The Emerging Role of Epigenetics in Therapeutic Targeting of Cardiomyopathies
Abstract
:1. Introduction
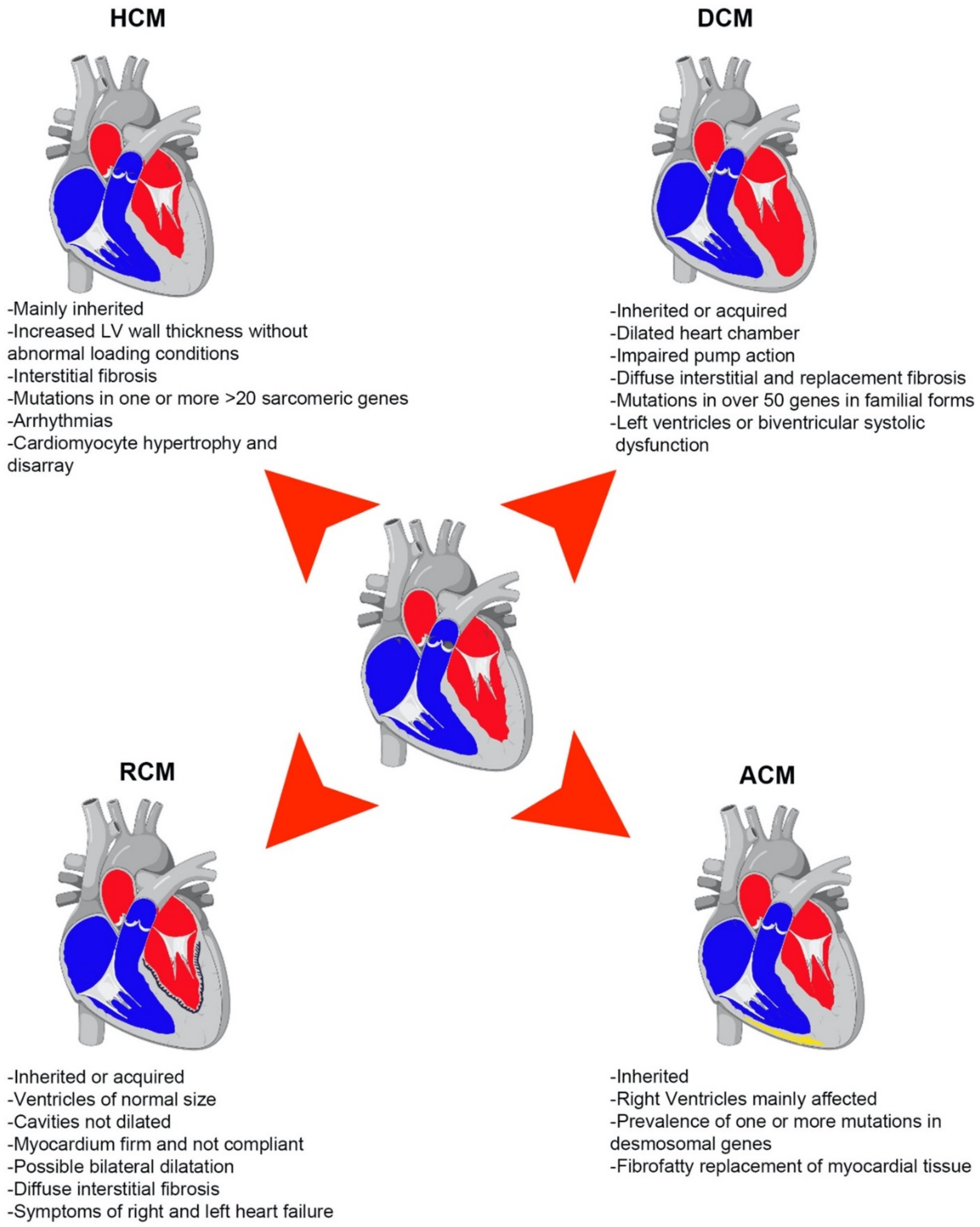
1.1. Types of Primary CMPs
1.1.1. Dilated Cardiomyopathy (DCM)
1.1.2. Hypertrophic Cardiomyopathy (HCM)
1.1.3. Restrictive Cardiomyopathy (RCM)
1.1.4. Arrhythmogenic Cardiomyopathy (ACM)
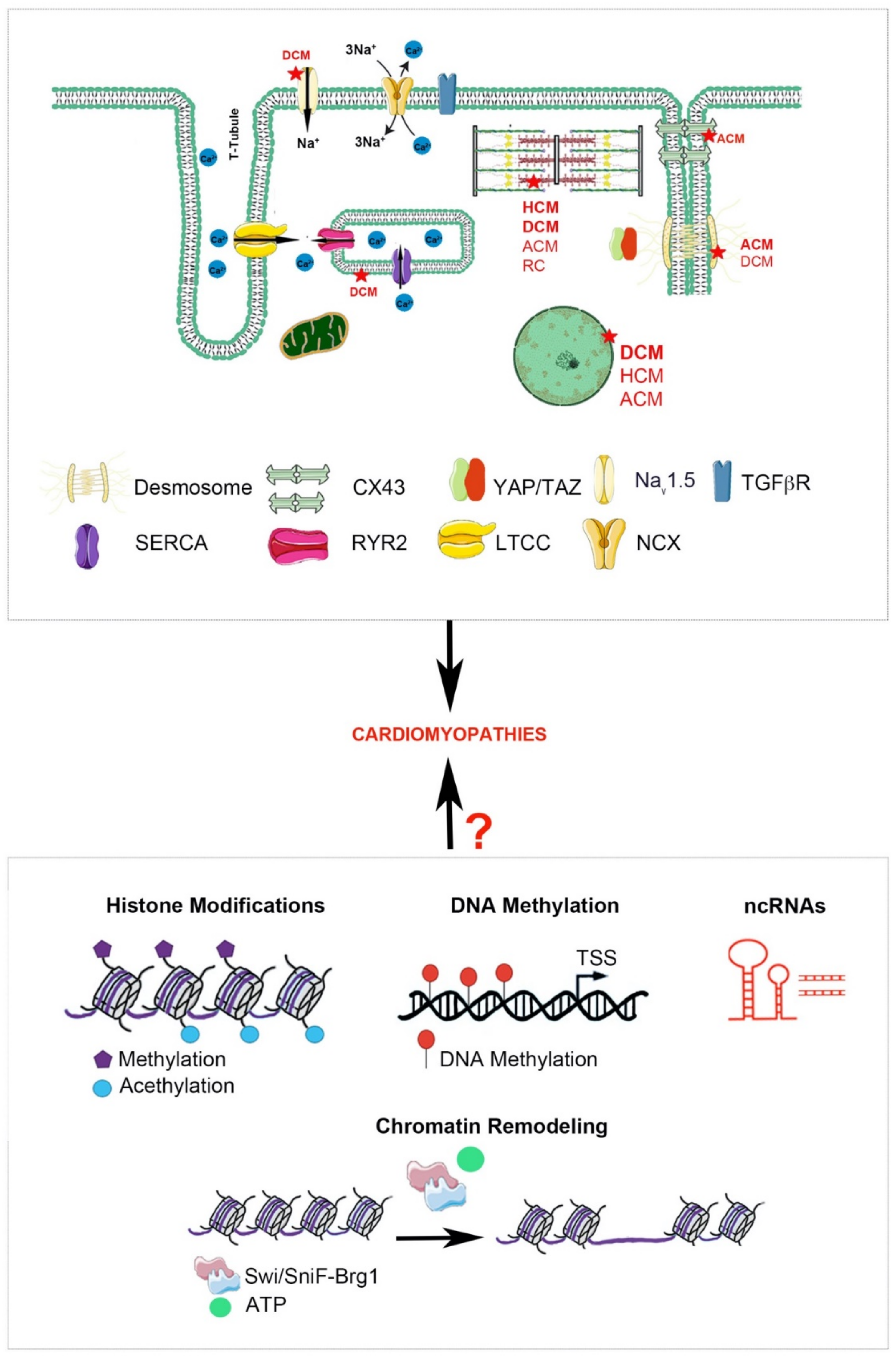
1.2. Epigenetic Mechanisms
1.2.1. DNA Methylation
1.2.2. Histone Modifications
1.2.3. Chromatin Remodeling
1.2.4. Noncoding RNAs
2. Molecular and Epigenetic Mechanisms in CMPs
2.1. Dilated Cardiomyopathy
2.1.1. Molecular Mechanisms in DCM: An Overview
2.1.2. DNA Methylation in DCM
2.1.3. Histone Modifications in DCM
2.1.4. Chromatin Remodeling in DCM
2.1.5. Noncoding RNAs in DCM
2.2. Hypertrophic Cardiomyopathy
2.2.1. Molecular Mechanisms in HCM: An Overview
2.2.2. DNA Methylation in HCM
2.2.3. Histone Modifications in HCM
2.2.4. Chromatin Remodeling in HCM
2.2.5. Noncoding RNAs in HCM
2.3. Restrictive Cardiomyopathy
2.3.1. Molecular Mechanisms in RCM: An Overview
2.3.2. DNA Methylation in RCM
2.3.3. Histone Modifications in RCM
2.3.4. Chromatin Remodeling in RCM
2.3.5. Noncoding RNAs in RCM
2.4. Arrhythmogenic Cardiomyopathy
2.4.1. Molecular Mechanisms in ACM: An Overview
2.4.2. DNA Methylation in ACM
2.4.3. Histone Modifications in ACM
2.4.4. Chromatin Remodeling in ACM
2.4.5. Noncoding RNAs in ACM
3. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Li, C.J.; Chen, C.S.; Yiang, G.T.; Tsai, A.P.; Liao, W.T.; Wu, M.Y. Advanced Evolution of Pathogenesis Concepts in Cardiomyopathies. J. Clin. Med. 2019, 8, 520. [Google Scholar] [CrossRef] [Green Version]
- Dadson, K.; Hauck, L.; Billia, F. Molecular mechanisms in cardiomyopathy. Clin. Sci. 2017, 131, 1375–1392. [Google Scholar] [CrossRef]
- Brieler, J.; Breeden, M.A.; Tucker, J. Cardiomyopathy: An Overview. Am. Fam. Phys. 2017, 96, 640–646. [Google Scholar]
- Elliott, P.; Andersson, B.; Arbustini, E.; Bilinska, Z.; Cecchi, F.; Charron, P.; Dubourg, O.; Kuhl, U.; Maisch, B.; McKenna, W.J.; et al. Classification of the cardiomyopathies: A position statement from the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur. Heart J. 2008, 29, 270–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luk, A.; Ahn, E.; Soor, G.S.; Butany, J. Dilated cardiomyopathy: A review. J. Clin. Pathol. 2009, 62, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, R.G.; Semsarian, C.; Macdonald, P. Dilated cardiomyopathy. Lancet 2017, 390, 400–414. [Google Scholar] [CrossRef]
- Elliott, P. Cardiomyopathy. Diagnosis and management of dilated cardiomyopathy. Heart 2000, 84, 106–112. [Google Scholar] [CrossRef] [Green Version]
- Schultheiss, H.P.; Fairweather, D.; Caforio, A.L.P.; Escher, F.; Hershberger, R.E.; Lipshultz, S.E.; Liu, P.P.; Matsumori, A.; Mazzanti, A.; McMurray, J.; et al. Dilated cardiomyopathy. Nat. Rev. Dis. Primers 2019, 5, 32. [Google Scholar] [CrossRef]
- Grogan, M.; Redfield, M.M.; Bailey, K.R.; Reeder, G.S.; Gersh, B.J.; Edwards, W.D.; Rodeheffer, R.J. Long-term outcome of patients with biopsy-proved myocarditis: Comparison with idiopathic dilated cardiomyopathy. J. Am. Coll. Cardiol. 1995, 26, 80–84. [Google Scholar] [CrossRef] [Green Version]
- Maron, B.J.; Rowin, E.J.; Maron, M.S. Global Burden of Hypertrophic Cardiomyopathy. JACC Heart Fail. 2018, 6, 376–378. [Google Scholar] [CrossRef]
- Elliott, P.M.; Anastasakis, A.; Borger, M.A.; Borggrefe, M.; Cecchi, F.; Charron, P.; Hagege, A.A.; Lafont, A.; Limongelli, G. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: The Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J. 2014, 35, 2733–2779. [Google Scholar] [CrossRef]
