miRNAs as Regulators of the Early Local Response to Burn Injuries
Abstract
:1. Introduction
2. Results
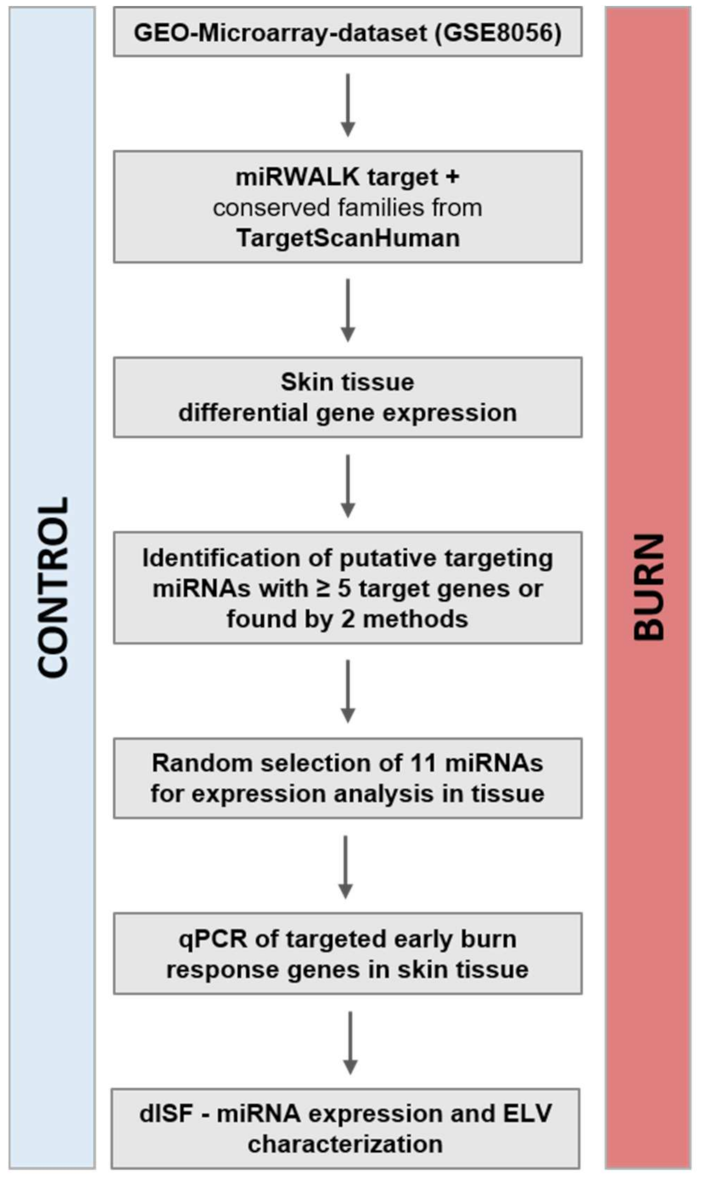
2.1. In the Early Response to Burn Injuries Putatively Involved miRNAs Are Identified through a Bioinformatics Approach
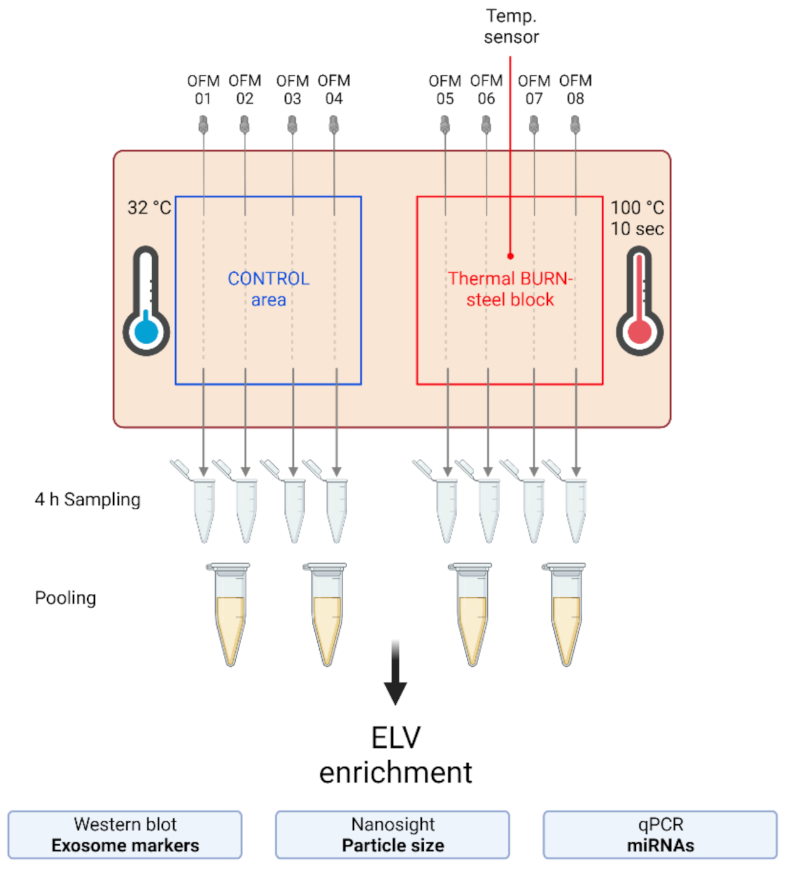
2.2. Extracellular Vesicles and Their miRNA Cargo in dISF of Burned Skin Have Been Characterized
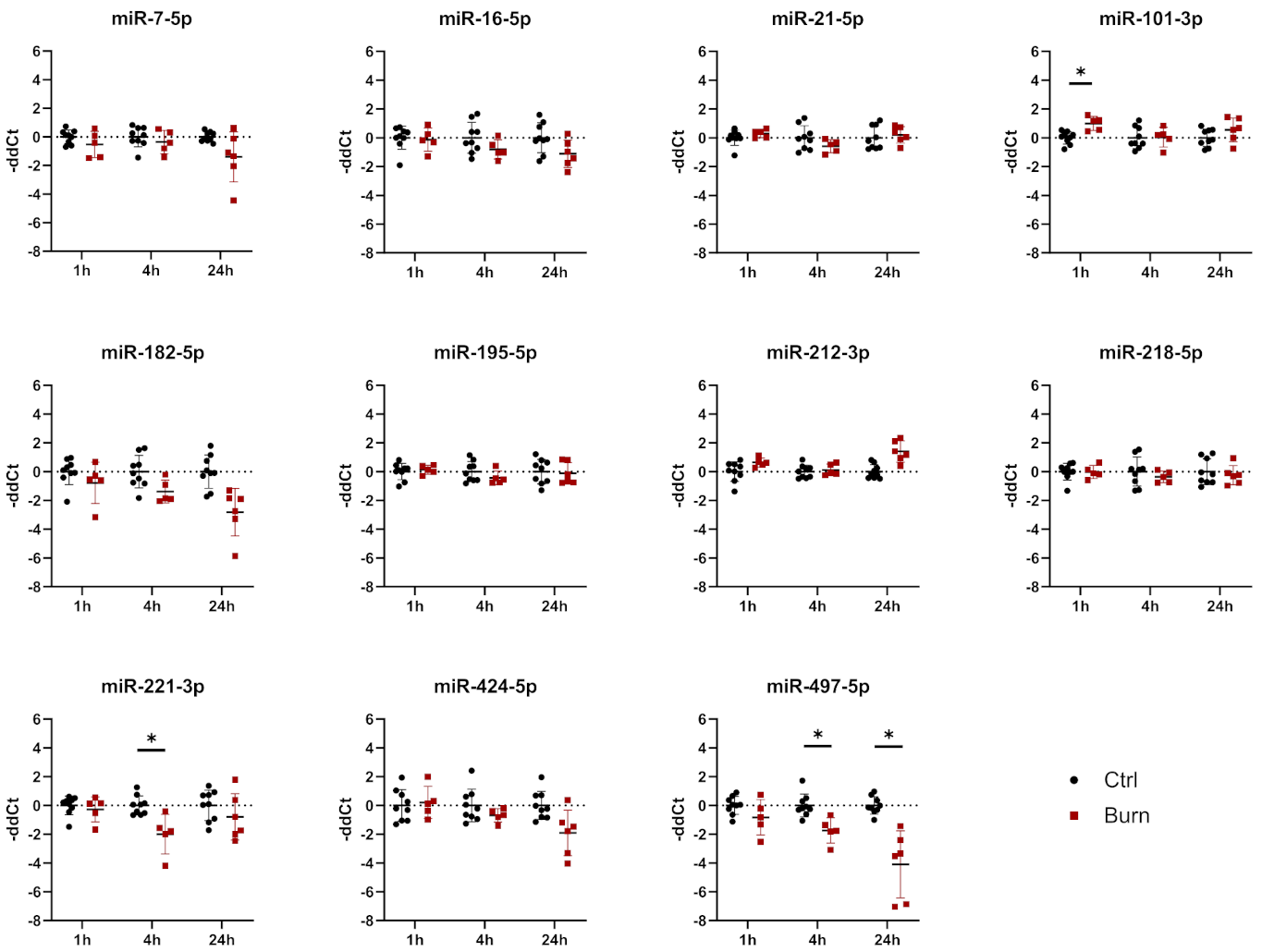
2.3. After a Burn Injury miR-497-5p Is Downregulated in dISF
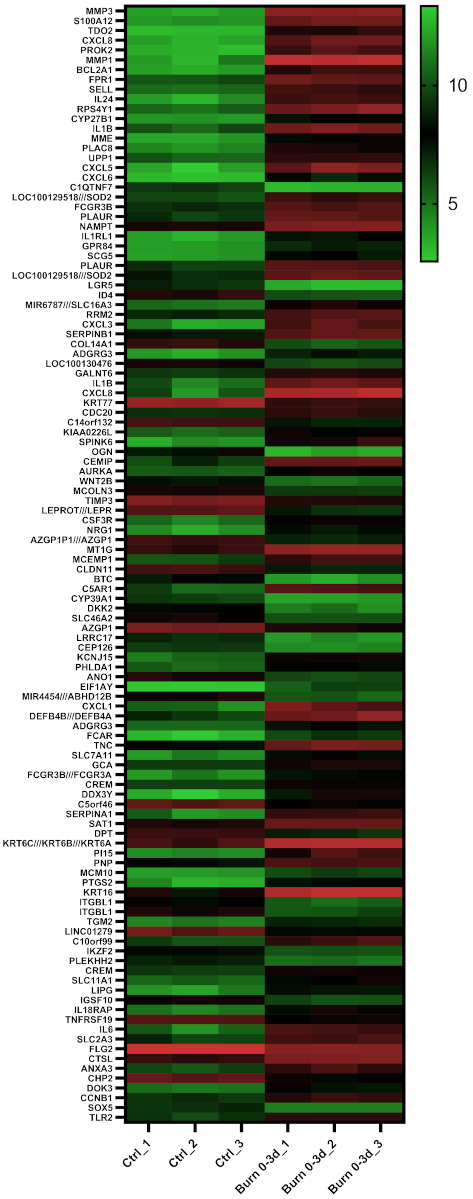
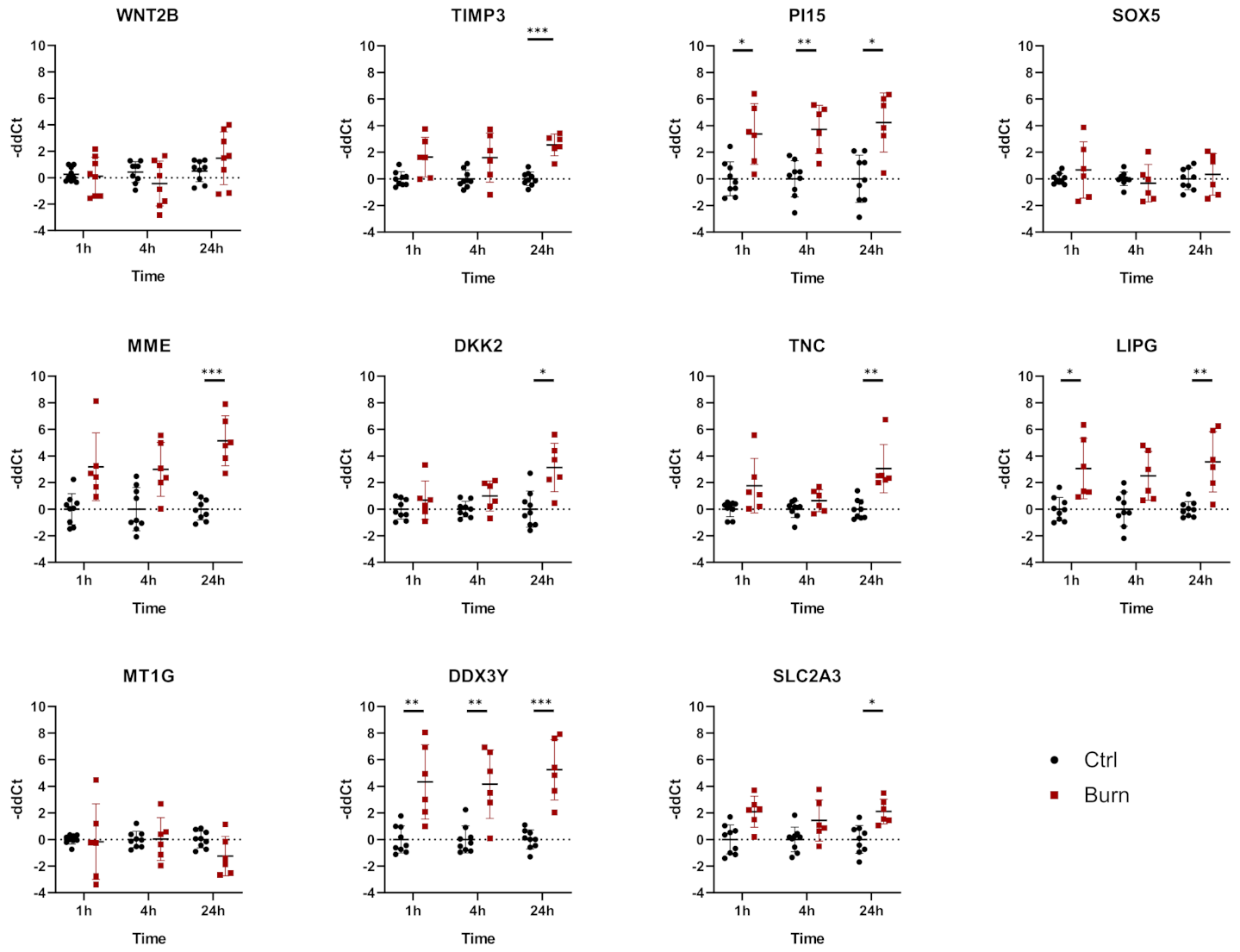
2.4. A Subset of miRNAs and Genes Is Differentially Expressed within the First 24 h in Burned Tissue
2.5. In Burned Skin Tissue miRNAs and Their Potential Targets Are Regulated in a Time-Dependent Manner
3. Discussion
4. Materials and Methods
4.1. Tissue Samples
4.2. Human Skin-Burn Injury Ex Vivo Model and Dermal Open Flow Microperfusion (dOFM)
4.3. Gene-Expression Data Analysis and Selection of miRNA-Targets
4.4. RNA Extraction and qPCR
4.5. qPCR Analysis
4.6. Sample Processing and ELV Enrichment
4.7. Nanoparticle Tracking Analysis
4.8. Protein Concentration and Immunoblot Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwacha, M.G. Macrophages and post-burn immune dysfunction. Burns 2003, 29, 1–14. [Google Scholar] [CrossRef]
- Plackett, T.P.; Colantoni, A.; Heinrich, S.A.; Messingham, K.A.N.; Gamelli, R.L.; Kovacs, E.J. The early acute phase response after burn injury in mice. J. Burn Care Res. 2007, 28, 167–172. [Google Scholar] [CrossRef]
- Nielson, C.B.; Duethman, N.C.; Howard, J.M.; Moncure, M.; Wood, J.G. Burns: Pathophysiology of Systemic Complications and Current Management. J. Burn Care Res. 2017, 38, e469–e481. [Google Scholar] [CrossRef]
- Kallinen, O.; Maisniemi, K.; Böhling, T.; Tukiainen, E.; Koljonen, V. Multiple organ failure as a cause of death in patients with severe burns. J. Burn Care Res. 2012, 33, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, M.G. Postburn hypermetabolism: Past, present, and future. J. Burn Care Res. 2016, 37, 86–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auger, C.; Samadi, O.; Jeschke, M.G. The biochemical alterations underlying post-burn hypermetabolism. Biochim. Biophys. Acta-Mol. Basis Dis. 2017, 1863, 2633–2644. [Google Scholar] [CrossRef]
