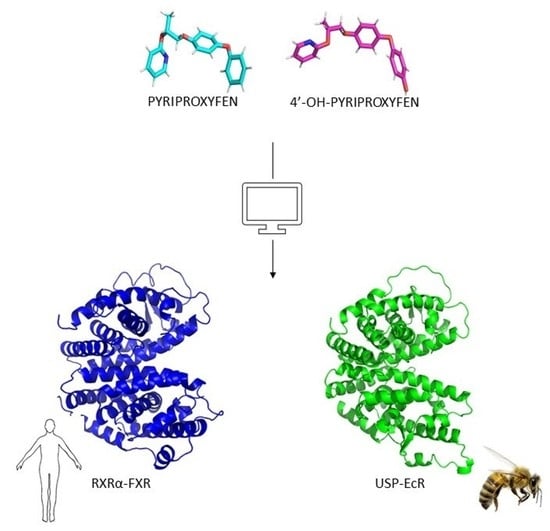
In Silico Prediction of the Mechanism of Action of Pyriproxyfen and 4′-OH-Pyriproxyfen against A. mellifera and H. sapiens Receptors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Molecular Model of USP and EcR
2.2. Comparison of A. mellifera and H. sapiens Models
2.3. Molecular Docking
2.4. Molecular Dynamic Simulations
3. Materials and Methods
3.1. Molecular Model of USP and EcR
3.2. Preparation of Proteins
3.3. Preparation of Ligands
3.4. GOLD Docking
3.5. AutoDock Docking
3.6. USP-EcR and RXRα-FXR Dimers and the Interfaces Key Interactions
3.7. Molecular Dynamic Simulations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patel, V.; Pauli, N.; Biggs, E.; Barbour, L.; Boruff, B. Why bees are critical for achieving sustainable development. Ambio 2021, 50, 49–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celli, G.; Maccagnani, B. Honey bees as bioindicators of environmental pollution. Bull. Insectol. 2003, 56, 137–139. [Google Scholar]
- Kielmanowicz, M.G.; Inberg, A.; Lerner, I.M.; Golani, Y.; Brown, N.; Turner, C.L.; Hayes, G.J.R.; Ballam, J.M. Prospective Large-Scale Field Study Generates Predictive Model Identifying Major Contributors to Colony Losses. PLoS Pathog. 2015, 11, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hristov, P.; Shumkova, R.; Palova, N.; Neov, B. Factors associated with honey bee colony losses: A mini-review. Vet. Sci. 2020, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Neov, B.; Georgieva, A.; Shumkova, R.; Radoslavov, G.; Hristov, P. Biotic and abiotic factors associated with colonies mortalities of managed honey bee (Apis mellifera). Diversity 2019, 11, 237. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, F.P. Pesticides, environment, and food safety. Food Energy Secur. 2017, 6, 48–60. [Google Scholar] [CrossRef]
- Bilal, M.; Iqbal, H.M.N.; Barceló, D. Persistence of pesticides-based contaminants in the environment and their effective degradation using laccase-assisted biocatalytic systems. Sci. Total Environ. 2019, 695. [Google Scholar] [CrossRef]
- Kim, K.H.; Kabir, E.; Jahan, S.A. Exposure to pesticides and the associated human health effects. Sci. Total Environ. 2017, 575, 525–535. [Google Scholar] [CrossRef]
- Md Meftaul, I.; Venkateswarlu, K.; Dharmarajan, R.; Annamalai, P.; Megharaj, M. Pesticides in the urban environment: A potential threat that knocks at the door. Sci. Total Environ. 2020, 711, 134612. [Google Scholar] [CrossRef] [PubMed]
- Faber, D. Poisoning the World for Profit: Petro-Chemical Capital and the Global Pesticide Crisis. Capital. Nat. Social. 2020, 31, 1–17. [Google Scholar] [CrossRef]
- Sabarwal, A.; Kumar, K.; Singh, R.P. Hazardous effects of chemical pesticides on human health–Cancer and other associated disorders. Environ. Toxicol. Pharmacol. 2018, 63, 103–114. [Google Scholar] [CrossRef]
- Rosas, L.G.; Eskenazi, B. Pesticides and child neurodevelopment. Curr. Opin. Pediatr. 2008, 20, 191–197. [Google Scholar] [CrossRef] [Green Version]
- Kaur, R.; Mavi, G.K.; Raghav, S.; Khan, I. Pesticides Classification and its Impact on Environment. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 1889–1897. [Google Scholar] [CrossRef]
