Interactions of Truncated Menaquinones in Lipid Monolayers and Bilayers
Abstract
:1. Introduction
2. Results
2.1. Compression Isotherm Studies of MKs in Langmuir Monolayers
2.2. Compression Isotherms of Normalized Mixed MK and DPPC or DPPE Films
2.3. Ideal Mixing of MK and DPPC or DPPE
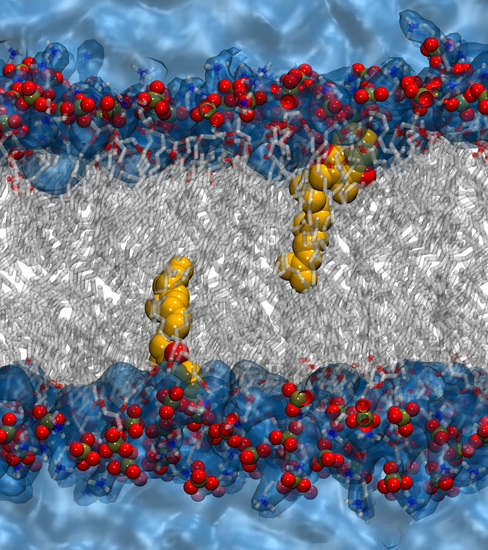
2.4. Molecular Dynamics Simulations of MKs in a Membrane Bilayer
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collins, M.D.; Jones, D. Distribution of Isoprenoid Quinone Structural Types in Bacteria and Their Taxonomic Implications. Microbiol. Rev. 1981, 45, 316–354. [Google Scholar] [CrossRef] [PubMed]
- Brennan, P.J. Mycobacterium and other actinomycetes. In Microbial Lipids; Ratledge, C., Wilkinson, S.G., Eds.; Academic Press: San Diego, CA, USA, 1988; Volume 1, pp. 203–298. [Google Scholar]
- Upadhyay, A.; Kumar, S.; Rooker, S.A.; Koehn, J.T.; Crans, D.C.; McNeil, M.R.; Lott, J.S.; Crick, D.C. Mycobacterial MenJ: An Oxidoreductase Involved in Menaquinone Biosynthesis. ACS Chem. Biol. 2018, 13, 2498–2507. [Google Scholar] [CrossRef]
- Braasch-Turi, M.; Crans, D.C. Synthesis of Naphthoquinone Derivatives: Menaquinones, Lipoquinones and Other Vitamin K Derivatives. Molecules 2020, 25, 4477. [Google Scholar] [CrossRef] [PubMed]
- Popa, D.-S.; Bigman, G.; Rusu, M.E. The Role of Vitamin K in Humans: Implication in Aging and Age-Associated Diseases. Antioxidants 2021, 10, 566. [Google Scholar] [CrossRef] [PubMed]
- Koehn, J.T.; Beuning, C.N.; Peters, B.J.; Dellinger, S.K.; Van Cleave, C.; Crick, D.C.; Crans, D.C. Investigating Substrate Analogues for Mycobacterial MenJ: Truncated and Partially Saturated Menaquinones. Biochemistry 2019, 58, 1596–1615. [Google Scholar] [CrossRef]
- Koehn, J.T.; Magallanes, E.S.; Peters, B.J.; Beuning, C.N.; Haase, A.A.; Zhu, M.J.; Rithner, C.D.; Crick, D.C.; Crans, D.C. A Synthetic Isoprenoid Lipoquinone, Menaquinone-2, Adopts a Folded Conformation in Solution and at a Model Membrane Interface. J. Org. Chem. 2018, 83, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Gupta, C.; Khaniya, U.; Chan, C.K.; Dehez, F.; Shekhar, M.; Gunner, M.R.; Sazanov, L.; Chipot, C.; Singharoy, A. Charge Transfer and Chemo-Mechanical Coupling in Respiratory Complex I. J. Am. Chem. Soc. 2020, 142, 9220–9230. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Hugenholtz, J.; van Halbeek, H.; Ljungdahl, L.G. Structure and Function of a Menaquinone Involved in Electron Transport in Membranes of Clostridium thermoautotrophicium and Clostridium thermoaceticum. J. Bacteriol. 1989, 171, 5823–5829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenaz, G.; Samori, B.; Fato, R.; Battino, M.; Castelli, C.P.; Domini, I. Localization and preferred orientations of ubiquinone homologs in model bilayers. Biochem. Cell. Biol. 1992, 70, 504–514. [Google Scholar] [CrossRef]
- Feng, S.; Wang, R.; Pastor, R.W.; Klauda, J.B.; Im, W. Location and Conformational Ensemble of Menaquinone and Menaquinol, and Protein-Lipid Modulations in Archaeal Membranes. J. Phys. Chem. B 2021, 125, 4714–4725. [Google Scholar] [CrossRef]
