Contribution of the Commensal Microflora to the Immunological Homeostasis and the Importance of Immune-Related Drug Development for Clinical Applications
Abstract
:1. Introduction
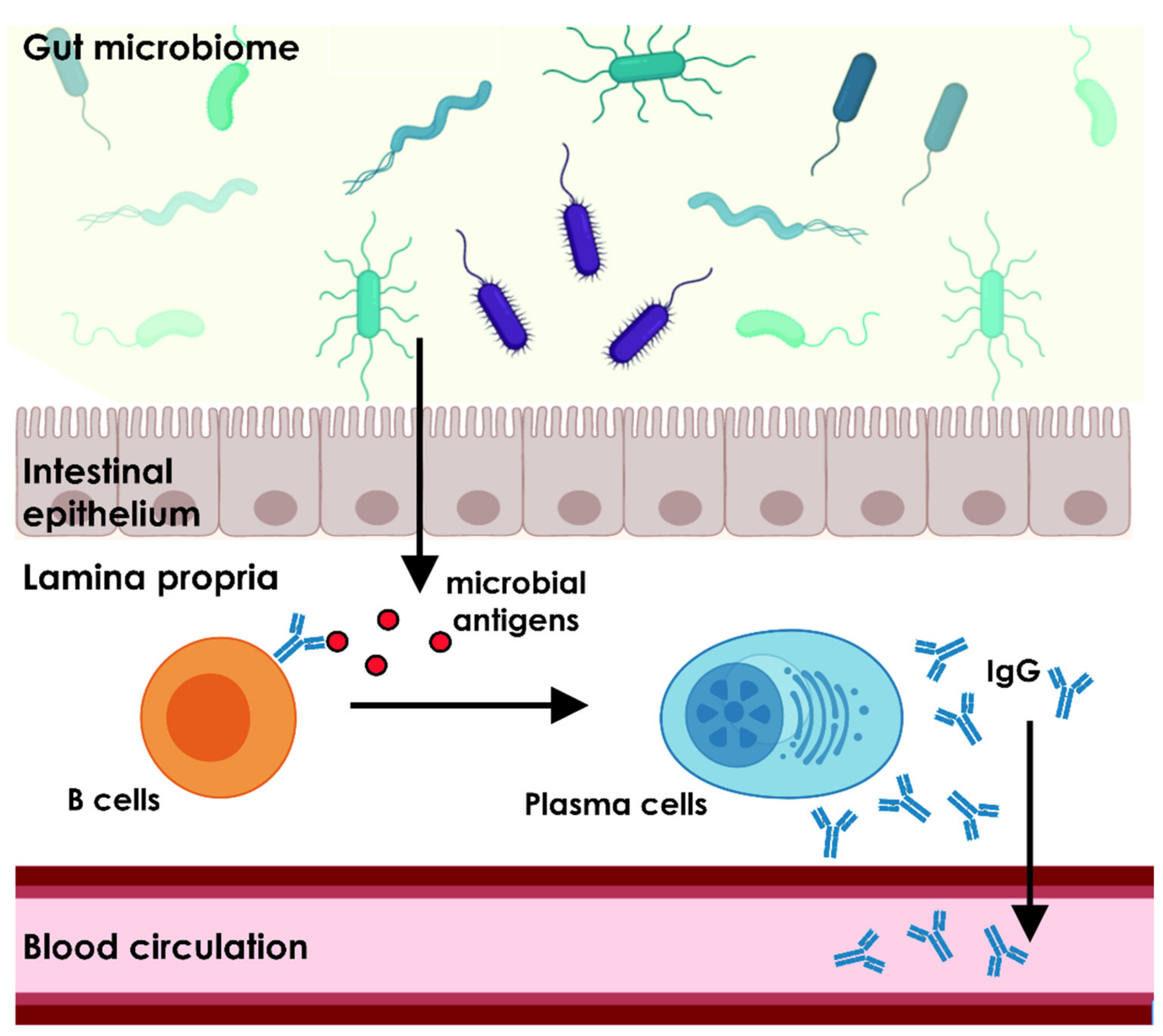
2. Commensal Microbiome Shapes the Host Immune System
3. The Gut Microbiome Is Essential for the Homeostasis of the Natural Antibody Repertoire
4. Perturbations of the Gut Microbiome and Autoimmunity in Different Disorders
5. Immune-Related Drug Development in Clinical Applications
5.1. Systemic Lupus Erythematosus (SLE)
5.2. Neurodegenerative Diseases
5.2.1. Alzheimer’s and Parkinson’s Disease
5.2.2. Glaucoma
5.3. Cancer and Melanoma
6. Conclusions
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Pasolli, E.; Asnicar, F.; Manara, S.; Zolfo, M.; Karcher, N.; Armanini, F.; Beghini, F.; Manghi, P.; Tett, A.; Ghensi, P.; et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 2019, 176, 649. [Google Scholar] [CrossRef] [Green Version]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5. [Google Scholar] [CrossRef]
- Wampach, L.; Heintz-Buschart, A.; Hogan, A.; Muller, E.E.L.; Narayanasamy, S.; Laczny, C.C.; Hugerth, L.W.; Bindl, L.; Bottu, J.; Andersson, A.F.; et al. Colonization and Succession within the Human Gut Microbiome by Archaea, Bacteria, and Microeukaryotes during the First Year of Life. Front. Microbiol. 2017, 8, 738. [Google Scholar] [CrossRef]
- Zuo, T.; Sun, Y.; Wan, Y.; Yeoh, Y.K.; Zhang, F.; Cheung, C.P.; Chen, N.; Luo, J.; Wang, W.; Sung, J.J.Y.; et al. Human-Gut-DNA Virome Variations across Geography, Ethnicity, and Urbanization. Cell Host Microbe 2020, 28, 741–751.e744. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015, 160, 447–460. [Google Scholar] [CrossRef] [Green Version]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [Green Version]
- Proctor, L.M.; Creasy, H.H.; Fettweis, J.M.; Lloyd-Price, J.; Mahurkar, A.; Zhou, W.Y.; Buck, G.A.; Snyder, M.P.; Strauss, J.F.; Weinstock, G.M.; et al. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef] [Green Version]
- Voth, E.; Khanna, S. The Integrative Human microbiome project: A mile stone in the understanding of the gut microbiome. Expert Rev. Gastroent. 2020, 14, 639–642. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- Franzosa, E.A.; Huang, K.; Meadow, J.F.; Gevers, D.; Lemon, K.P.; Bohannan, B.J.M.; Huttenhower, C. Identifying personal microbiomes using metagenomic codes. Proc. Natl. Acad. Sci. USA 2015, 112, E2930–E2938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laudadio, I.; Fulci, V.; Palone, F.; Stronati, L.; Cucchiara, S.; Carissimi, C. Quantitative Assessment of Shotgun Metagenomics and 16S rDNA Amplicon Sequencing in the Study of Human Gut Microbiome. Omics-A J. Integr. Biol. 2018, 22, 248–254. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [Green Version]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.E.; Kwak, S.E.; Lee, J.H.; Zhang, D.D.; Bae, J.H.; Song, W. Exercise, the Gut Microbiome, and Frailty. Ann. Geriatr. Med. Res. 2019, 23, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Cronin, O.; Barton, W.; Skuse, P.; Penney, N.C.; Garcia-Perez, I.; Murphy, E.F.; Woods, T.; Nugent, H.; Fanning, A.; Melgar, S.; et al. A Prospective Metagenomic and Metabolomic Analysis of the Impact of Exercise and/or Whey Protein Supplementation on the Gut Microbiome of Sedentary Adults. Msystems 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Konstantinidis, T.; Tsigalou, C.; Karvelas, A.; Stavropoulou, E.; Voidarou, C.; Bezirtzoglou, E. Effects of Antibiotics upon the Gut Microbiome: A Review of the Literature. Biomedicines 2020, 8, 502. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Elson, C.O. Adaptive immune education by gut microbiota antigens. Immunology 2018, 154, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Nunez, G. Role of the gut microbiota in the development and function of lymphoid cells. J. Immunol. 2013, 190, 1389–1395. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Limenitakis, J.P.; Greiff, V.; Yilmaz, B.; Scharen, O.; Urbaniak, C.; Zund, M.; Lawson, M.A.E.; Young, I.D.; Rupp, S.; et al. Mucosal or systemic microbiota exposures shape the B cell repertoire. Nature 2020, 584, 274–278. [Google Scholar] [CrossRef]
- Main, B.S.; Minter, M.R. Microbial Immuno-Communication in Neurodegenerative Diseases. Front. Neurosci. 2017, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M.M. Microbiota-Brain-Gut Axis and Neurodegenerative Diseases. Curr. Neurol. Neurosci. Rep. 2017, 17, 94. [Google Scholar] [CrossRef] [PubMed]
- DeMarshall, C.; Sarkar, A.; Nagele, E.P.; Goldwaser, E.; Godsey, G.; Acharya, N.K.; Nagele, R.G. Utility of autoantibodies as biomarkers for diagnosis and staging of neurodegenerative diseases. Int Rev. Neurobiol. 2015, 122, 1–51. [Google Scholar] [CrossRef]
- Kim, S.; Rigatto, K.; Gazzana, M.B.; Knorst, M.M.; Richards, E.M.; Pepine, C.J.; Raizada, M.K. Altered Gut Microbiome Profile in Patients With Pulmonary Arterial Hypertension. Hypertension 2020, 75, 1063–1071. [Google Scholar] [CrossRef]