- Van Driest, S.L.; Ommen, S.R.; Tajik, A.J.; Gersh, B.J.; Ackerman, M.J. Yield of genetic testing in hypertrophic cardiomyopathy. Mayo Clin. Proc. 2005, 80, 739–744. [Google Scholar] [CrossRef]
- Antunes, M.O.; Scudeler, T.L. Hypertrophic cardiomyopathy. Int. J. Cardiol. Heart Vasc. 2020, 27, 100503. [Google Scholar] [CrossRef]
- Pereira, N.L.; Grogan, M.; Dec, G.W. Spectrum of Restrictive and Infiltrative Cardiomyopathies: Part 2 of a 2-Part Series. J. Am. Coll. Cardiol. 2018, 71, 1149–1166. [Google Scholar] [CrossRef]
- Felker, G.M.; Thompson, R.E.; Hare, J.M.; Hruban, R.H.; Clemetson, D.E.; Howard, D.L.; Baughman, K.L.; Kasper, E.K. Underlying causes and long-term survival in patients with initially unexplained cardiomyopathy. N. Engl. J. Med. 2000, 342, 1077–1084. [Google Scholar] [CrossRef] [PubMed]
- Katritsis, D.; Wilmshurst, P.T.; Wendon, J.A.; Davies, M.J.; Webb-Peploe, M.M. Primary restrictive cardiomyopathy: Clinical and pathologic characteristics. J. Am. Coll. Cardiol. 1991, 18, 1230–1235. [Google Scholar] [CrossRef] [Green Version]
- Muchtar, E.; Blauwet, L.A.; Gertz, M.A. Restrictive Cardiomyopathy: Genetics, Pathogenesis, Clinical Manifestations, Diagnosis, and Therapy. Circ. Res. 2017, 121, 819–837. [Google Scholar] [CrossRef]
- Nihoyannopoulos, P.; Dawson, D. Restrictive cardiomyopathies. Eur. J. Echocardiogr. 2009, 10, iii23–iii33. [Google Scholar] [CrossRef] [Green Version]
- Towbin, J.A.; McKenna, W.J.; Abrams, D.J.; Ackerman, M.J.; Calkins, H.; Darrieux, F.C.C.; Daubert, J.P.; de Chillou, C.; DePasquale, E.C.; Desai, M.Y.; et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy: Executive summary. Heart Rhythm 2019, 16, e373–e407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevens, T.L.; Wallace, M.J.; Refaey, M.E.; Roberts, J.D.; Koenig, S.N.; Mohler, P.J. Arrhythmogenic Cardiomyopathy: Molecular Insights for Improved Therapeutic Design. J. Cardiovasc. Dev. Dis. 2020, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- McRae, A.T., 3rd; Chung, M.K.; Asher, C.R. Arrhythmogenic right ventricular cardiomyopathy: A cause of sudden death in young people. Cleve Clin. J. Med. 2001, 68, 459–467. [Google Scholar] [CrossRef] [Green Version]
- McGregor, S.M.; Husain, A.N. A Brief Review and Update of the Clinicopathologic Diagnosis of Arrhythmogenic Cardiomyopathy. Arch. Pathol. Lab. Med. 2015, 139, 1181–1186. [Google Scholar] [CrossRef]
- Van der Voorn, S.M.; Te Riele, A.; Basso, C.; Calkins, H.; Remme, C.A.; van Veen, T.A.B. Arrhythmogenic cardiomyopathy: Pathogenesis, pro-arrhythmic remodelling, and novel approaches for risk stratification and therapy. Cardiovasc. Res. 2020, 116, 1571–1584. [Google Scholar] [CrossRef]
- Corrado, D.; Basso, C.; Judge, D.P. Arrhythmogenic Cardiomyopathy. Circ. Res. 2017, 121, 784–802. [Google Scholar] [CrossRef] [Green Version]
- Papait, R.; Corrado, N.; Rusconi, F.; Serio, S.; M, V.G.L. It’s Time for An Epigenomics Roadmap of Heart Failure. Curr. Genom. 2015, 16, 237–244. [Google Scholar] [CrossRef] [Green Version]
- Papait, R.; Cattaneo, P.; Kunderfranco, P.; Greco, C.; Carullo, P.; Guffanti, A.; Vigano, V.; Stirparo, G.G.; Latronico, M.V.; Hasenfuss, G.; et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 2013, 110, 20164–20169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papait, R.; Kunderfranco, P.; Stirparo, G.G.; Latronico, M.V.; Condorelli, G. Long noncoding RNA: A new player of heart failure? J. Cardiovasc. Transl. Res. 2013, 6, 876–883. [Google Scholar] [CrossRef] [Green Version]
- Costantino, S.; Paneni, F.; Cosentino, F. Targeting chromatin remodeling to prevent cardiovascular disease in diabetes. Curr. Pharm. Biotechnol. 2015, 16, 531–543. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Greco, C.M.; Kunderfranco, P.; Rubino, M.; Larcher, V.; Carullo, P.; Anselmo, A.; Kurz, K.; Carell, T.; Angius, A.; Latronico, M.V.; et al. DNA hydroxymethylation controls cardiomyocyte gene expression in development and hypertrophy. Nat. Commun. 2016, 7, 12418. [Google Scholar] [CrossRef] [PubMed]