- Barret, J.P.; Herndon, D.N. Modulation of inflammatory and catabolic responses in severely burned children by early burn wound excision in the first 24 hours. Arch. Surg. 2003, 138, 127–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoeksema, H.; Van de Sijpe, K.; Tondu, T.; Hamdi, M.; Van Landuyt, K.; Blondeel, P.; Monstrey, S. Accuracy of early burn depth assessment by laser Doppler imaging on different days post burn. Burns 2009, 35, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [Green Version]
- Solvin, Å.Ø.; Chawla, K.; Olsen, L.C.; Danielsen, K.; Jenssen, M.; Furberg, A.S.; Saunes, M.; Hveem, K.; Sætrom, P.; Løset, M. RNA sequencing of a large number of psoriatic patients identifies 131 novel miRNAs and 11 miRNAs associated with disease severity. medRxiv 2021. [Google Scholar] [CrossRef]
- Srivastava, A.; Meisgen, F.; Pasquali, L.; Munkhammar, S.; Xia, P.; Ståhle, M.; Landén, N.X.; Pivarcsi, A.; Sonkoly, E. Next-Generation Sequencing Identifies the Keratinocyte-Specific miRNA Signature of Psoriasis. J. Investig. Dermatol. 2019, 139, 2547–2550.e12. [Google Scholar] [CrossRef]
- Lerman, G.; Avivi, C.; Mardoukh, C.; Barzilai, A.; Tessone, A.; Gradus, B.; Pavlotsky, F.; Barshack, I.; Polak-Charcon, S.; Orenstein, A.; et al. MiRNA expression in psoriatic skin: Reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS ONE 2011, 6, e20916. [Google Scholar] [CrossRef]
- Suárez-Fariñas, M.; Ungar, B.; Correa Da Rosa, J.; Ewald, D.A.; Rozenblit, M.; Gonzalez, J.; Xu, H.; Zheng, X.; Peng, X.; Estrada, Y.D.; et al. RNA sequencing atopic dermatitis transcriptome profiling provides insights into novel disease mechanisms with potential therapeutic implications. J. Allergy Clin. Immunol. 2015, 135, 1218–1227. [Google Scholar] [CrossRef]