- Caligiuri, G.; Stahl, D.; Kaveri, S.; Irinopoulous, T.; Savoie, F.; Mandet, C.; Vandaele, M.; Kazatchkine, M.D.; Michel, J.B.; Nicoletti, A. Autoreactive antibody repertoire is perturbed in atherosclerotic patients. Lab. Investig. 2003, 83, 939–947. [Google Scholar] [CrossRef] [Green Version]
- Bell, K.; Beutgen, V.M.; Nickels, S.; Lorenz, K.; Scheller, Y.; Elbaz, H.; Peto, T.; Ponto, K.A.; Schulz, A.; Wild, P.S.; et al. Results from the Population-Based Gutenberg Health Study Revealing Four Altered Autoantibodies in Retinal Vein Occlusion Patients. J. Ophthalmol. 2020, 2020, 8386160. [Google Scholar] [CrossRef]
- Fereidan-Esfahani, M.; Nayfeh, T.; Warrington, A.; Howe, C.L.; Rodriguez, M. IgM Natural Autoantibodies in Physiology and the Treatment of Disease. Methods Mol. Biol. 2019, 1904, 53–81. [Google Scholar] [CrossRef] [PubMed]
- Nagele, E.P.; Han, M.; Acharya, N.K.; DeMarshall, C.; Kosciuk, M.C.; Nagele, R.G. Natural IgG Autoantibodies Are Abundant and Ubiquitous in Human Sera, and Their Number Is Influenced By Age, Gender, and Disease. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Reyneveld, G.I.; Savelkoul, H.F.J.; Parmentier, H.K. Current Understanding of Natural Antibodies and Exploring the Possibilities of Modulation Using Veterinary Models. A Review. Front. Immunol. 2020, 11, 2139. [Google Scholar] [CrossRef] [PubMed]
- Palma, J.; Tokarz-Deptula, B.; Deptula, J.; Deptula, W. Natural antibodies—facts known and unknown. Cent. Eur. J. Immunol. 2018, 43, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Baig, M.H.; Ahmad, K.; Saeed, M.; Alharbi, A.M.; Barreto, G.E.; Ashraf, G.M.; Choi, I. Peptide based therapeutics and their use for the treatment of neurodegenerative and other diseases. Biomed. Pharmacother. 2018, 103, 574–581. [Google Scholar] [CrossRef]
- Lee, A.C.-L.; Harris, J.L.; Khanna, K.K.; Hong, J.-H. A comprehensive review on current advances in peptide drug development and design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef] [Green Version]
- Gomez de Aguero, M.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef]
- Rogier, E.W.; Frantz, A.L.; Bruno, M.E.; Wedlund, L.; Cohen, D.A.; Stromberg, A.J.; Kaetzel, C.S. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc. Natl. Acad. Sci. USA 2014, 111, 3074–3079. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Zhao, W.; Wu, M.; Song, X.; Caro, F.; Sun, X.; Gazzaniga, F.; Stefanetti, G.; Oh, S.; Mekalanos, J.J.; et al. Microbiota-targeted maternal antibodies protect neonates from enteric infection. Nature 2020, 577, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Pandiyan, P.; Bhaskaran, N.; Zou, M.; Schneider, E.; Jayaraman, S.; Huehn, J. Microbiome Dependent Regulation of Tregs and Th17 Cells in Mucosa. Front. Immunol. 2019, 10, 426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Chen, B.D.; Zhao, L.D.; Li, H. The Gut Microbiota: Emerging Evidence in Autoimmune Diseases. Trends Mol. Med. 2020, 26, 862–873. [Google Scholar] [CrossRef]
- Geuking, M.B.; Cahenzli, J.; Lawson, M.A.; Ng, D.C.; Slack, E.; Hapfelmeier, S.; McCoy, K.D.; Macpherson, A.J. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 2011, 34, 794–806. [Google Scholar] [CrossRef] [Green Version]
- Tan, T.G.; Sefik, E.; Geva-Zatorsky, N.; Kua, L.; Naskar, D.; Teng, F.; Pasman, L.; Ortiz-Lopez, A.; Jupp, R.; Wu, H.J.; et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. USA 2016, 113, E8141–E8150. [Google Scholar] [CrossRef] [Green Version]
- Ostman, S.; Rask, C.; Wold, A.E.; Hultkrantz, S.; Telemo, E. Impaired regulatory T cell function in germ-free mice. Eur. J. Immunol. 2006, 36, 2336–2346. [Google Scholar] [CrossRef]
- Bird, J.A.; Spergel, J.M.; Jones, S.M.; Rachid, R.; Assa’ad, A.H.; Wang, J.; Leonard, S.A.; Laubach, S.S.; Kim, E.H.; Vickery, B.P.; et al. Efficacy and Safety of AR101 in Oral Immunotherapy for Peanut Allergy: Results of ARC001, a Randomized, Double-Blind, Placebo-Controlled Phase 2 Clinical Trial. J. Allergy Clin. Immunol. Pract. 2018, 6, 476–485.e473. [Google Scholar] [CrossRef] [PubMed]
- Nussenblatt, R.B.; Whitcup, S.M.; de Smet, M.D.; Caspi, R.R.; Kozhich, A.T.; Weiner, H.L.; Vistica, B.; Gery, I. Intraocular inflammatory disease (uveitis) and the use of oral tolerance: A status report. Ann. N. Y. Acad. Sci. 1996, 778, 325–337. [Google Scholar] [CrossRef]
- Mao, R.F.; Chen, Y.Y.; Zhang, J.; Chang, X.; Wang, Y.F. Type 1 diabetes mellitus and its oral tolerance therapy. World J. Diabetes 2020, 11, 400–415. [Google Scholar] [CrossRef]
- Kumar, S.R.P.; Wang, X.; Avuthu, N.; Bertolini, T.B.; Terhorst, C.; Guda, C.; Daniell, H.; Herzog, R.W. Role of Small Intestine and Gut Microbiome in Plant-Based Oral Tolerance for Hemophilia. Front. Immunol. 2020, 11, 844. [Google Scholar] [CrossRef]
- Ahmed, T.; Sumazaki, R.; Nagai, Y.; Shibasaki, M.; Takita, H. Immune response to food antigens: Kinetics of food-specific antibodies in the normal population. Acta Paediatr. Jpn. 1997, 39, 322–328. [Google Scholar] [CrossRef]