- Lind, L.; Ingelsson, E.; Sundstrom, J.; Siegbahn, A.; Lampa, E. Methylation-based estimated biological age and cardiovascular disease. Eur. J. Clin. Investig. 2018, 48. [Google Scholar] [CrossRef]
- Pagiatakis, C.; Musolino, E.; Gornati, R.; Bernardini, G.; Papait, R. Epigenetics of aging and disease: A brief overview. Aging Clin. Exp. Res. 2019, 33, 737–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papait, R.; Serio, S.; Pagiatakis, C.; Rusconi, F.; Carullo, P.; Mazzola, M.; Salvarani, N.; Miragoli, M.; Condorelli, G. Histone Methyltransferase G9a Is Required for Cardiomyocyte Homeostasis and Hypertrophy. Circulation 2017, 136, 1233–1246. [Google Scholar] [CrossRef]
- Sun, X.; Hota, S.K.; Zhou, Y.Q.; Novak, S.; Miguel-Perez, D.; Christodoulou, D.; Seidman, C.E.; Seidman, J.G.; Gregorio, C.C.; Henkelman, R.M.; et al. Cardiac-enriched BAF chromatin-remodeling complex subunit Baf60c regulates gene expression programs essential for heart development and function. Biol. Open 2018, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, I.L.; Gao, X.L.; Sham, M.H.; Wang, Z. SWI/SNF Protein Component BAF250a Regulates Cardiac Progenitor Cell Differentiation by Modulating Chromatin Accessibility during Second Heart Field Development. J. Biol. Chem. 2012, 287, 24255–24262. [Google Scholar] [CrossRef] [Green Version]
- Vieira, J.M.; Howard, S.; Villa Del Campo, C.; Bollini, S.; Dube, K.N.; Masters, M.; Barnette, D.N.; Rohling, M.; Sun, X.; Hankins, L.E.; et al. BRG1-SWI/SNF-dependent regulation of the Wt1 transcriptional landscape mediates epicardial activity during heart development and disease. Nat. Commun. 2017, 8, 16034. [Google Scholar] [CrossRef] [Green Version]
- Bultman, S.J.; Holley, D.W.; de Ridder, G.G.; Pizzo, S.V.; Sidorova, T.N.; Murray, K.T.; Jensen, B.C.; Wang, Z.J.; Bevilacqua, A.; Chen, X.; et al. BRG1 and BRM SWI/SNF ATPases redundantly maintain cardiomyocyte homeostasis by regulating cardiomyocyte mitophagy and mitochondrial dynamics in vivo. Cardiovasc. Pathol. 2016, 25, 258–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Mauro, V.; Crasto, S.; Colombo, F.S.; Di Pasquale, E.; Catalucci, D. Wnt signalling mediates miR-133a nuclear re-localization for the transcriptional control of Dnmt3b in cardiac cells. Sci. Rep. 2019, 9, 9320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Xu, W.W.; Hu, S.J. Heart failure: Advanced development in genetics and epigenetics. Biomed. Res. Int. 2015, 2015, 352734. [Google Scholar] [CrossRef]
- Lakdawala, N.K.; Winterfield, J.R.; Funke, B.H. Dilated cardiomyopathy. Circ. Arrhythm. Electrophysiol. 2013, 6, 228–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jefferies, J.L.; Towbin, J.A. Dilated cardiomyopathy. Lancet 2010, 375, 752–762. [Google Scholar] [CrossRef]
- Reichart, D.; Magnussen, C.; Zeller, T.; Blankenberg, S. Dilated cardiomyopathy: From epidemiologic to genetic phenotypes: A translational review of current literature. J. Intern. Med. 2019, 286, 362–372. [Google Scholar] [CrossRef] [Green Version]
- Movassagh, M.; Choy, M.K.; Knowles, D.A.; Cordeddu, L.; Haider, S.; Down, T.; Siggens, L.; Vujic, A.; Simeoni, I.; Penkett, C.; et al. Distinct epigenomic features in end-stage failing human hearts. Circulation 2011, 124, 2411–2422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meder, B.; Haas, J.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Frese, K.; Lai, A.; Nietsch, R.; Scheiner, C.; Mester, S.; Bordalo, D.M.; et al. Epigenome-Wide Association Study Identifies Cardiac Gene Patterning and a Novel Class of Biomarkers for Heart Failure. Circulation 2017, 136, 1528–1544. [Google Scholar] [CrossRef]