- Polak, M.E. Early life regulation of inflammation in atopic dermatitis by microRNA. Br. J. Dermatol. 2021, 184, 391–392. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, D.; Xiao, Z.; Zhang, M. MiRNA expression profiles in keloid tissue and corresponding normal skin tissue. Aesthetic Plast. Surg. 2012, 36, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Lv, C.; Jiang, B.; Long, X.; Zhang, P.; Zhang, M.; Xie, T.; Huang, X. MicroRNA profiling in denatured dermis of deep burn patients. Burns 2012, 38, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Singhvi, G.; Manchanda, P.; Krishna Rapalli, V.; Kumar Dubey, S.; Gupta, G.; Dua, K. MicroRNAs as biological regulators in skin disorders. Biomed. Pharmacother. 2018, 108, 996–1004. [Google Scholar] [CrossRef]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.R.; Taylor, R.M.; Tran, B.Q.; Boyd, G.; Glaros, T.; Chavez, V.H.; Krishnakumar, R.; Sinha, A.; Poorey, K.; Williams, K.P.; et al. Extraction and biomolecular analysis of dermal interstitial fluid collected with hollow microneedles. Commun. Biol. 2018, 1, 173. [Google Scholar] [CrossRef] [Green Version]
- Bodenlenz, M.; Aigner, B.; Dragatin, C.; Liebenberger, L.; Zahiragic, S.; Höfferer, C.; Birngruber, T.; Priedl, J.; Feichtner, F.; Schaupp, L.; et al. Clinical applicability of dOFM devices for dermal sampling. Ski. Res. Technol. 2013, 19, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Terlecki-Zaniewicz, L.; Pils, V.; Bobbili, M.R.; Lämmermann, I.; Perrotta, I.; Grillenberger, T.; Schwestka, J.; Weiß, K.; Pum, D.; Arcalis, E.; et al. Extracellular Vesicles in Human Skin: Cross-Talk from Senescent Fibroblasts to Keratinocytes by miRNAs. J. Investig. Dermatol. 2019, 139, 2425–2436.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Yan, Z.; Yang, F.; Huang, Y.; Yu, Y.; Zhou, L.; Sun, Z.; Cui, D.; Yan, Y. Exosomes Derived from Human Umbilical Cord Mesenchymal Stem Cells Accelerate Cutaneous Wound Healing by Enhancing Angiogenesis through Delivering Angiopoietin-2. Stem Cell Rev. Rep. 2021, 17, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Greco, J.A.; Pollins, A.C.; Boone, B.E.; Levy, S.E.; Nanney, L.B. A microarray analysis of temporal gene expression profiles in thermally injured human skin. Burns 2010, 36, 192–204. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, E.; Fink, J.; Eberl, A.; Prugger, E.M.; Kolb, D.; Luze, H.; Schwingenschuh, S.; Birngruber, T.; Magnes, C.; Mautner, S.I.; et al. A novel human ex vivo skin model to study early local responses to burn injuries. Sci. Rep. 2021, 11, 364. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [Green Version]