- Nixon, G.L.; Pidathala, C.; Shone, A.E.; Antoine, T.; Fisher, N.; O’Neill, P.M.; Ward, S.A.; Biagini, G.A. Targeting the mitochondrial electron transport chain of Plasmodium falciparum: New strategies towards the development of improved antimalarials for the elimination era. Future Med. Chem. 2013, 5, 1573–1591. [Google Scholar] [CrossRef]
- Berube, B.J.; Russell, D.; Castro, L.; Choi, S.-R.; Narayanasamy, P.; Parish, T. Novel Men A Inhibitors Are Bactericidal against Mycobacterium tuberculosis and Synergize with Electron Transport Chain Inhibitors. Antimicrob. Agent Chemother. 2019, 63, e02661-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debnath, J.; Siricilla, S.; Wan, B.; Crick, D.C.; Lenaerts, A.J.; Franzblau, S.G.; Kurosu, M. Discovery of Selective Menaquinone Biosynthesis Inhibitors against Mycobacterium tuberculosis. J. Med. Chem. 2012, 55, 3739–3755. [Google Scholar] [CrossRef] [PubMed]
- Kurosu, M.; Begari, E. Vitamin K2 in Electron Transport System: Are Enzymes Involved in Vitamin K2 Biosynthesis Promising Drug Targets? Molecules 2012, 15, 1531–1553. [Google Scholar] [CrossRef] [Green Version]
- Afri, M.; Ehrenberg, B.; Talmon, Y.; Schmidt, J.; Cohen, Y.; Frimer, A.A. Active oxygen chemistry within the liposomal bilayer Part III: Locating Vitamin E, ubiquinol and ubiquinone and their derivatives in the lipid bilayer. Chem. Phys. Lipids 2004, 131, 107–121. [Google Scholar] [CrossRef]
- Cornell, B.A.; Keniry, M.A.; Post, A.; Roberston, R.N.; Weir, L.E.; Westerman, P.W. Location and activity of ubiquinone 10 and ubiquinone analogues in model and biological membranes. Biochemistry 1987, 26, 7702–7707. [Google Scholar] [CrossRef]
- Upadhyay, A.; Fontes, F.L.; Gonzalez-Juarrero, M.; McNeil, M.R.; Crans, D.C.; Jackson, M.; Crick, D.C. Partial Saturation of Menaquinone in Mycobacterium tuberculosis: Function and Essentiality of a Novel Reductase, MenJ. ACS Cent. Sci. 2015, 1, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Koehn, J.T.; Gonzalez-Juarrero, M.; Crans, D.C.; Crick, D.C. Mycobacterium tuberculosis survival in J774A.1 Cells Is Dependent on MenJ Moonlighting Activity, Not Its Enzymatic Activity. ACS Infect. Dis. 2020, 6, 2661–2671. [Google Scholar] [CrossRef] [PubMed]
- Möhwald, H.; Brezesinski, G. From Langmuir Monolayers to Multilayer Films. Langmuir 2016, 32, 10445–10458. [Google Scholar] [CrossRef]
- Stefaniu, C.; Brezesinski, G.; Möhwald, H. Langmuir monolayers as models to study processes at membrane surfaces. Adv. Colloid Interface Sci. 2014, 208, 197–213. [Google Scholar] [CrossRef]
- Hoyo, J.; Guaus, E.; Torrent-Burgués, J.; Sanz, F. Biomimetic monolayer films of digalactosyldiacylglycerol incorporating plastoquinone. Biochim. Biophys. Acta 2015, 1848, 1341–1351. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.N.; Chapman, D. Micelles, Monolayers, and Biomembranes; Wiley-Liss: New York, NY, USA, 1995. [Google Scholar]
- Roche, Y.; Peretti, P.; Bernard, S. Influence of the chain length of ubiquinones on their interaction with DPPC mixed monolayers. Biochim. Biophys. Acta 2006, 1758, 468–478. [Google Scholar] [CrossRef] [Green Version]