- Burton, O.T.; Tamayo, J.M.; Stranks, A.J.; Koleoglou, K.J.; Oettgen, H.C. Allergen-specific IgG antibody signaling through FcgammaRIIb promotes food tolerance. J. Allergy Clin. Immunol. 2018, 141, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Rachid, R.; Umetsu, D.T. Immunological mechanisms for desensitization and tolerance in food allergy. Semin Immunopathol. 2012, 34, 689–702. [Google Scholar] [CrossRef] [Green Version]
- Lupinek, C.; Hochwallner, H.; Johansson, C.; Mie, A.; Rigler, E.; Scheynius, A.; Alm, J.; Valenta, R. Maternal allergen-specific IgG might protect the child against allergic sensitization. J. Allergy Clin. Immunol. 2019, 144, 536–548. [Google Scholar] [CrossRef] [Green Version]
- Ohsaki, A.; Venturelli, N.; Buccigrosso, T.M.; Osganian, S.K.; Lee, J.; Blumberg, R.S.; Oyoshi, M.K. Maternal IgG immune complexes induce food allergen-specific tolerance in offspring. J. Exp. Med. 2018, 215, 91–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Chaudhary, N.; Yang, N.; Granato, A.; Turner, J.A.; Howard, S.L.; Devereaux, C.; Zuo, T.; Shrestha, A.; Goel, R.R.; et al. Microbial symbionts regulate the primary Ig repertoire. J. Exp. Med. 2018, 215, 1397–1415. [Google Scholar] [CrossRef] [PubMed]
- Wilmore, J.R.; Gaudette, B.T.; Gomez Atria, D.; Hashemi, T.; Jones, D.D.; Gardner, C.A.; Cole, S.D.; Misic, A.M.; Beiting, D.P.; Allman, D. Commensal Microbes Induce Serum IgA Responses that Protect against Polymicrobial Sepsis. Cell Host Microbe 2018, 23, 302–311.e303. [Google Scholar] [CrossRef] [Green Version]
- Kawamoto, S.; Maruya, M.; Kato, L.M.; Suda, W.; Atarashi, K.; Doi, Y.; Tsutsui, Y.; Qin, H.; Honda, K.; Okada, T.; et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 2014, 41, 152–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, M.Y.; Cisalpino, D.; Varadarajan, S.; Hellman, J.; Warren, H.S.; Cascalho, M.; Inohara, N.; Nunez, G. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity 2016, 44, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Gronwall, C.; Silverman, G.J. Natural IgM: Beneficial autoantibodies for the control of inflammatory and autoimmune disease. J. Clin. Immunol. 2014, 34 (Suppl. 1), S12–S21. [Google Scholar] [CrossRef] [Green Version]
- Bos, N.A.; Kimura, H.; Meeuwsen, C.G.; De Visser, H.; Hazenberg, M.P.; Wostmann, B.S.; Pleasants, J.R.; Benner, R.; Marcus, D.M. Serum immunoglobulin levels and naturally occurring antibodies against carbohydrate antigens in germ-free BALB/c mice fed chemically defined ultrafiltered diet. Eur. J. Immunol. 1989, 19, 2335–2339. [Google Scholar] [CrossRef]
- Mayasari, N.; Van Knegsel, A.T.; de Vries Reilingh, G.; Kemp, B.; Parmentier, H.K. Natural autoantibodies in Bos taurus calves during the first twelve weeks of life. Vet. Immunol. Immunopathol. 2016, 178, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Holodick, N.E.; Rodriguez-Zhurbenko, N.; Hernandez, A.M. Defining Natural Antibodies. Front. Immunol. 2017, 8, 872. [Google Scholar] [CrossRef] [Green Version]
- Griffin, D.O.; Holodick, N.E.; Rothstein, T.L. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20(+)CD27(+)CD43(+)CD70(-). J. Exp. Med. 2011, 208, 67–80. [Google Scholar] [CrossRef]
- Bello-Gil, D.; Audebert, C.; Olivera-Ardid, S.; Perez-Cruz, M.; Even, G.; Khasbiullina, N.; Gantois, N.; Shilova, N.; Merlin, S.; Costa, C.; et al. The Formation of Glycan-Specific Natural Antibodies Repertoire in GalT-KO Mice Is Determined by Gut Microbiota. Front. Immunol. 2019, 10, 342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landers, C.J.; Cohavy, O.; Misra, R.; Yang, H.; Lin, Y.C.; Braun, J.; Targan, S.R. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto- and microbial antigens. Gastroenterology 2002, 123, 689–699. [Google Scholar] [CrossRef]
- Christmann, B.S.; Abrahamsson, T.R.; Bernstein, C.N.; Duck, L.W.; Mannon, P.J.; Berg, G.; Bjorksten, B.; Jenmalm, M.C.; Elson, C.O. Human seroreactivity to gut microbiota antigens. J. Allergy Clin. Immunol. 2015, 136, 1378–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadlallah, J.; Sterlin, D.; Fieschi, C.; Parizot, C.; Dorgham, K.; El Kafsi, H.; Autaa, G.; Ghillani-Dalbin, P.; Juste, C.; Lepage, P.; et al. Synergistic convergence of microbiota-specific systemic IgG and secretory IgA. J. Allergy Clin. Immunol. 2019, 143, 1575–1585.e1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiebiger, U.; Bereswill, S.; Heimesaat, M.M. Dissecting the Interplay between Intestinal Microbiota and Host Immunity in Health and Disease: Lessons Learned from Germfree and Gnotobiotic Animal Models. Eur. J. Microbiol. Immunol. 2016, 6, 253–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mankarious, S.; Lee, M.; Fischer, S.; Pyun, K.H.; Ochs, H.D.; Oxelius, V.A.; Wedgwood, R.J. The Half-Lives of Igg Subclasses and Specific Antibodies in Patients with Primary Immunodeficiency Who Are Receiving Intravenously Administered Immunoglobulin. J. Lab. Clin. Med. 1988, 112, 634–640. [Google Scholar]
- Bonilla, F.A. Pharmacokinetics of immunoglobulin administered via intravenous or subcutaneous routes. Immunol. Allergy Clin. N. Am. 2008, 28, 803–819. [Google Scholar] [CrossRef]