- Monneret, C. What is an endocrine disruptor? Comptes Rendus Biol. 2017, 340, 403–405. [Google Scholar] [CrossRef]
- Mnif, W.; Hassine, A.I.H.; Bouaziz, A.; Bartegi, A.; Thomas, O.; Roig, B. Effect of endocrine disruptor pesticides: A review. Int. J. Environ. Res. Public Health 2011, 8, 2265–2303. [Google Scholar] [CrossRef] [Green Version]
- Diamanti-Kandarakis, E.; Bourguignon, J.P.; Giudice, L.C.; Hauser, R.; Prins, G.S.; Soto, A.M.; Zoeller, R.T.; Gore, A.C. Endocrine-disrupting chemicals: An Endocrine Society scientific statement. Endocr. Rev. 2009, 30, 293–342. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Wu, P.S.; Yang, E.C.; Nai, Y.S.; Huang, Z.Y. The impact of pyriproxyfen on the development of honey bee (Apis mellifera L.) colony in field. J. Asia Pac. Entomol. 2016, 19, 589–594. [Google Scholar] [CrossRef]
- Truman, J.W. The Evolution of Insect Metamorphosis. Curr. Biol. 2019, 29, R1252–R1268. [Google Scholar] [CrossRef]
- Wilson, T.G. The molecular site of action of juvenile hormone and juvenile hormone insecticides during metamorphosis: How these compounds kill insects. J. Insect Physiol. 2004, 50, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Abdourahime, H.; Anastassiadou, M.; Arena, M.; Auteri, D.; Barmaz, S.; Brancato, A.; Bura, L.; Carrasco Cabrera, L.; Chaideftou, E.; Chiusolo, A.; et al. Peer review of the pesticide risk assessment of the active substance pyriproxyfen. EFSA J. 2019, 17, e05732. [Google Scholar] [CrossRef] [PubMed]
- Fiaz, M.; Martínez, L.C.; Plata-Rueda, A.; Goncalves, W.G.; De Souza, D.L.L.; Cossolin, J.F.S.; Carvalho, P.E.G.R.; Martins, G.F.; Serrão, J.E. Pyriproxyfen, a juvenile hormone analog, damages midgut cells and interferes with behaviors of Aedes aegypti larvae. PeerJ 2019, 2019, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Devillers, J.; Devillers, H. Lethal and sublethal effects of pyriproxyfen on apis and non-apis bees. Toxics 2020, 8, 104. [Google Scholar] [CrossRef]
- Fisher, A.; Colman, C.; Hoffmann, C.; Fritz, B.; Rangel, J. The Effects of the Insect Growth Regulators Methoxyfenozide and Pyriproxyfen and the Acaricide Bifenazate on Honey Bee (Hymenoptera: Apidae) Forager Survival. J. Econ. Entomol. 2018, 111, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, P.; Wang, P.; Liu, D.; Zhou, Z. Toxicity risk assessment of pyriproxyfen and metabolites in the rat liver: A vitro study. J. Hazard. Mater. 2019, 121835. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.J.; Goh, K.S. Environmental Fate and Properties of Pyriproxyfen. J. Pestic. Sci. 2008, 33, 339–350. [Google Scholar] [CrossRef] [Green Version]
- Yoshino, H.; Kaneko, H.; Nakatsuka, I.; Yamada, H. Metabolism of pyriproxyfen. 3. In vitro metabolism in rats and mice. J. Agric. Food Chem. 1996, 44, 1578–1581. [Google Scholar] [CrossRef]
- Clayton, G.M.; Peak-Chew, S.Y.; Evans, R.M.; Schwabe, J.W.R. The structure of the ultraspiracle ligand-binding domain reveals a nuclear receptor locked in an inactive conformation. Proc. Natl. Acad. Sci. USA 2001, 98, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Oro, A.E.; McKeown, M.; Evans, R.M. Relationship between the product of the Drosophila ultraspiracle locus and the vertebrate retinoid X receptor. Nature 1990, 347, 298–301. [Google Scholar] [CrossRef]