- Katsikas, H.; Quinn, P. The polyisoprenoid chain length influences the interaction of ubiquinones with phospholipid bilayers. Biochim. Biophys. Acta 1982, 689, 363–369. [Google Scholar] [CrossRef]
- Roche, Y.; Peretti, P.; Bernard, S. DSC and Raman studies of the side chain length effect of ubiquinones on the thermotropic phase behavior of liposomes. Thermochim. Acta 2006, 447, 81–88. [Google Scholar] [CrossRef]
- Galassi, V.V.; Arantes, G.M. Partition, orientation and mobility of ubiquinones in a lipid bilayer. Biochim. Biophys. Acta 2015, 1847, 1560–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, M.H.; Arantes, G.M. Effects of lipid composition on membrane distribution and permeability of natural quinones. RSC Adv. 2019, 9, 16892–16899. [Google Scholar] [CrossRef] [Green Version]
- Aroti, A.; Leontidis, E.; Maltseva, E.; Brezesinski, G. Effects of Hofmeister anions on DPPC Langmuir monolayers at the air-water interface. J. Phys. Chem. B. 2004, 108, 15238–15245. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, T.; Kato, S. Detailed Analysis of the Surface Area and Elasticity in the Saturated 1,2-Diacylphosphatidylcholine/Cholesterol Binary Monolayer System. Langmuir 2015, 31, 9086–9096. [Google Scholar] [CrossRef]
- Quinn, P.J. Phase Behaviour of Binary Mixtures of Membrane Polar Lipids in Aqueous Sytems. Nat. Prod. Rep. 1987, 4, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Quinn, P.J.; Esfahani, M.A. Ubiquinones Have Surface-Active Properties Suited to Transport Electrons and Protons across Membranes. Biochem. J. 1980, 185, 715–722. [Google Scholar] [CrossRef] [Green Version]
- Zhan, T.J.; Eilers, M.; Guo, Z.; Ksebati, M.B.; Simon, M.; Scholten, J.D.; Smith, S.O.; Gibbs, R.A. Evaluation of Isoprenoid Conformation in Solution and in the Active Site of Protein-Farnesyl Transferase Using Carbon-13 Labeling in Conjunction with Solution- and Solid-State NMR. J. Am. Chem. Soc. 2000, 122, 7153–7164. [Google Scholar] [CrossRef]
- Hoyo, J.; Torrent-Burgués, J.; Guaus, E. Biomimetic monolayer films of monogalactosyldiacylglycerol incorporating ubiquinone. J. Colloid. Interface Sci. 2012, 384, 189–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyo, J.; Guaus, E.; Torrent-Burgués, J. Tuning ubiquinone position in biomimetic monolayer membranes. Eur. Phys. J. E: Soft Matter Biol. Phys. 2017, 40, 62. [Google Scholar] [CrossRef] [Green Version]
- Patterson, M.; Vogel, H.J.; Prenner, E.J. Biophysical characterization of monofilm model systems composed of selected tear film phospholipids. Biochim. Biophys. Acta Biomembr. 2016, 1858, 403–414. [Google Scholar] [CrossRef]
- Ma, G.; Allen, H.C. Condensing effect of palmitic acid on DPPC in mixed Langmuir monolayers. Langmuir 2007, 23, 589–597. [Google Scholar] [CrossRef]
- Veldhuizen, E.J.A.; Haagsman, H.P. Role of pulmonary surfactant components in surface film formation and dynamics. Biochim Biophys Acta-Biomembr. 2000, 1467, 255–270. [Google Scholar] [CrossRef] [Green Version]