- Wu, T.; Tanguay, R.M. Antibodies against heat shock proteins in environmental stresses and diseases: Friend or foe? Cell Stress Chaperones 2006, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mantej, J.; Polasik, K.; Piotrowska, E.; Tukaj, S. Autoantibodies to heat shock proteins 60, 70, and 90 in patients with rheumatoid arthritis. Cell Stress Chaperones 2019, 24, 283–287. [Google Scholar] [CrossRef]
- Beutgen, V.M.; Schmelter, C.; Pfeiffer, N.; Grus, F.H. Autoantigens in the trabecular meshwork and glaucoma-specific alterations in the natural autoantibody repertoire. Clin. Transl. Immunol. 2020, 9, e01101. [Google Scholar] [CrossRef] [Green Version]
- Rollenske, T.; Szijarto, V.; Lukasiewicz, J.; Guachalla, L.M.; Stojkovic, K.; Hartl, K.; Stulik, L.; Kocher, S.; Lasitschka, F.; Al-Saeedi, M.; et al. Cross-specificity of protective human antibodies against Klebsiella pneumoniae LPS O-antigen. Nat. Immunol. 2018, 19, 617–624. [Google Scholar] [CrossRef]
- Dimitrov, J.D.; Planchais, C.; Roumenina, L.T.; Vassilev, T.L.; Kaveri, S.V.; Lacroix-Desmazes, S. Antibody polyreactivity in health and disease: Statu variabilis. J. Immunol. 2013, 191, 993–999. [Google Scholar] [CrossRef] [Green Version]
- Haas, A.; Zimmermann, K.; Graw, F.; Slack, E.; Rusert, P.; Ledergerber, B.; Bossart, W.; Weber, R.; Thurnheer, M.C.; Battegay, M.; et al. Systemic antibody responses to gut commensal bacteria during chronic HIV-1 infection. Gut 2011, 60, 1506–1519. [Google Scholar] [CrossRef]
- Ohshima, N.; Iba, Y.; Kubota-Koketsu, R.; Asano, Y.; Okuno, Y.; Kurosawa, Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J. Virol. 2011, 85, 11048–11057. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.H.; Zhang, Y.; Hu, Y.F.; Wahl, L.M.; Cisar, J.O.; Notkins, A.L. The broad antibacterial activity of the natural antibody repertoire is due to polyreactive antibodies. Cell Host Microbe 2007, 1, 51–61. [Google Scholar] [CrossRef] [Green Version]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- Lechuga, S.; Ivanov, A.I. Disruption of the epithelial barrier during intestinal inflammation: Quest for new molecules and mechanisms. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Van Nieuwenhoven, M.A.; Brouns, F.; Brummer, R.J. Gastrointestinal profile of symptomatic athletes at rest and during physical exercise. Eur. J. Appl. Physiol. 2004, 91, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Chivero, E.T.; Ahmad, R.; Thangaraj, A.; Periyasamy, P.; Kumar, B.; Kroeger, E.; Feng, D.; Guo, M.L.; Roy, S.; Dhawan, P.; et al. Cocaine Induces Inflammatory Gut Milieu by Compromising the Mucosal Barrier Integrity and Altering the Gut Microbiota Colonization. Sci. Rep. 2019, 9, 12187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunne, J.L.; Triplett, E.W.; Gevers, D.; Xavier, R.; Insel, R.; Danska, J.; Atkinson, M.A. The intestinal microbiome in type 1 diabetes. Clin. Exp. Immunol. 2014, 177, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013, 2, e01202. [Google Scholar] [CrossRef]
- Ruff, W.E.; Dehner, C.; Kim, W.J.; Pagovich, O.; Aguiar, C.L.; Yu, A.T.; Roth, A.S.; Vieira, S.M.; Kriegel, C.; Adeniyi, O.; et al. Pathogenic Autoreactive T and B Cells Cross-React with Mimotopes Expressed by a Common Human Gut Commensal to Trigger Autoimmunity. Cell Host Microbe 2019, 26, 100–113.e108. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Ren, J.; Dai, C.; Kannapell, C.C.; Wang, H.; Gaskin, F.; Fu, S.M. Nature of T cell epitopes in lupus antigens and HLA-DR determines autoantibody initiation and diversification. Ann. Rheum Dis. 2019, 78, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Azzouz, D.; Omarbekova, A.; Heguy, A.; Schwudke, D.; Gisch, N.; Rovin, B.H.; Caricchio, R.; Buyon, J.P.; Alekseyenko, A.V.; Silverman, G.J. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 2019, 78, 947–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manfredo Vieira, S.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.; Ruff, W.; et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359, 1156–1161. [Google Scholar] [CrossRef] [Green Version]
- Greiling, T.M.; Dehner, C.; Chen, X.; Hughes, K.; Iniguez, A.J.; Boccitto, M.; Ruiz, D.Z.; Renfroe, S.C.; Vieira, S.M.; Ruff, W.E.; et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, Y.; Takeda, K. Host-microbiota interactions in rheumatoid arthritis. Exp. Mol. Med. 2019, 51, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Mendez, R.; Watane, A.; Farhangi, M.; Cavuoto, K.M.; Leith, T.; Budree, S.; Galor, A.; Banerjee, S. Gut microbial dysbiosis in individuals with Sjogren’s syndrome. Microb. Cell Fact. 2020, 19, 90. [Google Scholar] [CrossRef] [Green Version]
- Moon, J.; Choi, S.H.; Yoon, C.H.; Kim, M.K. Gut dysbiosis is prevailing in Sjogren’s syndrome and is related to dry eye severity. PLoS ONE 2020, 15, e0229029. [Google Scholar] [CrossRef]
- Miraglia, F.; Colla, E. Microbiome, Parkinson’s Disease and Molecular Mimicry. Cells 2019, 8, 222. [Google Scholar] [CrossRef] [Green Version]