- Kolbinger, F.; Loesche, C.; Valentin, M.A.; Jiang, X.; Cheng, Y.; Jarvis, P.; Peters, T.; Calonder, C.; Bruin, G.; Polus, F.; et al. β-Defensin 2 is a responsive biomarker of IL-17A–driven skin pathology in patients with psoriasis. J. Allergy Clin. Immunol. 2017, 139, 923–932.e8. [Google Scholar] [CrossRef] [Green Version]
- Bodenlenz, M.; Tiffner, K.I.; Raml, R.; Augustin, T.; Dragatin, C.; Birngruber, T.; Schimek, D.; Schwagerle, G.; Pieber, T.R.; Raney, S.G.; et al. Open Flow Microperfusion as a Dermal Pharmacokinetic Approach to Evaluate Topical Bioequivalence. Clin. Pharmacokinet. 2017, 56, 91–98. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Wang, P.; Zhang, J.; Zhao, D. MicroRNA-497-5p downregulation inhibits cell viability, reduces extracellular matrix deposition and induces apoptosis in human hyperplastic scar fibroblasts by regulating Smad7. Exp. Ther. Med. 2021, 21, 1–8. [Google Scholar] [CrossRef]
- Finnerty, C.C.; Jeschke, M.G.; Branski, L.K.; Barret, J.P.; Dziewulski, P.; Herndon, D.N. Hypertrophic scarring: The greatest unmet challenge after burn injury. Lancet 2016, 388, 1427–1436. [Google Scholar] [CrossRef] [Green Version]
- Chai, L.; Kang, X.J.; Sun, Z.Z.; Zeng, M.F.; Yu, S.R.; Ding, Y.; Liang, J.Q.; Li, T.T.; Zhao, J. MiR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag. Res. 2018, 10, 989–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Ma, R.; Yue, J.; Li, N.; Li, Z.; Qi, D. MiR-497 suppresses YAP1 and inhibits tumor growth in non-small cell lung cancer. Cell. Physiol. Biochem. 2015, 37, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Xiong, G.; Cao, Z.; Zheng, S.; You, L.; Zhang, T.; Zhao, Y. miR-497 expression, function and clinical application in cancer. Oncotarget 2016, 7, 55900–55911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, W.; Zhang, W.Y.; Bai, J.B.; Zhang, H.X.; Zhao, Y.Y.; Li, X.Y.; Zhao, S.H. The