- Yang, G.; Zhu, Y.; Dong, X.; Duan, Z.; Niu, X.; Wei, J. TLR2-ICAM1-Gadd45alpha axis mediates the epigenetic effect of selenium on DNA methylation and gene expression in Keshan disease. Biol. Trace Elem. Res. 2014, 159, 69–80. [Google Scholar] [CrossRef]
- Tabish, A.M.; Arif, M.; Song, T.; Elbeck, Z.; Becker, R.C.; Knoll, R.; Sadayappan, S. Association of intronic DNA methylation and hydroxymethylation alterations in the epigenetic etiology of dilated cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 2019, 317, H168–H180. [Google Scholar] [CrossRef] [PubMed]
- Ito, E.; Miyagawa, S.; Fukushima, S.; Yoshikawa, Y.; Saito, S.; Saito, T.; Harada, A.; Takeda, M.; Kashiyama, N.; Nakamura, Y.; et al. Histone Modification Is Correlated With Reverse Left Ventricular Remodeling in Nonischemic Dilated Cardiomyopathy. Ann. Thorac. Surg. 2017, 104, 1531–1539. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Wang, X.; Zhang, Y.; Ru, X.; Zhou, L.; Tian, Y. H3K9 histone methyltransferase G9a ameliorates dilated cardiomyopathy via the downregulation of cell adhesion molecules. Mol. Med. Rep. 2015, 11, 3872–3879. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Xiao, B.; Neppl, R.L.; Kallin, E.M.; Li, J.; Chen, T.; Wang, D.Z.; Xiao, X.; Zhang, Y. DOT1L regulates dystrophin expression and is critical for cardiac function. Genes. Dev. 2011, 25, 263–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montgomery, R.L.; Davis, C.A.; Potthoff, M.J.; Haberland, M.; Fielitz, J.; Qi, X.; Hill, J.A.; Richardson, J.A.; Olson, E.N. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes. Dev. 2007, 21, 1790–1802. [Google Scholar] [CrossRef] [Green Version]
- Miksiunas, R.; Rucinskas, K.; Janusauskas, V.; Labeit, S.; Bironaite, D. Histone Deacetylase Inhibitor Suberoylanilide Hydroxamic Acid Improves Energetic Status and Cardiomyogenic Differentiation of Human Dilated Myocardium-Derived Primary Mesenchymal Cells. Int. J. Mol. Sci. 2020, 21, 4845. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Huebsch, N.; Koo, S.; Mandegar, M.A.; Siemons, B.; Boggess, S.; Conklin, B.R.; Grigoropoulos, C.P.; Healy, K.E. Contractile deficits in engineered cardiac microtissues as a result of MYBPC3 deficiency and mechanical overload. Nat. Biomed. Eng. 2018, 2, 955–967. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.S.; Cserjesi, P.; Markham, B.E.; Molkentin, J.D. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J. Biol. Chem. 2002, 277, 24390–24398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuno, A.; Hori, Y.S.; Hosoda, R.; Tanno, M.; Miura, T.; Shimamoto, K.; Horio, Y. Resveratrol improves cardiomyopathy in dystrophin-deficient mice through SIRT1 protein-mediated modulation of p300 protein. J. Biol. Chem. 2013, 288, 5963–5972. [Google Scholar] [CrossRef] [Green Version]
- Han, P.; Li, W.; Yang, J.; Shang, C.; Lin, C.H.; Cheng, W.; Hang, C.T.; Cheng, H.L.; Chen, C.H.; Wong, J.; et al. Epigenetic response to environmental stress: Assembly of BRG1-G9a/GLP-DNMT3 repressive chromatin complex on Myh6 promoter in pathologically stressed hearts. Biochim. Biophys. Acta. 2016, 1863, 1772–1781. [Google Scholar] [CrossRef]
- Zhao, W.; Wu, X.; Wang, Z.; Pan, B.; Liu, L.; Liu, L.; Huang, X.; Tian, J. Epigenetic regulation of phosphodiesterase 4d in restrictive cardiomyopathy mice with cTnI mutations. Sci. China Life Sci. 2020, 63, 563–570. [Google Scholar] [CrossRef]
- Zhao, W.; Qian, L.; Luo, J.; Pan, B.; Liu, L.J.; Tian, J. Cardiac troponin I R193H mutant interacts with HDAC1 to repress phosphodiesterase 4D expression in cardiomyocytes. Genes Dis. 2021, 8, 569–579. [Google Scholar] [CrossRef]
