A SmelAAT Acyltransferase Variant Causes a Major Difference in Eggplant (Solanum melongena L.) Peel Anthocyanin Composition
Abstract
:1. Introduction
2. Results
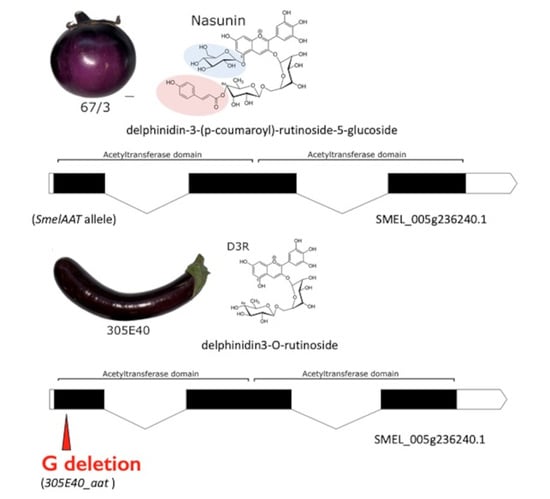
2.1. Identification of a Candidate Gene for NAS or D3R Accumulation
2.2. Identification of a Homozygous Loss-of-Function SmelAAT Allele in ‘305E40’
2.3. Complementation of SmelAAT Induces NAS Production in “Type 2” Genotypes Homozygous for the 305E40_aat305E40_aat Allele
2.4. The Expression of Functional SmelAAT Increases the Transcription of Smel5GT1
2.5. An AAT-HRM Indel Marker Correlates with the NAS/D3R Phenotype
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Peel Samplings
4.2. Molecular Phylogenetic Analysis by Neighbor Joining Method
4.3. DNA Extraction and HRM Analysis
4.4. Cloning of the SmelAAT cDNA Sequences
4.5. RNA Extraction and RT-qPCR Analysis
4.6. Eggplant Transformation
4.7. Anthocyanins’ Extractions and HPLC Analytical Conditions
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Chu, G.; Hu, Z.; Gao, Q.; Cui, B.; Tian, S.; Wang, B.; Chen, G. Genetically engineered anthocyanin pathway for high health-promoting pigment production in eggplant. Mol. Breed. 2016, 36, 54. [Google Scholar] [CrossRef]
- Cao, G.; Sofic, E.; Prior, R.L. Antioxidant Capacity of Tea and Common Vegetables. J. Agric. Food Chem. 1996, 44, 3426–3431. [Google Scholar] [CrossRef]
- Lila, M.A. Anthocyanins and Human Health: An In Vitro Investigative Approach. J. Biomed. Biotechnol. 2004, 2004, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Mateus, N.; de Freitas, V. Anthocyanins as Food Colorants. In Anthocyanins; Springer: New York, NY, USA, 2008; pp. 284–304. [Google Scholar]
- Bendokas, V.; Stanys, V.; Mažeikienė, I.; Trumbeckaite, S.; Baniene, R.; Liobikas, J. Anthocyanins: From the Field to the Antioxidants in the Body. Antioxidants 2020, 9, 819. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A Conserved Network of Transcriptional Activators and Repressors Regulates Anthocyanin Pigmentation in Eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Martin, C.; Gerats, T. Control of Pigment Biosynthesis Genes during Petal Development. Plant Cell 1993, 5, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Holton, T.A.; Cornish, E.C. Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef]
- Pelletier, M.K.; Murrell, J.R.; Shirley, B.W. Characterization of Flavonol Synthase and Leucoanthocyanidin Dioxygenase Genes in Arabidopsis (Further Evidence for Differential Regulation of “Early” and “Late” Genes). Plant Physiol. 1997, 113, 1437–1445. [Google Scholar] [CrossRef] [Green Version]
- Kähkönen, M.P.; Heinonen, M. Antioxidant Activity of Anthocyanins and Their Aglycons. J. Agric. Food Chem. 2003, 51, 628–633. [Google Scholar] [CrossRef]
- Tanaka, Y.; Ohmiya, A. Seeing is believing: Engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotechnol. 2008, 19, 190–197. [Google Scholar] [CrossRef]
