.; Zhang, Q.J.; Tomasson, M.H.; DiPersio, J.F.; Vij, R. Bortezomib inhibits osteoclast activity in patients with multiple myeloma. Clin. Lymphoma Myeloma 2007, 7, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Raje, N.; Schoonmaker, J.A.; Liu, J.C.; Hideshima, T.; Wein, M.N.; Jones, D.C.; Vallet, S.; Bouxsein, M.L.; Pozzi, S.; et al. Pharmacologic targeting of a stem/progenitor population in vivo is associated with enhanced bone regeneration in mice. J. Clin. Investig. 2008, 118, 491–504. [Google Scholar] [CrossRef]
- Qiang, Y.W.; Hu, B.; Chen, Y.; Zhong, Y.; Shi, B.; Barlogie, B.; Shaughnessy, J.D., Jr. Bortezomib induces osteoblast differentiation via Wnt-independent activation of beta-catenin/TCF signaling. Blood 2009, 113, 4319–4330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrett, I.R.; Chen, D.; Gutierrez, G.; Zhao, M.; Escobedo, A.; Rossini, G.; Harris, S.E.; Gallwitz, W.; Kim, K.B.; Hu, S.; et al. Selective inhibitors of the osteoblast proteasome stimulate bone formation in vivo and in vitro. J. Clin. Investig. 2003, 111, 1771–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; De Veirman, K.; Fan, R.; Jian, Q.; Zhang, Y.; Lei, L.; Evans, H.; Wang, Y.; Lei, L.; Wang, B.; et al. ER stress arm XBP1s plays a pivotal role in proteasome inhibition-induced bone formation. Stem Cell Res. Ther. 2020, 11, 516. [Google Scholar] [CrossRef]
- Qiang, Y.-W.; Heuck, C.J.; Shaughnessy, J.D., Jr.; Barlogie, B.; Epstein, J. Proteasome inhibitors and bone disease. Semin Hematol. 2012, 49, 243–248. [Google Scholar] [CrossRef] [Green Version]
- Ang, E.; Pavlos, N.J.; Rea, S.L.; Qi, M.; Chai, T.; Walsh, J.P.; Ratajczak, T.; Zheng, M.H.; Xu, J. Proteasome inhibitors impair RANKL-induced NF-kappaB activity in osteoclast-like cells via disruption of p62, TRAF6, CYLD, and IkappaBalpha signaling cascades. J. Cell. Physiol. 2009, 220, 450–459. [Google Scholar] [CrossRef]
- Phimphilai, M.; Zhao, Z.; Boules, H.; Roca, H.; Franceschi, R.T. BMP signaling is required for RUNX2-dependent induction of the osteoblast phenotype. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2006, 21, 637–646. [Google Scholar] [CrossRef] [Green Version]
- Al-Salihi, M.A.; Herhaus, L.; Macartney, T.; Sapkota, G.P. USP11 augments TGFβ signalling by deubiquitylating ALK5. Open Biol. 2012, 2, 120063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichhorn, P.J.; Rodón, L.; Gonzàlez-Juncà, A.; Dirac, A.; Gili, M.; Martínez-Sáez, E.; Aura, C.; Barba, I.; Peg, V.; Prat, A.; et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat. Med. 2012, 18, 429–435. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, F.; Drabsch, Y.; Gao, R.; Snaar-Jagalska, B.E.; Mickanin, C.; Huang, H.; Sheppard, K.A.; Porter, J.A.; Lu, C.X.; et al. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nat. Cell Biol. 2012, 14, 717–726. [Google Scholar] [CrossRef]
- Xiao, L.; Peng, X.; Liu, F.; Tang, C.; Hu, C.; Xu, X.; Wang, M.; Luo, Y.; Yang, S.; Song, P.; et al. AKT regulation of mesothelial-to-mesenchymal transition in peritoneal dialysis is modulated by Smurf2 and deubiquitinating enzyme USP4. BMC Cell Biol. 2015, 16, 7. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Xie, F.; Jin, K.; Zhang, Z.; Clerici, M.; Gao, R.; van Dinther, M.; Sixma, T.K.; Huang, H.; Zhang, L.; et al. USP4 inhibits SMAD4 monoubiquitination and promotes activin and BMP signaling. EMBO J. 2017, 36, 1623–1639. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.H.; Yu, Y.; Mao, R.F.; Tan, X.J.; Xu, G.F.; Zhang, H.; Lu, X.B.; Fu, S.B.; Yang, J. USP4 targets TAK1 to downregulate TNFα-induced NF-κB activation. Cell Death Differ. 2011, 18, 1547–1560. [Google Scholar] [CrossRef]