- Jimenez, J.; Rentschler, S.L. Transcriptional and Epigenetic Regulation of Cardiac Electrophysiology. Pediatr. Cardiol. 2019, 40, 1325–1330. [Google Scholar] [CrossRef] [Green Version]
- Aiba, T.; Tomaselli, G.F. Electrical remodeling in the failing heart. Curr. Opin. Cardiol. 2010, 25, 29–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khandekar, A.; Springer, S.; Wang, W.; Hicks, S.; Weinheimer, C.; Diaz-Trelles, R.; Nerbonne, J.M.; Rentschler, S. Notch-Mediated Epigenetic Regulation of Voltage-Gated Potassium Currents. Circ. Res. 2016, 119, 1324–1338. [Google Scholar] [CrossRef] [Green Version]
- Rouhi, L.; Fan, S.; Cheedipudi, S.M.; Braza-Boils, A.; Molina, M.S.; Yao, Y.; Robertson, M.J.; Coarfa, C.; Gimeno, J.R.; Molina, P.; et al. The EP300/TP53 pathway, a suppressor of the Hippo and canonical WNT pathways, is activated in human hearts with arrhythmogenic cardiomyopathy in the absence of overt heart failure. Cardiovasc. Res. 2021. [Google Scholar] [CrossRef]
- Pagiatakis, C.; Gordon, J.W.; Ehyai, S.; McDermott, J.C. A novel RhoA/ROCK-CPI-17-MEF2C signaling pathway regulates vascular smooth muscle cell gene expression. J. Biol. Chem. 2012, 287, 8361–8370. [Google Scholar] [CrossRef] [Green Version]
- Calderon-Dominguez, M.; Belmonte, T.; Quezada-Feijoo, M.; Ramos-Sanchez, M.; Fernandez-Armenta, J.; Perez-Navarro, A.; Cesar, S.; Pena-Pena, L.; Vea, A.; Llorente-Cortes, V.; et al. Emerging role of microRNAs in dilated cardiomyopathy: Evidence regarding etiology. Transl. Res. 2020, 215, 86–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh, M.; Minami, Y.; Takahashi, Y.; Tabuchi, T.; Nakamura, M. Expression of microRNA-208 is associated with adverse clinical outcomes in human dilated cardiomyopathy. J. Card. Fail. 2010, 16, 404–410. [Google Scholar] [CrossRef]
- Ikeda, S.; Kong, S.W.; Lu, J.; Bisping, E.; Zhang, H.; Allen, P.D.; Golub, T.R.; Pieske, B.; Pu, W.T. Altered microRNA expression in human heart disease. Physiol. Genomics. 2007, 31, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zeng, C.; Wang, Y. Epigenetics in dilated cardiomyopathy. Curr. Opin. Cardiol. 2019, 34, 260–269. [Google Scholar] [CrossRef]
- Pagiatakis, C.; Hall, I.F.; Condorelli, G. Long non-coding RNA H19: A new avenue for RNA therapeutics in cardiac hypertrophy? Eur. Heart J. 2020, 41, 3475–3476. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, M.; Xu, W.; Chen, J.; Zhou, X. The long non-coding RNA H19 promotes cardiomyocyte apoptosis in dilated cardiomyopathy. Oncotarget 2017, 8, 28588–28594. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Jiang, H. Long non-coding RNA HAND2-AS1 downregulation predicts poor survival of patients with end-stage dilated cardiomyopathy. J. Int. Med. Res. 2019, 47, 3690–3698. [Google Scholar] [CrossRef]
- Huang, G.; Liu, J.; Yang, C.; Xiang, Y.; Wang, Y.; Wang, J.; Cao, M.; Yang, W. RNA sequencing discloses the genomewide profile of long noncoding RNAs in dilated cardiomyopathy. Mol. Med. Rep. 2019, 19, 2569–2580. [Google Scholar] [CrossRef] [PubMed]
- Mosqueira, D.; Mannhardt, I.; Bhagwan, J.R.; Lis-Slimak, K.; Katili, P.; Scott, E.; Hassan, M.; Prondzynski, M.; Harmer, S.C.; Tinker, A.; et al. CRISPR/Cas9 editing in human pluripotent stem cell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardiomyopathy. Eur. Heart J. 2018, 39, 3879–3892. [Google Scholar] [CrossRef]
- Monda, E.; Palmiero, G.; Rubino, M.; Verrillo, F.; Amodio, F.; Di Fraia, F.; Pacileo, R.; Fimiani, F.; Esposito, A.; Cirillo, A.; et al. Molecular Basis of Inflammation in the Pathogenesis of Cardiomyopathies. Int. J. Mol. Sci. 2020, 21, 6462. [Google Scholar] [CrossRef]