- Mennella, G.; Lo Scalzo, R.; Fibiani, M.; D’Alessandro, A.; Francese, G.; Toppino, L.; Acciarri, N.; De Almeida, A.E.; Rotino, G.L. Chemical and Bioactive Quality Traits During Fruit Ripening in Eggplant (S. melongena L.) and Allied Species. J. Agric. Food Chem. 2012, 60, 11821–11831. [Google Scholar] [CrossRef]
- Prohens, J.; Rodríguez-Burruezo, A.; Raigón, M.D.; Nuez, F. Total Phenolic Concentration and Browning Susceptibility in a Collection of Different Varietal Types and Hybrids of Eggplant: Implications for Breeding for Higher Nutritional Quality and Reduced Browning. J. Am. Soc. Hortic. Sci. 2007, 132, 638–646. [Google Scholar] [CrossRef] [Green Version]
- Plazas, M.; Andújar, I.; Vilanova, S.; Hurtado, M.; Gramazio, P.; Herraiz, F.J.; Prohens, J. Breeding for Chlorogenic Acid Content in Eggplant: Interest and Prospects. Not. Bot. Horti Agrobot. Cluj-Napoca 2013, 41, 26. [Google Scholar] [CrossRef] [Green Version]
- Docimo, T.; Francese, G.; Ruggiero, A.; Batelli, G.; De Palma, M.; Bassolino, L.; Toppino, L.; Rotino, G.L.; Mennella, G.; Tucci, M. Phenylpropanoids Accumulation in Eggplant Fruit: Characterization of Biosynthetic Genes and Regulation by a MYB Transcription Factor. Front. Plant Sci. 2016, 6, 1233. [Google Scholar] [CrossRef] [Green Version]
- Moglia, A.; Florio, F.E.; Iacopino, S.; Guerrieri, A.; Milani, A.M.; Comino, C.; Barchi, L.; Marengo, A.; Cagliero, C.; Rubiolo, P.; et al. Identification of a new R3 MYB type repressor and functional characterization of the members of the MBW transcriptional complex involved in anthocyanin biosynthesis in eggplant (S. melongena L.). PLoS ONE 2020, 15, e0232986. [Google Scholar] [CrossRef]
- Li, L.; He, Y.; Ge, H.; Liu, Y.; Chen, H. Functional characterization of SmMYB86, a negative regulator of anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Sci. 2021, 302, 110696. [Google Scholar] [CrossRef]
- Tigchelaar, E.C.; Janick, J.; Erickson, H.T. The genetics of anthocyanin coloration in eggplant (solanum melongena L.). Genetics 1968, 60, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Kroon, J.; Souer, E.; De Graaff, A.; Xue, Y.; Mol, J.; Koes, R. Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: Characterization of insertion sequences in two mutant alleles. Plant J. 1994, 5, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Tanaka, Y.; Fukuchi-Mizutani, M.; Fujiwara, H.; Fukui, Y.; Ashikari, T.; Murakami, Y.; Yamaguchi, M.; Kusumi, T. Molecular and biochemical characterization of a novel hydroxycinnamoyl-CoA: Anthocyanin 3-O-glucoside-6 ″-O-acyltransferase from Perilla frutescens. Plant Cell Physiol. 2000, 41, 495–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichiyanagi, T.; Kashiwada, Y.; Shida, Y.; Ikeshiro, Y.; Kaneyuki, T.; Konishi, T. Nasunin from Eggplant Consists of Cis−Trans Isomers of Delphinidin 3-[4-(p-Coumaroyl)-l-rhamnosyl (1→6)glucopyranoside]-5-glucopyranoside. J. Agric. Food Chem. 2005, 53, 9472–9477. [Google Scholar] [CrossRef]
- Azuma, K.; Ohyama, A.; Ippoushi, K.; Ichiyanagi, T.; Takeuchi, A.; Saito, T.; Fukuoka, H. Structures and Antioxidant Activity of Anthocyanins in Many Accessions of Eggplant and Its Related Species. J. Agric. Food Chem. 2008, 56, 10154–10159. [Google Scholar] [CrossRef] [PubMed]