- Hwang, S.J.; Lee, H.W.; Kim, H.R.; Lee, H.; Shin, C.H.; Yun, S.I.; Lee, D.H.; Kim, D.H.; Kim, K.K.; Joo, K.M.; et al. Ubiquitin-specific protease 4 controls metastatic potential through β-catenin stabilization in brain metastatic lung adenocarcinoma. Sci. Rep. 2016, 6, 21596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, S.I.; Kim, H.H.; Yoon, J.H.; Park, W.S.; Hahn, M.J.; Kim, H.C.; Chung, C.H.; Kim, K.K. Ubiquitin specific protease 4 positively regulates the WNT/β-catenin signaling in colorectal cancer. Mol. Oncol. 2015, 9, 1834–1851. [Google Scholar] [CrossRef] [PubMed]
- Herhaus, L.; Al-Salihi, M.A.; Dingwell, K.S.; Cummins, T.D.; Wasmus, L.; Vogt, J.; Ewan, R.; Bruce, D.; Macartney, T.; Weidlich, S.; et al. USP15 targets ALK3/BMPR1A for deubiquitylation to enhance bone morphogenetic protein signalling. Open Biol. 2014, 4, 140065. [Google Scholar] [CrossRef] [Green Version]
- Das, T.; Song, E.J.; Kim, E.E. The Multifaceted Roles of USP15 in Signal Transduction. Int. J. Mol. Sci. 2021, 22, 4728. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, M.B.; Shin, D.Y.; Oh, H.; Lee, K.-Y.; Zhai, B.; Gygi, S.P.; Lotinun, S.; Baron, R.; Liu, D.; Su, B.; et al. MEKK2 mediates an alternative β-catenin pathway that promotes bone formation. Proc. Natl. Acad. Sci. USA 2016, 113, E1226–E1235. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Langelotz, C.; Hetfeld-Pěchoč, B.K.J.; Schwenk, W.; Dubiel, W. The COP9 Signalosome Mediates β-Catenin Degradation by Deneddylation and Blocks Adenomatous Polyposis coli Destruction via USP15. J. Mol. Biol. 2009, 391, 691–702. [Google Scholar] [CrossRef]
- Guo, Y.C.; Wang, M.Y.; Zhang, S.W.; Wu, Y.S.; Zhou, C.C.; Zheng, R.X.; Shao, B.; Wang, Y.; Xie, L.; Liu, W.Q.; et al. Ubiquitin-specific protease USP34 controls osteogenic differentiation and bone formation by regulating BMP2 signaling. EMBO J. 2018, 37, e99398. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Lu, B.; Zamponi, R.; Charlat, O.; Aversa, R.; Yang, Z.; Sigoillot, F.; Zhu, X.; Hu, T.; Reece-Hoyes, J.S.; et al. USP7 inhibits Wnt/β-catenin signaling through promoting stabilization of Axin. Nat. Commun. 2019, 10, 4184. [Google Scholar] [CrossRef] [PubMed]
- Hariri, H.; Addison, W.N.; St-Arnaud, R. Ubiquitin specific peptidase Usp53 regulates osteoblast versus adipocyte lineage commitment. Sci. Rep. 2021, 11, 8418. [Google Scholar] [CrossRef]
- Hariri, H.; Pellicelli, M.; St-Arnaud, R. Nfil3, a target of the NACA transcriptional coregulator, affects osteoblast and osteocyte gene expression differentially. Bone 2020, 141, 115624. [Google Scholar] [CrossRef]
- Pellicelli, M.; Hariri, H.; Miller, J.A.; St-Arnaud, R. Lrp6 is a target of the PTH-activated αNAC transcriptional coregulator. Biochim. Biophys. Acta. Gene Regul. Mech. 2018, 1861, 61–71. [Google Scholar] [CrossRef]
- Pellicelli, M.; Miller, J.A.; Arabian, A.; Gauthier, C.; Akhouayri, O.; Wu, J.Y.; Kronenberg, H.M.; St-Arnaud, R. The PTH-Gαs-protein kinase A cascade controls αNAC localization to regulate bone mass. Mol. Cell Biol. 2014, 34, 1622–1633. [Google Scholar] [CrossRef] [Green Version]
- Jilka, R.L.; Weinstein, R.S.; Bellido, T.; Roberson, P.; Parfitt, A.M.; Manolagas, S.C. Increased bone formation by prevention of osteoblast apoptosis with parathyroid hormone. J. Clin. Investig. 1999, 104, 439–446. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.W.; Pajevic, P.D.; Selig, M.; Barry, K.J.; Yang, J.Y.; Shin, C.S.; Baek, W.Y.; Kim, J.E.; Kronenberg, H.M. Intermittent parathyroid hormone administration converts quiescent lining cells to active osteoblasts. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2012, 27, 2075–2084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, C.A.; Plotkin, L.I.; Galli, C.; Goellner, J.J.; Gortazar, A.R.; Allen, M.R.; Robling, A.G.; Bouxsein, M.; Schipani, E.; Turner, C.H.; et al. Control of bone mass and remodeling by PTH receptor signaling in osteocytes. PLoS ONE 2008, 3, e2942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hariri, H.; Pellicelli, M.; St-Arnaud, R. New PTH Signals Mediating Bone Anabolism. Curr. Mol. Biol. Rep. 2017, 3, 133–141. [Google Scholar] [CrossRef]