- Kuusisto, J.; Karja, V.; Sipola, P.; Kholova, I.; Peuhkurinen, K.; Jaaskelainen, P.; Naukkarinen, A.; Yla-Herttuala, S.; Punnonen, K.; Laakso, M. Low-grade inflammation and the phenotypic expression of myocardial fibrosis in hypertrophic cardiomyopathy. Heart 2012, 98, 1007–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.T.; Ma, Y.W.; Zhang, X.; Dong, W.; Gao, S.; Wang, J.Z.; Zhang, L.F.; Lu, D. Myocardial tissue-specific Dnmt1 knockout in rats protects against pathological injury induced by Adriamycin. Lab. Investig. 2020, 100, 974–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eom, G.H.; Nam, Y.S.; Oh, J.G.; Choe, N.; Min, H.K.; Yoo, E.K.; Kang, G.; Nguyen, V.H.; Min, J.J.; Kim, J.K.; et al. Regulation of acetylation of histone deacetylase 2 by p300/CBP-associated factor/histone deacetylase 5 in the development of cardiac hypertrophy. Circ. Res. 2014, 114, 1133–1143. [Google Scholar] [CrossRef] [Green Version]
- Hang, C.T.; Yang, J.; Han, P.; Cheng, H.L.; Shang, C.; Ashley, E.; Zhou, B.; Chang, C.P. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 2010, 466, 62–67. [Google Scholar] [CrossRef]
- Shi, H.; Chen, L.; Wang, H.; Zhu, S.; Dong, C.; Webster, K.A.; Wei, J. Synergistic induction of miR-126 by hypoxia and HDAC inhibitors in cardiac myocytes. Biochem. Biophys. Res. Commun. 2013, 430, 827–832. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Collyer, J.; Wang, M.; Sun, F.; Xu, F. Genetic Dissection of Hypertrophic Cardiomyopathy with Myocardial RNA-Seq. Int. J. Mol. Sci. 2020, 21, 3040. [Google Scholar] [CrossRef]
- Scolari, F.L.; Faganello, L.S.; Garbin, H.I.; Piva, E.M.B.; Biolo, A. A systematic review of microRNAs in patients with hypertrophic cardiomyopathy. Int. J. Cardiol. 2021, 327, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Gurha, P.; Chen, X.; Lombardi, R.; Willerson, J.T.; Marian, A.J. Knockdown of Plakophilin 2 Downregulates miR-184 Through CpG Hypermethylation and Suppression of the E2F1 Pathway and Leads to Enhanced Adipogenesis In Vitro. Circ. Res. 2016, 119, 731–750. [Google Scholar] [CrossRef] [Green Version]
- Calore, M.; Lorenzon, A.; Vitiello, L.; Poloni, G.; Khan, M.A.F.; Beffagna, G.; Dazzo, E.; Sacchetto, C.; Polishchuk, R.; Sabatelli, P.; et al. A novel murine model for arrhythmogenic cardiomyopathy points to a pathogenic role of Wnt signalling and miRNA dysregulation. Cardiovasc. Res. 2019, 115, 739–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Zhou, Y.; Wang, C.X. LncRNA-MIAT regulates fibrosis in hypertrophic cardiomyopathy (HCM) by mediating the expression of miR-29a-3p. J. Cell Biochem. 2018, 120, 7265–7275. [Google Scholar] [CrossRef]
- Gomez, J.; Lorca, R.; Reguero, J.R.; Martin, M.; Moris, C.; Alonso, B.; Iglesias, S.; Diaz-Molina, B.; Avanzas, P.; Coto, E. Genetic variation at the long noncoding RNA H19 gene is associated with the risk of hypertrophic cardiomyopathy. Epigenomics 2018, 10, 865–873. [Google Scholar] [CrossRef]
- Glezeva, N.; Moran, B.; Collier, P.; Moravec, C.S.; Phelan, D.; Donnellan, E.; Russell-Hallinan, A.; O’Connor, D.P.; Gallagher, W.M.; Gallagher, J.; et al. Targeted DNA Methylation Profiling of Human Cardiac Tissue Reveals Novel Epigenetic Traits and Gene Deregulation Across Different Heart Failure Patient Subtypes. Circ. Heart Fail. 2019, 12, e005765. [Google Scholar] [CrossRef] [Green Version]
- Simon, D.N.; Zastrow, M.S.; Wilson, K.L. Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus 2010, 1, 264–272. [Google Scholar] [CrossRef]