- Toppino, L.; Barchi, L.; Lo Scalzo, R.; Palazzolo, E.; Francese, G.; Fibiani, M.; D’Alessandro, A.; Papa, V.; Laudicina, V.A.; Sabatino, L.; et al. Mapping Quantitative Trait Loci Affecting Biochemical and Morphological Fruit Properties in Eggplant (Solanum melongena L.). Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Falcón, J.E.; Prohens, J.; Vilanova, S.; Nuez, F. Diversity in commercial varieties and landraces of black eggplants and implications for broadening the breeders’ gene pool. Ann. Appl. Biol. 2009, 154, 453–465. [Google Scholar] [CrossRef]
- Tatebe, T. On inheritance of color in Solanum melongena L. Jpn. J. Genet. 1939, 15, 261–271. [Google Scholar] [CrossRef] [Green Version]
- Barchi, L.; Lanteri, S.; Portis, E.; Valè, G.; Volante, A.; Pulcini, L.; Ciriaci, T.; Acciarri, N.; Barbierato, V.; Toppino, L.; et al. A RAD Tag Derived Marker Based Eggplant Linkage Map and the Location of QTLs Determining Anthocyanin Pigmentation. PLoS ONE 2012, 7, e43740. [Google Scholar] [CrossRef] [Green Version]
- Portis, E.; Barchi, L.; Toppino, L.; Lanteri, S.; Acciarri, N.; Felicioni, N.; Fusari, F.; Barbierato, V.; Cericola, F.; Valè, G.; et al. QTL Mapping in Eggplant Reveals Clusters of Yield-Related Loci and Orthology with the Tomato Genome. PLoS ONE 2014, 9, e89499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, H.; Liu, Y.; Jiang, M.; Zhang, J.; Han, H.; Chen, H. Analysis of genetic diversity and structure of eggplant populations (Solanum melongena L.) in China using simple sequence repeat markers. Sci. Hortic. 2013, 162, 71–75. [Google Scholar] [CrossRef]
- Cericola, F.; Portis, E.; Lanteri, S.; Toppino, L.; Barchi, L.; Acciarri, N.; Pulcini, L.; Sala, T.; Rotino, G.L. Linkage disequilibrium and genome-wide association analysis for anthocyanin pigmentation and fruit color in eggplant. BMC Genom. 2014, 15, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Toppino, L.; Barchi, L.; Mercati, F.; Acciarri, N.; Perrone, D.; Martina, M.; Gattolin, S.; Sala, T.; Fadda, S.; Mauceri, A.; et al. A New Intra-Specific and High-Resolution Genetic Map of Eggplant Based on a RIL Population, and Location of QTLs Related to Plant Anthocyanin Pigmentation and Seed Vigour. Genes 2020, 11, 745. [Google Scholar] [CrossRef]
- Sulli, M.; Barchi, L.; Toppino, L.; Diretto, G.; Sala, T.; Lanteri, S.; Rotino, G.L.; Giuliano, G. An Eggplant Recombinant Inbred Population Allows the Discovery of Metabolic QTLs Controlling Fruit Nutritional Quality. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- D’Auria, J.C. Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 2006, 9, 331–340. [Google Scholar] [CrossRef]