NF-κB-modulated microRNAs miR-195 and miR-497 inhibit myoblast proliferation by targeting Igf1r, Insr and cyclin genes. J. Cell Sci. 2016, 129, 39–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Wang, L.E.I.; Liu, W.E.I.; Li, F.E.I. MicroRNA-497-5p inhibits proliferation and invasion of non-small cell lung cancer by regulating FGF2. Oncol. Lett. 2019, 17, 3425–3431. [Google Scholar] [CrossRef] [Green Version]
- Jeong Kim, Y.; Jin Hwang, S.; Chan Bae, Y.; Sup Jung, J. MiR-21 regulates adipogenic differentiation through the modulation of TGF-β signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells 2009, 27, 3093–3102. [Google Scholar] [CrossRef]
- Wang, T.; Feng, Y.; Sun, H.; Zhang, L.; Hao, L.; Shi, C.; Wang, J.; Li, R.; Ran, X.; Su, Y.; et al. MiR-21 regulates skin wound healing by targeting multiple aspects of the healing process. Am. J. Pathol. 2012, 181, 1911–1920. [Google Scholar] [CrossRef]
- Wu, Y.; Song, Y.; Xiong, Y.; Wang, X.; Xu, K.; Han, B.; Bai, Y.; Li, L.; Zhang, Y.; Zhou, L. MicroRNA-21 (Mir-21) Promotes Cell Growth and Invasion by Repressing Tumor Suppressor PTEN in Colorectal Cancer. Cell. Physiol. Biochem. 2017, 43, 945–958. [Google Scholar] [CrossRef]
- Ourô, S.; Mourato, C.; Velho, S.; Cardador, A.; Ferreira, M.P.; Albergaria, D.; Castro, R.E.; Maio, R.; Rodrigues, C.M.P. Potential of miR-21 to Predict Incomplete Response to Chemoradiotherapy in Rectal Adenocarcinoma. Front. Oncol. 2020, 10, 2212. [Google Scholar] [CrossRef]
- Zhao, B.; Chen, Y.; Yang, N.; Chen, Q.; Bao, Z.; Liu, M.; Hu, S.; Li, J.; Wu, X. miR-218-5p regulates skin and hair follicle development through Wnt/β-catenin signaling pathway by targeting SFRP2. J. Cell. Physiol. 2019, 234, 20329–20341. [Google Scholar] [CrossRef]
- Shi, Z.M.; Wang, L.; Shen, H.; Jiang, C.F.; Ge, X.; Li, D.M.; Wen, Y.Y.; Sun, H.R.; Pan, M.H.; Li, W.; et al. Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene 2017, 36, 2577–2588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, C. microRNA-211-3p has a Role in the Effects of Lipopolysaccharide on Endoplasmic Reticulum Stress in Cultured Human Skin Fibroblasts. Med. Sci. Monit. Basic Res. 2019, 25, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Qiu, J.; Zhang, H. MiR-221-3p as a Potential Biomarker for Patients with Psoriasis and Its Role in Inflammatory Responses in Keratinocytes. Skin Pharmacol. Physiol. 2021, 1–7. [Google Scholar] [CrossRef]