- Schiano, C.; Costa, V.; Aprile, M.; Grimaldi, V.; Maiello, C.; Esposito, R.; Soricelli, A.; Colantuoni, V.; Donatelli, F.; Ciccodicola, A.; et al. Heart failure: Pilot transcriptomic analysis of cardiac tissue by RNA-sequencing. Cardiol. J. 2017, 24, 539–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rainer, J.; Meraviglia, V.; Blankenburg, H.; Piubelli, C.; Pramstaller, P.P.; Paolin, A.; Cogliati, E.; Pompilio, G.; Sommariva, E.; Domingues, F.S.; et al. The arrhythmogenic cardiomyopathy-specific coding and non-coding transcriptome in human cardiac stromal cells. BMC Genom. 2018, 19, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.N.; Gurha, P.; Lombardi, R.; Ruggiero, A.; Willerson, J.T.; Marian, A.J. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ. Res. 2014, 114, 454–468. [Google Scholar] [CrossRef] [PubMed]
- Pagiatakis, C.; Sun, D.; Tobin, S.W.; Miyake, T.; McDermott, J.C. TGFbeta-TAZ/SRF signalling regulates vascular smooth muscle cell differentiation. FEBS J. 2017, 284, 1644–1656. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, M.A.; Warley, A.; Roberts, R.G.; Ehler, E.; Ellis, J.A. Identification of an emerin-beta-catenin complex in the heart important for intercalated disc architecture and beta-catenin localisation. Cell Mol. Life Sci. 2010, 67, 781–796. [Google Scholar] [CrossRef]
- Pilichou, K.; Thiene, G.; Bauce, B.; Rigato, I.; Lazzarini, E.; Migliore, F.; Perazzolo Marra, M.; Rizzo, S.; Zorzi, A.; Daliento, L.; et al. Arrhythmogenic cardiomyopathy. Orphanet. J. Rare Dis. 2016, 11, 33. [Google Scholar] [CrossRef] [Green Version]
- Passaro, F.; De Martino, I.; Zambelli, F.; Di Benedetto, G.; Barbato, M.; D’Erchia, A.M.; Manzari, C.; Pesole, G.; Mutarelli, M.; Cacchiarelli, D.; et al. YAP contributes to DNA methylation remodeling upon mouse embryonic stem cell differentiation. J. Biol. Chem. 2020, 296, 100138. [Google Scholar] [CrossRef]
- Singh, V.; Singh, A.P.; Sharma, I.; Singh, L.C.; Sharma, J.; Borthakar, B.B.; Rai, A.K.; Kataki, A.C.; Kapur, S.; Saxena, S. Epigenetic deregulations of Wnt/beta-catenin and transforming growth factor beta-Smad pathways in esophageal cancer: Outcome of DNA methylation. J. Cancer Res. Ther. 2019, 15, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Di Mauro, V.; Ceriotti, P.; Lodola, F.; Salvarani, N.; Modica, J.; Bang, M.L.; Mazzanti, A.; Napolitano, C.; Priori, S.G.; Catalucci, D. Peptide-Based Targeting of the L-Type Calcium Channel Corrects the Loss-of-Function Phenotype of Two Novel Mutations of the CACNA1 Gene Associated With Brugada Syndrome. Front. Physiol. 2020, 11, 616819. [Google Scholar] [CrossRef]
- Monte, E.; Rosa-Garrido, M.; Karbassi, E.; Chen, H.; Lopez, R.; Rau, C.D.; Wang, J.; Nelson, S.F.; Wu, Y.; Stefani, E.; et al. Reciprocal Regulation of the Cardiac Epigenome by Chromatin Structural Proteins Hmgb and Ctcf: Implications for Transcriptional Regulation. J. Biol. Chem. 2016, 291, 15428–15446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanconato, F.; Forcato, M.; Battilana, G.; Azzolin, L.; Quaranta, E.; Bodega, B.; Rosato, A.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015, 17, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Stadiotti, I.; Pompilio, G.; Maione, A.S.; Pilato, C.A.; D’Alessandra, Y.; Sommariva, E. Arrhythmogenic cardiomyopathy: What blood can reveal? Heart Rhythm 2019, 16, 470–477. [Google Scholar] [CrossRef] [Green Version]
- Pagiatakis, C.; Condorelli, G. The RNA Methylome Blackboard. Circulation 2019, 139, 546–548. [Google Scholar] [CrossRef] [PubMed]
- Stege, N.M.; de Boer, R.A.; van den Berg, M.P.; Sillje, H.H.W. The Time Has Come to Explore Plasma Biomarkers in Genetic Cardiomyopathies. Int. J. Mol. Sci. 2021, 22, 2955. [Google Scholar] [CrossRef]
- Roncarati, R.; Viviani Anselmi, C.; Losi, M.A.; Papa, L.; Cavarretta, E.; Da Costa Martins, P.; Contaldi, C.; Saccani Jotti, G.; Franzone, A.; Galastri, L.; et al. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2014, 63, 920–927. [Google Scholar] [CrossRef] [Green Version]