- Schilmiller, A.L.; Charbonneau, A.L.; Last, R.L. Identification of a BAHD acetyltransferase that produces protective acyl sugars in tomato trichomes. Proc. Natl. Acad. Sci. USA 2012, 109, 16377–16382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.; Meyer, R.S.; Yang, T.; Whitaker, B.D.; Trouth, F.; Shangguan, L.; Huang, J.; Litt, A.; Little, D.P.; Ke, H.; et al. A novel hydroxycinnamoyl transferase for synthesis of hydroxycinnamoyl spermine conjugates in plants. BMC Plant Biol. 2019, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Zhang, Y.; Peterek, S.; Matros, A.; Rallapalli, G.; Tandrón, Y.A.; Butelli, E.; Kallam, K.; Hertkorn, N.; Mock, H.-P.; et al. Ectopic expression of snapdragon transcription factors facilitates the identification of genes encoding enzymes of anthocyanin decoration in tomato. Plant J. 2015, 83, 686–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallam, K.; Appelhagen, I.; Luo, J.; Albert, N.; Zhang, H.; Deroles, S.; Hill, L.; Findlay, K.; Andersen, Ø.M.; Davies, K.; et al. Aromatic Decoration Determines the Formation of Anthocyanic Vacuolar Inclusions. Curr. Biol. 2017, 27, 945–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaul, O. Unique Aspects of Plant Nonsense-Mediated mRNA Decay. Trends Plant Sci. 2015, 20, 767–779. [Google Scholar] [CrossRef]
- Yamazaki, M.; Yamagishi, E.; Gong, Z.; Fukuchi-Mizutani, M.; Fukui, Y.; Tanaka, Y.; Kusumi, T.; Yamaguchi, M.; Saito, K. Two flavonoid glucosyltransferases from Petunia hybrida: Molecular cloning, biochemical properties and developmentally regulated expression. Plant Mol. Biol. 2002, 48, 401–411. [Google Scholar] [CrossRef]
- Barchi, L.; Pietrella, M.; Venturini, L.; Minio, A.; Toppino, L.; Acquadro, A.; Andolfo, G.; Aprea, G.; Avanzato, C.; Bassolino, L.; et al. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Giusti, M.M. Anthocyanins: Natural Colorants with Health-Promoting Properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef]
- Wallace, T.C.; Slavin, M.; Frankenfeld, C.L. Systematic Review of Anthocyanins and Markers of Cardiovascular Disease. Nutrients 2016, 8, 32. [Google Scholar] [CrossRef]
- Matzke, A.J.; Matzke, M.A. Position effects and epigenetic silencing of plant transgenes. Curr. Opin. Plant Biol. 1998, 1, 142–148. [Google Scholar] [CrossRef]
- Andersen, Ø.M.; Jordheim, M. Chemistry of Flavonoid-Based Colors in Plants. Compr. Nat. Prod. II 2010, 3, 547–614. [Google Scholar] [CrossRef]
- Rizza, F.; Mennella, G.; Collonnier, C.; Sihachakr, D.; Kashyap, V.; Rajam, M.; Presterà, M.; Rotino, G. Androgenic dihaploids from somatic hybrids between Solanum melongena and S. aethiopicum group gilo as a source of resistance to Fusarium oxysporum f. sp. melongenae. Plant Cell Rep. 2002, 20, 1022–1032. [Google Scholar] [CrossRef]
- Rinaldo, A.R.; Cavallini, E.; Jia, Y.; Moss, S.M.A.; McDavid, D.A.; Hooper, L.C.; Robinson, S.P.; Tornielli, G.B.; Zenoni, S.; Ford, C.M.; et al. A grapevine anthocyanin acyltransferase, transcriptionally regulated by VvMYBA, can produce most acylated anthocyanins present in grape skins. Plant Physiol. 2015, 169, 1897–1916. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Reed, G.H.; Gundry, C.N.; Vandersteen, J.G.; Pryor, R.J. High-Resolution Genotyping by Amplicon Melting Analysis Using LCGreen. Clin. Chem. 2003, 49, 853–860. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbierato, V.; Sala, T.; Rinaldi, P.; Bassolino, L.; Barchi, L.; Rotino, G.L.; Toppino, L. A spiking strategy facilitates housekeeping selection for RT-qPCR analysis under different biotic stresses in eggplant. Protoplasma 2017, 254, 2215–2223. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Rotino, G.L.; Gleddie, S. Transformation of eggplant (Solanum melongena L.) using a binary Agrobacterium tumefaciens vector. Plant Cell Rep. 1990, 9, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, S.; Mennella, G.; Onofaro, V.; Perri, E.; Sunseri, F.; Rotino, G.L. Production of transgenic eggplant (Solanum melongena L.) resistant to Colorado Potato Beetle (Leptinotarsa decemlineata Say). Theor. Appl. Genet. 1997, 95, 329–334. [Google Scholar] [CrossRef]
- Chambonnet, D. Culture d’antheres in vitro chez trois Solanacees maraicheres: Le piment (Capsicum annuum L.), l’aubergine (Solanum melongena L.), la tomate (Lycopersicon esculentum Mill.) et obtention de plantes haploides. Ph.D. Thesis, Academie de Montpellier, Montpellier, France, 1985. [Google Scholar]
- Sunseri, F.; Fiore, M.C.; Mastrovito, F.; Tramontano, E.; Rotino, G. In vivo selection and genetic analysis for kanamycin resistance in transgenic eggplant (Solanum melongena L.). J Genet. Breed. 1993, 47, 229. [Google Scholar]
- Braga, P.C.; Scalzo, R.L.; Sasso, M.D.; Lattuada, N.; Greco, V.; Fibiani, M. Characterization and antioxidant activity of semi-purified extracts and pure delphinidin-glycosides from eggplant peel (Solanum melongena L.). J. Funct. Foods 2016, 20, 411–421. [Google Scholar] [CrossRef]
- Matsubara, K.; Kaneyuki, T.; Miyake, T.; Mori, M. Antiangiogenic Activity of Nasunin, an Antioxidant Anthocyanin, in Eggplant Peels. J. Agric. Food Chem. 2005, 53, 6272–6275. [Google Scholar] [CrossRef]







| Genotype | D3R | ±sd | NAS | ±sd | NAS | |
|---|---|---|---|---|---|---|
| mM | mM | % | ||||
| DR2 | Ctr | 37.41 | 4.00 b | 0.00 | 0.00 f | 0 |
| DR2 | 14-1 | 8.65 | 0.75 ef | 0.29 | 0.42 f | 2.91 |
| DR2 | 18-1 * | 9.12 | 0.64 ef | 7.46 | 0.45 b | 45.03 |
| DR2 | 3-1 * | 43.96 | 1.28 a | 3.20 | 0.05 de | 6.78 |
| DR2 | 35-1 | 20.94 | 0.25 c | 5.88 | 0.14 c | 21.91 |
| DR2 | 49-1 * | 14.66 | 1.84 d | 2.18 | 0.27 e | 12.93 |
| DR2 | 5-5 | 6.74 | 0.15 fg | 3.88 | 0.32 d | 36.52 |
| DR2 | 51-1 | 3.95 | 0.17 gh | 0.04 | 0.03 f | 0.93 |
| DR2 | 6-3 | 1.38 | 0.16 h | 0.11 | 0.02 f | 7.61 |
| DR2 | 8-1 * | 12.63 | 1.22 de | 3.14 | 0.51 de | 19.85 |
| DR2 | 8-2 | 25.12 | 0.70 c | 5.22 | 0.34 c | 17.19 |
| DR2 | 32-2 * | 23.32 | 1.59 c | 9.18 | 1.02 a | 28.21 |
| 305E40 | 3-2 | 9.07 | 0.24 | 2.97 | 0.03 | 24.67 |
| 305E40 ++ | Ctr | 12.69 | 0.27 | 0 | 0 | 0 |
| 67/3 ++ | Ctr | 0 | 0 | 3.28 | 0.05 | 100 |
| Name | Code | HRM Genotype | Peel Visual Color Phenotype | HPLC Phenotype |
|---|---|---|---|---|
| DADALI | AM 001 | AA | L | - |
| CIMA VIOLA | AM 004 | aa | P | - |
| 1F5(9) | AM 010 | aa | P | - |
| CCR3 | AM 013 | aa | P | - |
| VIOLETTA SAIS | AM 014 | AA | L | - |