- Wilmink, G.J.; Roth, C.L.; Ibey, B.L.; Ketchum, N.; Bernhard, J.; Cerna, C.Z.; Roach, W.P. Identification of microRNAs associated with hyperthermia-induced cellular stress response. Cell Stress Chaperones 2010, 15, 1027–1038. [Google Scholar] [CrossRef] [Green Version]
- Mueller, S.N.; Zaid, A.; Carbone, F.R. Tissue-Resident T Cells: Dynamic Players in Skin Immunity. Front. Immunol. 2014, 5, 332. [Google Scholar] [CrossRef] [Green Version]
- Bianchi, F.; Nicassio, F.; Di Fiore, P.P. Unbiased vs. biased approaches to the identification of cancer signatures: The case of lung cancer. Cell Cycle 2008, 7, 729–734. [Google Scholar] [CrossRef]
- Rooda, I.; Hensen, K.; Kaselt, B.; Kasvandik, S.; Pook, M.; Kurg, A.; Salumets, A.; Velthut-Meikas, A. Target prediction and validation of microRNAs expressed from FSHR and aromatase genes in human ovarian granulosa cells. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Spies, M.; Dasu, M.R.K.; Svrakic, N.; Nesic, O.; Barrow, R.E.; Perez-Polo, J.R.; Herndon, D.N. Gene expression analysis in burn wounds of rats. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2002, 283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef]
- Szabowski, A.; Maas-Szabowski, N.; Andrecht, S.; Kolbus, A.; Schorpp-Kistner, M.; Fusenig, N.E.; Angel, P. c-Jun and JunB antagonistically control cytokine-regulated mesenchymal-epidermal interaction in skin. Cell 2000, 103, 745–755. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Awoyemi, A.A.; Fahy, K.E.; Thapa, P.; Borchers, C.; Wu, B.Y.; McGlone, C.L.; Schmeusser, B.; Sattouf, Z.; Rohan, C.A.; et al. Keratinocyte-derived microvesicle particles mediate ultraviolet B radiation–induced systemic immunosuppression. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef] [PubMed]