- Yarandi, S.S.; Peterson, D.A.; Treisman, G.J.; Moran, T.H.; Pasricha, P.J. Modulatory Effects of Gut Microbiota on the Central Nervous System: How Gut Could Play a Role in Neuropsychiatric Health and Diseases. J. Neurogastroenterol. Motil. 2016, 22, 201–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ransohoff, R.M. How neuroinflammation contributes to neurodegeneration. Science 2016, 353, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.F.; Shen, Y.Q. Dysbiosis of gut microbiota and microbial metabolites in Parkinson’s Disease. Ageing Res. Rev. 2018, 45, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, K.; Mulak, A. Brain-Gut-Microbiota Axis in Alzheimer’s Disease. J. Neurogastroenterol. Motil. 2019, 25, 48–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noble, J.M.; Scarmeas, N.; Celenti, R.S.; Elkind, M.S.; Wright, C.B.; Schupf, N.; Papapanou, P.N. Serum IgG antibody levels to periodontal microbiota are associated with incident Alzheimer disease. PLoS ONE 2014, 9, e114959. [Google Scholar] [CrossRef] [Green Version]
- Sparks Stein, P.; Steffen, M.J.; Smith, C.; Jicha, G.; Ebersole, J.L.; Abner, E.; Dawson, D., 3rd. Serum antibodies to periodontal pathogens are a risk factor for Alzheimer’s disease. Alzheimers Dement. 2012, 8, 196–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kountouras, J.; Boziki, M.; Gavalas, E.; Zavos, C.; Deretzi, G.; Grigoriadis, N.; Tsolaki, M.; Chatzopoulos, D.; Katsinelos, P.; Tzilves, D.; et al. Increased cerebrospinal fluid Helicobacter pylori antibody in Alzheimer’s disease. Int. J. Neurosci. 2009, 119, 765–777. [Google Scholar] [CrossRef]
- Dobbs, S.M.; Dobbs, R.J.; Weller, C.; Charlett, A. Link between Helicobacter pylori infection and idiopathic parkinsonism. Med. Hypotheses 2000, 55, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Suwarnalata, G.; Tan, A.H.; Isa, H.; Gudimella, R.; Anwar, A.; Loke, M.F.; Mahadeva, S.; Lim, S.Y.; Vadivelu, J. Augmentation of Autoantibodies by Helicobacter pylori in Parkinson’s Disease Patients May Be Linked to Greater Severity. PLoS ONE 2016, 11, e0153725. [Google Scholar] [CrossRef] [Green Version]
- Sim, K.Y.; Im, K.C.; Park, S.G. The Functional Roles and Applications of Immunoglobulins in Neurodegenerative Disease. Int. J. Mol. Sci. 2020, 21, 5295. [Google Scholar] [CrossRef] [PubMed]
- Floyd, J.L.; Grant, M.B. The Gut-Eye Axis: Lessons Learned from Murine Models. Ophthalmol. Ther. 2020, 9, 499–513. [Google Scholar] [CrossRef]
- Gong, H.; Zhang, S.; Li, Q.; Zuo, C.; Gao, X.; Zheng, B.; Lin, M. Gut microbiota compositional profile and serum metabolic phenotype in patients with primary open-angle glaucoma. Exp. Eye Res. 2020, 191, 107921. [Google Scholar] [CrossRef]
- Chen, H.; Cho, K.S.; Vu, T.H.K.; Shen, C.H.; Kaur, M.; Chen, G.; Mathew, R.; McHam, M.L.; Fazelat, A.; Lashkari, K.; et al. Commensal microflora-induced T cell responses mediate progressive neurodegeneration in glaucoma. Nat. Commun. 2018, 9, 3209. [Google Scholar] [CrossRef]
- Wu, Y.; Yao, Q.; Jiang, G.X.; Wang, G.; Cheng, Q. Identification of distinct blood-based biomarkers in early stage of Parkinson’s disease. Neurol. Sci. 2020, 41, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.Z.; Zailan, F.Z.; Wong, B.Y.X.; Ng, K.P.; Kandiah, N. Identification of novel candidate autoantibodies in Alzheimer’s disease. Eur. J. Neurol. 2020, 27, 2292–2296. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Zhurbenko, N.; Quach, T.D.; Hopkins, T.J.; Rothstein, T.L.; Hemandez, A.M. Human B-1 Cells and B-1 Cell Antibodies Change With Advancing Age. Front. Immunol. 2019, 10, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Britschgi, M.; Olin, C.E.; Johns, H.T.; Takeda-Uchimura, Y.; LeMieux, M.C.; Rufibach, K.; Rajadas, J.; Zhang, H.; Tomooka, B.; Robinson, W.H.; et al. Neuroprotective natural antibodies to assemblies of amyloidogenic peptides decrease with normal aging and advancing Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2009, 106, 12145–12150. [Google Scholar] [CrossRef] [Green Version]
- Bell, K.; Wilding, C.; Funke, S.; Pfeiffer, N.; Grus, F.H. Protective effect of 14-3-3 antibodies on stressed neuroretinal cells via the mitochondrial apoptosis pathway. BMC Ophthalmol. 2015, 15, 64. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.-M.; Hwang, Y.-C.; Liu, I.-J.; Lee, C.-C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020, 27, 1–30. [Google Scholar] [CrossRef]
- Samaranayake, H.; Wirth, T.; Schenkwein, D.; Räty, J.K.; Ylä-Herttuala, S. Challenges in monoclonal antibody-based therapies. Ann. Med. 2009, 41, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Sayyed, Z.; Ameer, M.A.; Arif, A.W.; Kiran, F.; Iftikhar, A.; Iftikhar, W.; Ahmad, M.Q.; Malik, M.B.; Kumar, V. Systemic lupus erythematosus: An overview of the disease pathology and its management. Cureus 2018, 10, e3288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, F.; Tucker, L.; Rubinstein, D.; Guillaume, T.; Andre-Schwartz, J.; Barrett, K.; Schwartz, R.; Logtenberg, T. Molecular analysis of a germ line-encoded idiotypic marker of pathogenic human lupus autoantibodies. J. Immunol. 1990, 145, 2545–2553. [Google Scholar]
- Blank, M.; Shoenfeld, Y. The story of the 16/6 idiotype and systemic lupus erythematosus. IMAJ 2008, 10, 37. [Google Scholar]
- Walsman, A.; Mendlovic, S.; Ruiz, P.J.; Zinger, H.; Meshorer, A.; Mozes, E. The role of the 16/6 idiotype network in the induction and manifestations of systemic lupus erythematosus. Int. Immunol. 1993, 5, 1293–1300. [Google Scholar] [CrossRef] [PubMed]