- Roura, S.; Gamez-Valero, A.; Lupon, J.; Galvez-Monton, C.; Borras, F.E.; Bayes-Genis, A. Proteomic signature of circulating extracellular vesicles in dilated cardiomyopathy. Lab. Invest. 2018, 98, 1291–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anselmo, A.; Frank, D.; Papa, L.; Viviani Anselmi, C.; Di Pasquale, E.; Mazzola, M.; Panico, C.; Clemente, F.; Soldani, C.; Pagiatakis, C.; et al. Myocardial hypoxic stress mediates functional cardiac extracellular vesicle release. Eur. Heart J. 2021, 42, 2780–2792. [Google Scholar] [CrossRef] [PubMed]
- Jansen, F.; Nickenig, G.; Werner, N. Extracellular Vesicles in Cardiovascular Disease: Potential Applications in Diagnosis, Prognosis, and Epidemiology. Circ. Res. 2017, 120, 1649–1657. [Google Scholar] [CrossRef] [PubMed]
| Gene | Protein | Disease | References |
|---|---|---|---|
| ACTC1 | Actin alpha cardiac muscle 1 | DCM | [8] |
| TTN | Titin | DCM | [8] |
| MYH6 | Myosin heavy chain 6 | DCM | [8] |
| TNNT2 | Troponin T | DCM | [8] |
| LMNA | Prelamin A/C | DCM | [8] |
| PLNB | Cardiac phospolamban | DCM | [8] |
| MYH7 | Myosin heavy chain 7 | HCM | [11,12] |
| MYBPC | myosin-binding protein C | HCM | [11,12] |
| TNNI3, TNNT2 | Cardiac troponin I and T | HCM | [11,12] |
| TPM1 | tropomyosin α-1 chain | HCM | [11,12] |
| MYL3 | Myosin light chain 3 | HCM | [11,12] |
| TNNT2 | Cardiac troponin T | RCM | [18] |
| TNNI3 | Cardiac troponin I | RCM | [18] |
| ACTC1 | Actin alpha cardiac muscle 1 | RCM | [18] |
| MYH7 | Myosin heavy chain 7 | RCM | [18] |
| JUP | Plakoglobin | ACM | [22] |
| DSP | Desmoplakin | ACM | [22] |
| PKP2 | Plakophilin-2 | ACM | [22] |
| DSG2 | Desmoglein-2 | ACM | [22] |
| DSC2 | Desmocollin-2 | ACM | [22] |
| PLNB | Cardiac phospolamban | ACM | [23] |
| RYR2 | Cardiac ryanodine receptor | ACM | [23] |
| TGF-b3 | Transforming growth factor b 3 | ACM | [23] |
| TMEM43 | Transmembrane protein-43 | ACM | [23] |
| SCN5A | Cardiac sodium channel | ACM | [23] |
| Disease | Modification | Enzymes | References |
|---|---|---|---|
| DCM | H3K4me3, H3K9me2, H3K9me3, H3K79me3 | EP300, G9A, HDAC1, HDAC2, DOT1L | [47,48,49,50,52] |
| HCM | H3K9me2 | G9A, HDACs, EP300 | [54,55] |
| RCM | H3K4Ac, H3K9Ac, H3K4me3 | SMYD1, HDAC1 | [56,57] |
| ACM | H3K4me3, H3K4me2, H3K9Ac, H3K9me2, H3K9me3 | EP300 | [58,59,60,61] |
| miRNAs | Disease | Up or Down Regulated | Cardiac/Circulating | References |
|---|---|---|---|---|
| miR-17-5p, miR-28, miR-106 | DCM | Down | Cardiac | [63] |
| miR-208 | DCM | Up | Cardiac | [64] |
| miR-1, miR-19a/b | DCM | Down | Cardiac | [65] |
| miR-21, miR-26, miR-29, miR-30 and miR-133a | DCM | Up | Circulating | [66] |
| miR-3193, miR-3671 | HCM | Up | Cardiac | [74] |
| miR-487a, miR-654, miR-30d, miR-154 | HCM | Down | Cardiac | [78] |
| 1-3p, mir-19b, mir-21, mir-29a, mir-155, mir-221 | HCM | Up | Circulating | [79] |
| miR-184, miR-499-5p | ACM | Down | Cardiac | [80,81] |
| miR-217-5p, miR-708-5p | ACM | Up | Cardiac | [81] |
| miR-320a | ACM | Down | Circulating | [81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pagiatakis, C.; Di Mauro, V. The Emerging Role of Epigenetics in Therapeutic Targeting of Cardiomyopathies. Int. J. Mol. Sci. 2021, 22, 8721. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22168721
Pagiatakis C, Di Mauro V. The Emerging Role of Epigenetics in Therapeutic Targeting of Cardiomyopathies. International Journal of Molecular Sciences. 2021; 22(16):8721. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22168721
Chicago/Turabian StylePagiatakis, Christina, and Vittoria Di Mauro. 2021. "The Emerging Role of Epigenetics in Therapeutic Targeting of Cardiomyopathies" International Journal of Molecular Sciences 22, no. 16: 8721. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22168721