- Sasorith, S.; Billas, I.M.L.; Iwema, T.; Moras, D.; Wurtz, J.M. Structure-based analysis of the ultraspiracle protein and docking studies of putative ligands. J. Insect Sci. 2002, 2. [Google Scholar] [CrossRef]
- Jones, G.; Sharp, P.A. Ultraspiracle: An invertebrate nuclear receptor for juvenile hormones. Proc. Natl. Acad. Sci. USA 1997, 94, 13499–13503. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, Y.; Henrich, V.C. Arthropod nuclear receptors and their role in molting. FEBS J. 2009, 276, 6128–6157. [Google Scholar] [CrossRef] [Green Version]
- Henrich, V.C.; Burns, E.; Yelverton, D.P.; Christensen, E.; Weinberger, C. Juvenile hormone potentiates ecdysone receptor-dependent transcription in a mammalian cell culture system. Insect Biochem. Mol. Biol. 2003, 33, 1239–1247. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Lu, Y.; Tian, S.; Ma, F.; Wei, Y.; Xu, S.; Li, Y. Structural insights into the heterodimeric complex of the nuclear receptors FXR and RXR. J. Biol. Chem. 2018, 293, 12535–12541. [Google Scholar] [CrossRef] [Green Version]
- Kemper, J.K. Regulation of FXR transcriptional activity in health and disease: Emerging roles of FXR cofactors and post-translational modifications. Biochim. Biophys. Acta Mol. Basis Dis. 2011, 1812, 842–850. [Google Scholar] [CrossRef] [Green Version]
- Jones, G.; Jones, D.; Teal, P.; Sapa, A.; Wozniak, M. The retinoid-X receptor ortholog, ultraspiracle, binds with nanomolar affinity to an endogenous morphogenetic ligand. FEBS J. 2006, 273, 4983–4996. [Google Scholar] [CrossRef] [PubMed]
- Velarde, R.A.; Robinson, G.E.; Fahrbach, S.E. Nuclear receptors of the honey bee: Annotation and expression in the adult brain. Insect Mol. Biol. 2006, 15, 583–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, L. Insect Endocrinology; Academic Press: Cambridge, MA, USA, 2012; ISBN 9780123847492. [Google Scholar]
- Kojetin, D.J.; Matta-Camacho, E.; Hughes, T.S.; Srinivasan, S.; Nwachukwu, J.C.; Cavett, V.; Nowak, J.; Chalmers, M.J.; Marciano, D.P.; Kamenecka, T.M.; et al. Structural mechanism for signal transduction in RXR nuclear receptor heterodimers. Nat. Commun. 2015, 6, 8013. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Zou, Q.; Xu, J.; Zhang, J.; Liu, J. Ligand binding and heterodimerization with retinoid X receptor (RXR) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding. J. Biol. Chem. 2018, 293, 18180–18191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bissantz, C.; Folkers, G.; Rognan, D. Protein-based virtual screening of chemical databases. 1. Evaluation of different docking/scoring combinations. J. Med. Chem. 2000, 43, 4759–4767. [Google Scholar] [CrossRef]
- Charifson, P.S.; Corkery, J.J.; Murcko, M.A.; Walters, W.P. Consensus scoring: A method for obtaining improved hit rates from docking databases of three-dimensional structures into proteins. J. Med. Chem. 1999, 42, 5100–5109. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.J.; Blanchard, S.G.; Bledsoe, R.K.; Chandra, G.; Consler, T.G.; Kliewer, S.A.; Stimmel, J.B.; Willson, T.M.; Zavacki, A.M.; Moore, D.D.; et al. Bile acids: Natural ligands for an orphan nuclear receptor. Science 1999, 284, 1365–1368. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, J.; Hollister, K.; Sowers, L.C.; Forman, B.M. Endogenous Bile Acids Are Ligandsfor the Nuclear Receptor FXR/BAR. Mol. Cell 1999, 3, 543–553. [Google Scholar] [CrossRef]