| LUGA 063 | AM 015 | aa | P | - |
| PROSPEROSA | AM 016 | AA | L | - |
| LUNGA VIOLETTA | AM 018 | AA | P * | NAS |
| TAL1/1 | AM 021 | aa | P | - |
| ANGIO | AM 022 | AA | L | - |
| DR2 | AM 026 | aa | P | - |
| FANT E13D | AM 029 | AA | P * | D3R * |
| SNL 600 | AM 034 | AA | L | - |
| CIN 01/24-6 | AM 035 | AA | P * | D3R * |
| VIOLA CIN | AM 036 | AA | L | - |
| V. TOSCANA | AM 037 | AA | L | - |
| 44074 | AM 042 | AA | L | - |
| 55-08 | AM 045 | AA | L | - |
| 16-09 | AM 046 | AA | L | - |
| P621-08 (74-4) | AM 047 | AA | L | - |
| P328 | AM 053 | AATv | P | D3R * |
| S1052-08 | AM 056 | AATv | P | D3R * |
| LS 3805 MINDEN | AM 086 | AA | L | - |
| LS611 | AM 103 | aa | P | - |
| NAGA UNGU | AM 106 | AA | L | - |
| N286 | AM 107 | aa | P | - |
| N24 | AM 110 | aa | P | - |
| N243 | AM 111 | aa | P | - |
| N286 | AM 112 | aa | P | - |
| N321-14 | AM 113 | Aa | L | NAS |
| PI17 | AM 124 | aa | P | - |
| LUNGA MARINA | AM 139 | aa | P | - |
| BUIA | AM 156 | aa | P | - |
| ANK2 | AM 158 | aa | P | - |
| ANGIO3 | AM 167 | AA | L | - |
| SM 19/14 | AM 170 | aa | P | - |
| PALERMITANA | AM 171 | AA | L | - |
| JM | AM 174 | AA | L | - |
| THAI TH472 | AM 179 | aa | P | - |
| LISTADA | AM 180 | aa | P | - |
| THAI TH449 | AM 190 | aa | L * | D3R |
| THAI TH4760 | AM 199 | aa | P | -- |
| TOPAK | AM 217 | aa | P | -- |
| PI169648 | AM 221 | aa | P | - |
| TOPATAN | AM 222 | aa | P | - |
| PI171859 | AM 232 | aa | P | - |
| L129 | AM 268 | AA | L | - |
| DRS4 | AM 274 | aa | P | - |
| CAAS 6 | AM 280 | AA | L | - |
| CAAS 16 | AM 290 | AA | L | - |
| CAAS 17 | AM 291 | AA | P * | NAS |
| LONGO | AM 300 | aa | P | - |
| TALINDO purple | AM 302 | aa | P | - |
| BANGLADESH | AM 318 | aa | P | - |
| USTICA | AM 322 | AA | L | - |
| INDIA1 | AM 360 | AA | L | - |
| L316 | AM 371 | aa | P | - |
| LP742 | AM 378 | AA | L | - |
| L. VIOLA MEDIA | - | aa | P | - |
| ANOMINORI | - | AA | L | - |
| VIOLA OVALE | - | AA | L | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Florio, F.E.; Gattolin, S.; Toppino, L.; Bassolino, L.; Fibiani, M.; Lo Scalzo, R.; Rotino, G.L. A SmelAAT Acyltransferase Variant Causes a Major Difference in Eggplant (Solanum melongena L.) Peel Anthocyanin Composition. Int. J. Mol. Sci. 2021, 22, 9174. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22179174
Florio FE, Gattolin S, Toppino L, Bassolino L, Fibiani M, Lo Scalzo R, Rotino GL. A SmelAAT Acyltransferase Variant Causes a Major Difference in Eggplant (Solanum melongena L.) Peel Anthocyanin Composition. International Journal of Molecular Sciences. 2021; 22(17):9174. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22179174
Chicago/Turabian StyleFlorio, Francesco Elia, Stefano Gattolin, Laura Toppino, Laura Bassolino, Marta Fibiani, Roberto Lo Scalzo, and Giuseppe Leonardo Rotino. 2021. "A SmelAAT Acyltransferase Variant Causes a Major Difference in Eggplant (Solanum melongena L.) Peel Anthocyanin Composition" International Journal of Molecular Sciences 22, no. 17: 9174. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22179174