- Dayan, M.; Segal, R.; Sthoeger, Z.; Waisman, A.; Brosh, N.a.; Elkayam, O.; Eilat, E.; Fridkin, M.; Mozes, E. Immune response of SLE patients to peptides based on the complementarity determining regions of a pathogenic anti-DNA monoclonal antibody. J. Clin. Immunol. 2000, 20, 187–194. [Google Scholar] [CrossRef]
- Eilat, E.; Zinger, H.; Nyska, A.; Mozes, E. Prevention of systemic lupus erythematosus-like disease in (NZBxNZW) F1 mice by treating with CDR1-and CDR3-based peptides of a pathogenic autoantibody. J. Clin. Immunol. 2000, 20, 268–278. [Google Scholar] [CrossRef]
- Sharabi, A.; Zinger, H.; Zborowsky, M.; Sthoeger, Z.M.; Mozes, E. A peptide based on the complementarity-determining region 1 of an autoantibody ameliorates lupus by up-regulating CD4+ CD25+ cells and TGF-β. Proc. Natl. Acad. Sci. USA 2006, 103, 8810–8815. [Google Scholar] [CrossRef] [Green Version]
- Sthoeger, Z.; Zinger, H.; Sharabi, A.; Asher, I.; Mozes, E. The tolerogenic peptide, hCDR1, down-regulates the expression of interferon-α in murine and human systemic lupus erythematosus. PLoS ONE 2013, 8, e60394. [Google Scholar] [CrossRef]
- Sthoeger, Z.M.; Sharabi, A.; Dayan, M.; Zinger, H.; Asher, I.; Sela, U.; Mozes, E. The tolerogenic peptide hCDR1 downregulates pathogenic cytokines and apoptosis and upregulates immunosuppressive molecules and regulatory T cells in peripheral blood mononuclear cells of lupus patients. Hum. Immunol. 2009, 70, 139–145. [Google Scholar] [CrossRef]
- Sela, U.; Sharabi, A.; Dayan, M.; Hershkoviz, R.; Mozes, E. The role of dendritic cells in the mechanism of action of a peptide that ameliorates lupus in murine models. Immunology 2009, 128, e395–e405. [Google Scholar] [CrossRef] [PubMed]
- Eilat, E.; Fridkin, M.; Mozes, E. A peptide based on the CDR1 of a pathogenic anti-DNA antibody is more efficient than its analogs in inhibiting autoreactive T cells. Immunobiology 2000, 202, 383–393. [Google Scholar] [CrossRef]
- Urowitz, M.B.; Isenberg, D.A.; Wallace, D.J. Safety and efficacy of hCDR1 (Edratide) in patients with active systemic lupus erythematosus: Results of phase II study. Lupus Sci. Med. 2015, 2, e000104. [Google Scholar] [CrossRef]
- Gladman, D.D.; Ibanez, D.; Urowitz, M.B. Systemic lupus erythematosus disease activity index 2000. J. Rheumatol. 2002, 29, 288–291. [Google Scholar] [PubMed]
- Uribe, A.G.; Vilá, L.M.; McGwin, G.; Sanchez, M.L.; Reveille, J.D.; Alarcón, G.S. The Systemic Lupus Activity Measure-revised, the Mexican Systemic Lupus Erythematosus Disease Activity Index (SLEDAI), and a modified SLEDAI-2K are adequate instruments to measure disease activity in systemic lupus erythematosus. J. Rheumatol. 2004, 31, 1934–1940. [Google Scholar]
- Cresswell, L.; Yee, C.-S.; Farewell, V.; Rahman, A.; Teh, L.-S.; Griffiths, B.; Bruce, I.N.; Ahmad, Y.; Prabu, A.; Akil, M. Numerical scoring for the Classic BILAG index. Rheumatology 2009, 48, 1548–1552. [Google Scholar] [CrossRef] [Green Version]
- Sthoeger, Z.; Sharabi, A.; Asher, I.; Zinger, H.; Segal, R.; Shearer, G.; Elkayam, O.; Mozes, E. The tolerogenic peptide hCDR1 immunomodulates cytokine and regulatory molecule gene expression in blood mononuclear cells of primary Sjogren’s syndrome patients. Clin. Immunol. 2018, 192, 85–91. [Google Scholar] [CrossRef]
- Van Bulck, M.; Sierra-Magro, A.; Alarcon-Gil, J.; Perez-Castillo, A.; Morales-Garcia, J.A. Novel approaches for the treatment of Alzheimer’s and Parkinson’s disease. Int. J. Mol. Sci. 2019, 20, 719. [Google Scholar] [CrossRef] [Green Version]
- Irvine, G.B.; El-Agnaf, O.M.; Shankar, G.M.; Walsh, D.M. Protein aggregation in the brain: The molecular basis for Alzheimer’s and Parkinson’s diseases. Mol. Med. 2008, 14, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Perchiacca, J.M.; Ladiwala, A.R.A.; Bhattacharya, M.; Tessier, P.M. Structure-based design of conformation-and sequence-specific antibodies against amyloid β. Proc. Natl. Acad. Sci. USA 2012, 109, 84–89. [Google Scholar] [CrossRef] [Green Version]
- Ladiwala, A.R.A.; Bhattacharya, M.; Perchiacca, J.M.; Cao, P.; Raleigh, D.P.; Abedini, A.; Schmidt, A.M.; Varkey, J.; Langen, R.; Tessier, P.M. Rational design of potent domain antibody inhibitors of amyloid fibril assembly. Proc. Natl. Acad. Sci. USA 2012, 109, 19965–19970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perchiacca, J.M.; Tessier, P.M. Engineering aggregation-resistant antibodies. Annu. Rev. Chem. Biomol. Eng. 2012, 3, 263–286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zheng, J.; Nussinov, R.; Ma, B. Molecular recognition between Aβ-specific single-domain antibody and Aβ Misfolded aggregates. Antibodies 2018, 7, 25. [Google Scholar] [CrossRef] [Green Version]
- Castelletto, V.; Ryumin, P.; Cramer, R.; Hamley, I.; Taylor, M.; Allsop, D.; Reza, M.; Ruokolainen, J.; Arnold, T.; Hermida-Merino, D. Self-assembly and anti-amyloid cytotoxicity activity of amyloid beta peptide derivatives. Sci. Rep. 2017, 7, 43637. [Google Scholar] [CrossRef] [Green Version]