- Wang, Y.D.; Chen, W.D.; Moore, D.D.; Huang, W. FXR: A metabolic regulator and cell protector. Cell Res. 2008, 18, 1087–1095. [Google Scholar] [CrossRef] [Green Version]
- Chitranshi, N.; Dheer, Y.; Kumar, S.; Graham, S.L.; Gupta, V. Molecular docking, dynamics, and pharmacology studies on bexarotene as an agonist of ligand-activated transcription factors, retinoid X receptors. J. Cell. Biochem. 2019, 120, 11745–11760. [Google Scholar] [CrossRef]
- Dawson, M.I.; Xia, Z. The retinoid X receptors and their ligands. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2012, 1821, 21–56. [Google Scholar] [CrossRef] [Green Version]
- Gampe, R.T.; Montana, V.G.; Lambert, M.H.; Miller, A.B.; Bledsoe, R.K.; Milburn, M.V.; Kliewer, S.A.; Willson, T.M.; Xu, H.E. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol. Cell 2000, 5, 545–555. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J.; Pennsylvania, T.; Park, U. Basic Local Alignment Search Tool 2Department of Computer Science. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Bienert, S.; Waterhouse, A.; De Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [Green Version]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. Europe PMC Funders Group Phyre2 web portal for protein modelling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2014, 12, 7–8. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Sippl, M.J. Recognition of Errors in the Three-Dimensional Structures. Proteins Struct. Funct. Genet. 1993, 17, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, 407–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]








| RXRα | ||||
|---|---|---|---|---|
| Ligand | Gold Score | Chem Score | Hint Score | Affinity |
| 9-cis-retinoic acid | 72.96 | 39.72 | 1374.1 | −9.9 |
| Pyriproxyfen | 59.58 | 35.02 | 1376.3 | −9.8 |
| 4′-OH-pyriproxyfen | 64.52 | 35.82 | 1279.4 | −9.8 |
| FXR | ||||
| Chenodeoxycholic acid | 74.56 | 32.1 | 1920.2 | −11.2 |
| USP | ||||
| Juvenile hormone III | 49.02 | 26.73 | 841.04 | −6.9 |
| Pyriproxyfen | 60.29 | 32.14 | 1178.7 | −9.0 |
| 4′-OH-pyriproxyfen | 56.15 | 29.75 | 1136.0 | −9 |
| EcR | ||||
| 20-hydroxyecdysone | 77.58 | 25.98 | −2580.88 | −9.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spaggiari, G.; Iovine, N.; Cozzini, P. In Silico Prediction of the Mechanism of Action of Pyriproxyfen and 4′-OH-Pyriproxyfen against A. mellifera and H. sapiens Receptors. Int. J. Mol. Sci. 2021, 22, 7751. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147751
Spaggiari G, Iovine N, Cozzini P. In Silico Prediction of the Mechanism of Action of Pyriproxyfen and 4′-OH-Pyriproxyfen against A. mellifera and H. sapiens Receptors. International Journal of Molecular Sciences. 2021; 22(14):7751. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147751
Chicago/Turabian StyleSpaggiari, Giulia, Nadia Iovine, and Pietro Cozzini. 2021. "In Silico Prediction of the Mechanism of Action of Pyriproxyfen and 4′-OH-Pyriproxyfen against A. mellifera and H. sapiens Receptors" International Journal of Molecular Sciences 22, no. 14: 7751. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147751