- Wein, M.N. Parathyroid Hormone Signaling in Osteocytes. JBMR Plus 2017, 2, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Shirakawa, J.; Harada, H.; Noda, M.; Ezura, Y. PTH-Induced Osteoblast Proliferation Requires Upregulation of the Ubiquitin-Specific Peptidase 2 (Usp2) Expression. Calcif. Tissue Int. 2016, 98, 306–315. [Google Scholar] [CrossRef]
- Alhebbi, H.; Peer-Zada, A.A.; Al-Hussaini, A.A.; Algubaisi, S.; Albassami, A.; AlMasri, N.; Alrusayni, Y.; Alruzug, I.M.; Alharby, E.; Samman, M.A.; et al. New paradigms of USP53 disease: Normal GGT cholestasis, BRIC, cholangiopathy, and responsiveness to rifampicin. J. Hum. Genet. 2021, 66, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Kazmierczak, M.; Harris, S.L.; Kazmierczak, P.; Shah, P.; Starovoytov, V.; Ohlemiller, K.K.; Schwander, M. Progressive Hearing Loss in Mice Carrying a Mutation in Usp53. J. Neurosci. 2015, 35, 15582–15598. [Google Scholar] [CrossRef] [Green Version]
- Kurban, M.; Kim, C.A.; Kiuru, M.; Fantauzzo, K.; Cabral, R.; Abbas, O.; Levy, B.; Christiano, A.M. Copy number variations on chromosome 4q26-27 are associated with Cantu syndrome. Dermatology 2011, 223, 316–320. [Google Scholar] [CrossRef] [Green Version]
- Maddirevula, S.; Alhebbi, H.; Alqahtani, A.; Algoufi, T.; Alsaif, H.S.; Ibrahim, N.; Abdulwahab, F.; Barr, M.; Alzaidan, H.; Almehaideb, A.; et al. Identification of novel loci for pediatric cholestatic liver disease defined by KIF12, PPM1F, USP53, LSR, and WDR83OS pathogenic variants. Genet. Med. Off. J. Am. Coll. Med. Genet. 2019, 21, 1164–1172. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; Gong, J.Y.; Li, L.T.; Li, J.Q.; Zhang, M.H.; Lu, Y.; Xie, X.B.; Hong, Y.R.; Yu, Z.; et al. Low-GGT intrahepatic cholestasis associated with biallelic USP53 variants: Clinical, histological and ultrastructural characterization. Liver Int. Off. J. Int. Assoc. Study Liver 2020, 40, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Gui, D.; Dong, Z.; Peng, W.; Jiang, W.; Huang, G.; Liu, G.; Ye, Z.; Wang, Y.; Xu, Z.; Fu, J.; et al. Ubiquitin-specific peptidase 53 inhibits the occurrence and development of clear cell renal cell carcinoma through NF-κB pathway inactivation. Cancer Med. 2021, 10, 3674–3688. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wu, X.; Wang, H.; Yu, H.; Wang, J. USP53 promotes apoptosis and inhibits glycolysis in lung adenocarcinoma through FKBP51-AKT1 signaling. Mol. Carcinog. 2020, 59, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yao, X.; Wu, C.; Chen, S.; Fan, D. Knockdown of Ubiquitin-Specific Protease 53 Enhances the Radiosensitivity of Human Cervical Squamous Cell Carcinoma by Regulating DNA Damage-Binding Protein 2. Technol. Cancer Res. Treat. 2020, 19, 1533033820929792. [Google Scholar] [CrossRef]
- Baek, D.; Park, K.H.; Lee, K.-M.; Jung, S.; Joung, S.; Kim, J.; Lee, J.W. Ubiquitin-specific protease 53 promotes osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Cell Death Dis. 2021, 12, 238. [Google Scholar] [CrossRef]


| Gene | Log2 FC | p-Value |
|---|---|---|
| Usp2 | 4.65406 | 5.0 × 10−5 |
| Usp53 | 2.17645 | 5.0 × 10−5 |
| Usp36 | 0.851054 | 5.0 × 10−5 |
| Usp9x | 0.544075 | 5.0 × 10−5 |
| Usp18 | 0.369972 | 2.9 × 10−3 |
| Usp12 | 0.355183 | 9.5 × 10−4 |
| Usp35 | −1.33667 | 1.0 × 10−4 |
| Usp27x | −1.07571 | 3.0 × 10−4 |
| Usp30 | −0.436837 | 2.0 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hariri, H.; St-Arnaud, R. Expression and Role of Ubiquitin-Specific Peptidases in Osteoblasts. Int. J. Mol. Sci. 2021, 22, 7746. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147746
Hariri H, St-Arnaud R. Expression and Role of Ubiquitin-Specific Peptidases in Osteoblasts. International Journal of Molecular Sciences. 2021; 22(14):7746. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147746
Chicago/Turabian StyleHariri, Hadla, and René St-Arnaud. 2021. "Expression and Role of Ubiquitin-Specific Peptidases in Osteoblasts" International Journal of Molecular Sciences 22, no. 14: 7746. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147746