- Perchiacca, J.M.; Ladiwala, A.R.A.; Bhattacharya, M.; Tessier, P.M. Aggregation-resistant domain antibodies engineered with charged mutations near the edges of the complementarity-determining regions. Protein Eng. Des. Sel. 2012, 25, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Perchiacca, J.M.; Lee, C.C.; Tessier, P.M. Optimal charged mutations in the complementarity-determining regions that prevent domain antibody aggregation are dependent on the antibody scaffold. Protein Eng. Des. Sel. 2014, 27, 29–39. [Google Scholar] [CrossRef] [Green Version]
- Julian, M.C.; Rabia, L.A.; Desai, A.A.; Arsiwala, A.; Gerson, J.E.; Paulson, H.L.; Kane, R.S.; Tessier, P.M. Nature-inspired design and evolution of anti-amyloid antibodies. J. Biol. Chem. 2019, 294, 8438–8451. [Google Scholar] [CrossRef] [Green Version]
- Sormanni, P.; Aprile, F.A.; Vendruscolo, M. Third generation antibody discovery methods: In silico rational design. Chem. Soc. Rev. 2018, 47, 9137–9157. [Google Scholar] [CrossRef]
- Sormanni, P.; Aprile, F.A.; Vendruscolo, M. Rational design of antibodies targeting specific epitopes within intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2015, 112, 9902–9907. [Google Scholar] [CrossRef] [Green Version]
- Osborne, D.M.; Fitzgerald, D.P.; O’Leary, K.E.; Anderson, B.M.; Lee, C.C.; Tessier, P.M.; McNay, E.C. Intrahippocampal administration of a domain antibody that binds aggregated amyloid-beta reverses cognitive deficits produced by diet-induced obesity. Biochim. Biophys. Acta 2016, 1860, 1291–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmelter, C.; Fomo, K.N.; Perumal, N.; Manicam, C.; Bell, K.; Pfeiffer, N.; Grus, F.H. Synthetic polyclonal-derived CDR peptides as an innovative strategy in glaucoma therapy. J. Clin. Med. 2019, 8, 1222. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Lei, J.; Wang, K.; Ma, L.; Liu, D.; Du, Y.; Wu, Y.; Zhang, S.; Wang, W.; Ma, X. Mitochondrial Omi/HtrA2 promotes caspase activation through cleavage of HAX-1 in aging heart. Rejuvenation Res. 2017, 20, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Su, X.J.; Huang, L.; Qu, Y.; Mu, D. Progress in research on the role of Omi/HtrA2 in neurological diseases. Rev. Neurosci. 2019, 30, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Appleton, B.A.; Wu, P.; Wiesmann, C.; Sidhu, S.S. Structural and functional analysis of the ligand specificity of the HtrA2/Omi PDZ domain. Protein Sci. 2007, 16, 1738–1750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, L.M.; Turk, B.E.; Cowling, V.; Borg, A.; Jarrell, E.T.; Cantley, L.C.; Downward, J. Binding specificity and regulation of the serine protease and PDZ domains of HtrA2/Omi. J. Biol. Chem. 2003, 278, 49417–49427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walle, L.V.; Lamkanfi, M.; Vandenabeele, P. The mitochondrial serine protease HtrA2/Omi: An overview. Cell Death Differ. 2008, 15, 453–460. [Google Scholar] [CrossRef] [Green Version]
- Schmelter, C.; Fomo, K.N.; Perumal, N.; Pfeiffer, N.; Grus, F.H. Regulation of the HTRA2 Protease Activity by an Inhibitory Antibody-Derived Peptide Ligand and the Influence on HTRA2-Specific Protein Interaction Networks in Retinal Tissues. Biomedicines 2021, 9, 1013. [Google Scholar] [CrossRef]
- Schmelter, C.; Perumal, N.; Funke, S.; Bell, K.; Pfeiffer, N.; Grus, F.H. Peptides of the variable IgG domain as potential biomarker candidates in primary open-angle glaucoma (POAG). Hum. Mol. Genet. 2017, 26, 4451–4464. [Google Scholar] [CrossRef]
- Bandarchi, B.; Jabbari, C.A.; Vedadi, A.; Navab, R. Molecular biology of normal melanocytes and melanoma cells. J. Clin. Pathol. 2013, 66, 644–648. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Sheikh, M.S. Melanoma: Molecular pathogenesis and therapeutic management. Mol. Cell. Pharmacol. 2014, 6, 228. [Google Scholar]
- Ju, R.J.; Stehbens, S.J.; Haass, N.K. The role of melanoma cell-stroma interaction in cell motility, invasion, and metastasis. Front. Med. 2018, 5, 307. [Google Scholar] [CrossRef] [Green Version]
- Winder, M.; Virós, A. Mechanisms of drug resistance in melanoma. In Mechanisms of Drug Resistance in Cancer Therapy; Springer: Cham, Switzerland, 2017; pp. 91–108. [Google Scholar]
- Polonelli, L.; Pontón, J.; Elguezabal, N.; Moragues, M.D.; Casoli, C.; Pilotti, E.; Ronzi, P.; Dobroff, A.S.; Rodrigues, E.G.; Juliano, M.A. Antibody complementarity-determining regions (CDRs) can display differential antimicrobial, antiviral and antitumor activities. PLoS ONE 2008, 3, e2371. [Google Scholar] [CrossRef] [Green Version]
- Gabrielli, E.; Pericolini, E.; Cenci, E.; Ortelli, F.; Magliani, W.; Ciociola, T.; Bistoni, F.; Conti, S.; Vecchiarelli, A.; Polonelli, L. Antibody complementarity-determining regions (CDRs): A bridge between adaptive and innate immunity. PLoS ONE 2009, 4, e8187. [Google Scholar] [CrossRef] [Green Version]
- Dobroff, A.S.; Rodrigues, E.G.; Juliano, M.A.; Friaça, D.M.; Nakayasu, E.S.; Almeida, I.C.; Mortara, R.A.; Jacysyn, J.F.; Amarante-Mendes, G.P.; Magliani, W. Differential antitumor effects of IgG and IgM monoclonal antibodies and their synthetic complementarity-determining regions directed to new targets of B16F10-Nex2 melanoma cells. Transl. Oncol. 2010, 3, 204. [Google Scholar] [CrossRef]