- Holmgaard, R.; Benfeldt, E.; Nielsen, J.B.; Gatschelhofer, C.; Sorensen, J.A.; Höfferer, C.; Bodenlenz, M.; Pieber, T.R.; Sinner, F. Comparison of open-flow microperfusion and microdialysis methodologies when sampling topically applied fentanyl and benzoic acid in human dermis ex vivo. Pharm. Res. 2012, 29, 1808–1820. [Google Scholar] [CrossRef]
- Dweep, H.; Gretz, N. MiRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| No. | % | |
|---|---|---|
| Total genes annotated | 54,675 | 100% |
| padj < 0.05 | 3558 | 6.5% |
| padj < 0.01 | 992 | 1.8% |
| padj < 0.001 | 114 | 0.2% |
| upregulated | 79 | 70% |
| downregulated | 35 | 30% |
| ID | padj | P | t | B | logFC | Gene Symbol | Gene Title | GSM 198875 0 h | GSM 198876 0 h | GSM 198877 0 h | Mean 0 h | GSM 198866 3 d | GSM 198867 3 d | GSM 198868 3 d | Mean 3 d | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 203434_s_at | 0.0001246 | 3.19 × 10−8 | 19.65 | 9.10 | 4.28 | MME | membrane metalloendopeptidase | 12.02 | 11.80 | 16.98 | 13.60 | 211.84 | 278.65 | 300.48 | 263.66 |
| 49 | 238512_at | 0.0006066 | 5.67 × 10−7 | −13.73 | 6.78 | −2.38 | WNT2B | Wnt family member 2B | 190.56 | 162.10 | 190.51 | 181.06 | 35.37 | 30.40 | 38.82 | 34.87 |
| 51 | 201150_s_at | 0.0006066 | 5.88 × 10−7 | −13.67 | 6.75 | −2.35 | TIMP3 | TIMP metallopeptidase inhibitor 3 | 2684.61 | 2025.09 | 2336.93 | 2348.88 | 471.36 | 471.65 | 431.15 | 458.05 |
| 56 | 204745_x_at | 0.0006066 | 6.22 × 10−7 | 13.57 | 6.70 | 2.62 | MT1G | metallothionein 1G | 642.73 | 474.11 | 677.58 | 598.14 | 3296.03 | 3958.74 | 3704.69 | 3653.15 |
| 62 | 219908_at | 0.000613 | 7.14 × 10−7 | −13.34 | 6.58 | −3.11 | DKK2 | dickkopf WNT signaling pathway inhibitor 2 | 210.42 | 228.11 | 234.28 | 224.27 | 26.20 | 34.89 | 18.92 | 26.67 |
| 76 | 201645_at | 0.000613 | 8.52 × 10−7 | 13.04 | 6.42 | 3.08 | TNC | tenascin C | 293.96 | 214.10 | 190.91 | 232.99 | 1506.81 | 2321.70 | 2087.41 | 1971.97 |
| 81 | 205000_at | 0.0006873 | 1.02 × 10−6 | 12.75 | 6.26 | 4.96 | DDX3Y | DEAD-box helicase 3. Y-linked | 11.08 | 6.00 | 10.63 | 9.24 | 152.53 | 371.48 | 375.00 | 299.67 |
| 87 | 229947_at | 0.0007308 | 1.16 × 10−6 | 12.54 | 6.14 | 5.05 | PI15 | peptidase inhibitor 15 | 18.37 | 22.90 | 19.28 | 20.18 | 332.87 | 1159.94 | 760.88 | 751.23 |