- Rodrigues, E.G.; Dobroff, A.S.; Taborda, C.P.; Travassos, L.R. Antifungal and antitumor models of bioactive protective peptides. An. Acad. Bras. Ciências 2009, 81, 503–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arruda, D.C.; Santos, L.C.; Melo, F.M.; Pereira, F.V.; Figueiredo, C.R.; Matsuo, A.L.; Mortara, R.A.; Juliano, M.A.; Rodrigues, E.G.; Dobroff, A.S. β-Actin-binding complementarity-determining region 2 of variable heavy chain from monoclonal antibody C7 induces apoptosis in several human tumor cells and is protective against metastatic melanoma. J. Biol. Chem. 2012, 287, 14912–14922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueiredo, C.R.; Matsuo, A.L.; Massaoka, M.H.; Polonelli, L.; Travassos, L.R. Anti-tumor activities of peptides corresponding to conserved complementary determining regions from different immunoglobulins. Peptides 2014, 59, 14–19. [Google Scholar] [CrossRef]
- Figueiredo, C.R.; Matsuo, A.L.; Azevedo, R.A.; Massaoka, M.H.; Girola, N.; Polonelli, L.; Travassos, L.R. A novel microtubule de-stabilizing complementarity-determining region C36L1 peptide displays antitumor activity against melanoma in vitro and in vivo. Sci. Rep. 2015, 5, 14310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueiredo, C.R.; Azevedo, R.A.; Mousdell, S.; Resende-Lara, P.T.; Ireland, L.; Santos, A.; Girola, N.; Cunha, R.L.; Schmid, M.C.; Polonelli, L. Blockade of MIF–CD74 signalling on macrophages and dendritic cells restores the antitumour immune response against metastatic melanoma. Front. Immunol. 2018, 9, 1132. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.R.; Azevedo, R.A.; Mousdell, S.; Resende-Lara, P.T.; Ireland, L.; Santos, A.; Girola, N.; Cunha, R.L.; Schmid, M.C.; Polonelli, L. Interfering with MIF-CD74 signalling on macrophages and dendritic cells with a peptide-based approach restores the immune response against metastatic melanoma. bioRxiv 2018, 248807. [Google Scholar] [CrossRef] [Green Version]
- Yaddanapudi, K.; Rendon, B.E.; Lamont, G.; Kim, E.J.; Al Rayyan, N.; Richie, J.; Albeituni, S.; Waigel, S.; Wise, A.; Mitchell, R.A. MIF Is Necessary for Late-Stage Melanoma Patient MDSC Immune Suppression and Differentiation. Cancer Immunol. Res. 2016, 4, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girola, N.; Matsuo, A.L.; Figueiredo, C.R.; Massaoka, M.H.; Farias, C.F.; Arruda, D.C.; Azevedo, R.A.; Monteiro, H.P.; Resende-Lara, P.T.; Cunha, R.L. The Ig VH complementarity-determining region 3-containing Rb9 peptide, inhibits melanoma cells migration and invasion by interactions with Hsp90 and an adhesion G-protein coupled receptor. Peptides 2016, 85, 1–15. [Google Scholar] [CrossRef]
- Jafari, A.; Rezaei-Tavirani, M.; Farhadihosseinabadi, B.; Taranejoo, S.; Zali, H. HSP90 and Co-chaperones: Impact on Tumor Progression and Prospects for Molecular-Targeted Cancer Therapy. Cancer Investig. 2020, 38, 310–328. [Google Scholar] [CrossRef]
- Machado, F.C.; Girola, N.; Maia, V.S.; Bergami-Santos, P.C.; Morais, A.S.; Azevedo, R.A.; Figueiredo, C.R.; Barbuto, J.A.; Travassos, L.R. Immunomodulatory Protective Effects of Rb9 Cyclic-Peptide in a Metastatic Melanoma Setting and the Involvement of Dendritic Cells. Front. Immunol. 2020, 10, 3122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabaça, A.N.; Arruda, D.C.; Figueiredo, C.R.; Massaoka, M.H.; Farias, C.F.; Tada, D.B.; Maia, V.C.; Silva Junior, P.I.; Girola, N.; Real, F. AC-1001 H3 CDR peptide induces apoptosis and signs of autophagy in vitro and exhibits antimetastatic activity in a syngeneic melanoma model. FEBS Open Bio 2016, 6, 885–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girola, N.; Resende-Lara, P.T.; Figueiredo, C.R.; Massaoka, M.H.; Azevedo, R.A.; Cunha, R.L.; Polonelli, L.; Travassos, L.R. Molecular, Biological and Structural Features of VL CDR-1 Rb44 Peptide, Which Targets the Microtubule Network in Melanoma Cells. Front. Oncol. 2019, 9, 25. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beutgen, V.M.; Schmelter, C.; Pfeiffer, N.; Grus, F.H. Contribution of the Commensal Microflora to the Immunological Homeostasis and the Importance of Immune-Related Drug Development for Clinical Applications. Int. J. Mol. Sci. 2021, 22, 8896. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22168896
Beutgen VM, Schmelter C, Pfeiffer N, Grus FH. Contribution of the Commensal Microflora to the Immunological Homeostasis and the Importance of Immune-Related Drug Development for Clinical Applications. International Journal of Molecular Sciences. 2021; 22(16):8896. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22168896
Chicago/Turabian StyleBeutgen, Vanessa M., Carsten Schmelter, Norbert Pfeiffer, and Franz H. Grus. 2021. "Contribution of the Commensal Microflora to the Immunological Homeostasis and the Importance of Immune-Related Drug Development for Clinical Applications" International Journal of Molecular Sciences 22, no. 16: 8896. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22168896