| 101 | 219181_at | 0.0008774 | 1.62 × 10−6 | 12.02 | 5.83 | 3.30 | LIPG | lipase G. endothelial type | 16.70 | 12.60 | 27.13 | 18.81 | 196.16 | 170.26 | 164.54 | 176.99 |
| 106 | 202499_s_at | 0.0009022 | 1.77 × 10−6 | 11.89 | 5.75 | 3.23 | SLC2A3 | solute carrier family 2 member 3 | 133.38 | 70.10 | 72.10 | 91.86 | 785.79 | 729.58 | 970.77 | 828.71 |
| 113 | 238009_at | 0.0009914 | 2.07 × 10−6 | −11.65 | 5.61 | −2.13 | SOX5 | SRY-box 5 | 92.71 | 104.10 | 133.17 | 109.99 | 24.77 | 24.69 | 24.90 | 24.79 |
| WNT2B | TIMP3 | PI15 | SOX5 | MME | DKK2 | TNC | LIPG | MT1G | DDX3Y | SLC2A3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-7-5p | x | ||||||||||
| miR-16-5p | x | x | x | x | x | ||||||
| miR-21-5p | x | x | x | x | |||||||
| miR-101-3p | x | ||||||||||
| miR-182-5p | x | x | x | x | |||||||
| miR-195-5p | x | x | x | x | x | ||||||
| miR-212-3p | x | ||||||||||
| miR-218-5p | x | x | x | x | x | x | |||||
| miR-221-3p | x | x | |||||||||
| miR-424-5p | x | x | x | x | x | ||||||
| miR-497-5p | x | x | x | x | x |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foessl, I.; Haudum, C.W.; Vidakovic, I.; Prassl, R.; Franz, J.; Mautner, S.I.; Kainz, S.; Hofmann, E.; Obermayer-Pietsch, B.; Birngruber, T.; et al. miRNAs as Regulators of the Early Local Response to Burn Injuries. Int. J. Mol. Sci. 2021, 22, 9209. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22179209
Foessl I, Haudum CW, Vidakovic I, Prassl R, Franz J, Mautner SI, Kainz S, Hofmann E, Obermayer-Pietsch B, Birngruber T, et al. miRNAs as Regulators of the Early Local Response to Burn Injuries. International Journal of Molecular Sciences. 2021; 22(17):9209. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22179209
Chicago/Turabian StyleFoessl, Ines, Christoph Walter Haudum, Ivan Vidakovic, Ruth Prassl, Joakim Franz, Selma I. Mautner, Sonja Kainz, Elisabeth Hofmann, Barbara Obermayer-Pietsch, Thomas Birngruber, and et al. 2021. "miRNAs as Regulators of the Early Local Response to Burn Injuries" International Journal of Molecular Sciences 22, no. 17: 9209. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22179209