- Crane, J.M.; Puts, G.; Hall, S.B. Persistence of Phase Coexistence in Disaturated Phosphatidylcholine Monolayers at High Surface Pressures. Biophys. J. 1999, 77, 3134–4143. [Google Scholar] [CrossRef] [Green Version]
- Shaw, N. Lipid Composition as a Guide to the Classification of Bacteria. In Advances in Applied Microbioligy; Perlman, D., Ed.; Academic Press: New York, NY, USA, 1974; Volume 17, pp. 63–108. [Google Scholar]
- Fadeel, B.; Xue, D. The ins and outs of phospholipid asymmetry in the plasma membrane: Roles in health and disease. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 264–277. [Google Scholar] [CrossRef]
- Chakrabarti, A. Phospholipid Asymmetry in Biological Membranes: Is the Role of Phosphatidylethanolamine Underappreciated? J. Membr. Biol. 2021, 254, 127–132. [Google Scholar] [CrossRef]
- Olżyńska, A.; Zubek, M.; Roeselova, M.; Korchowiec, J.; Cwiklik, L. Mixed DPPC/POPC Monolayers: All-atom Molecular Dynamics Simulations and Langmuir Monolayer Experiments. Biochim. Biophys. Acta 2016, 1858, 3120–3130. [Google Scholar] [CrossRef]
- Träuble, H.; Eibl, H.; Sawada, H. Respiration—A Critical Phenomenon? Lipid Phase Transitions in the Lung Alveolar Surfactant. Naturwissenschaften 1974, 61, 344–354. [Google Scholar] [CrossRef]
- Javanainen, M.; Lamberg, A.; Cwiklik, L.; Vattulainen, I.; Ollila, O.H.S. Atomistic Model for Nearly Quantitative Simulations of Langmuir Monolayers. Langmuir 2018, 34, 2565–2572. [Google Scholar] [CrossRef] [PubMed]
- Marsh, D. Handbook of Lipid Bilayers, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Van Cleave, C.; Murakami, H.A.; Samart, N.; Koehn, J.T.; Maldonado, P.; Kreckel, H.D.; Cope, E.J.; Basile, A.; Crick, D.C.; Crans, D.C. Location of menaquinone and menaquinol headgroups in model membranes. Can. J. Chem. 2020, 98, 307–317. [Google Scholar] [CrossRef]
- Stidham, M.A.; McIntosh, T.; Siedow, J.N. On the localization of ubiquinone in phosphatidylcholine bilayers. Biochim. Biophys. Acta. 1984, 767, 423–431. [Google Scholar] [CrossRef]
- Koehn, J.T. Synthesis and Exploration of Biologically Important, Hydrophobic, Redox-Active Molecules: Investigation of Partial Saturation of Mycobacterial Electron Transport Lipids. Ph.D. Dissertation, Colorado State University, Fort Collins, CO, USA, 2019. [Google Scholar]
- Javanainen, M. Universal Method for Embedding Proteins into Complex Lipid Bilayers for Molecular Dynamics Simulations. J. Chem. Theory Comput. 2014, 10, 2577–2582. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apsotolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A High-Throughput and Highly Parallel Open Source Molecular Simulation Toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- MacKrell, A.D., Jr.; Bashford, D.; Bellott, M.; Dunbrack, R.L., Jr.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Achraya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-atom Additive Biological Force Fields. J. Comp. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 14101. [Google Scholar] [CrossRef] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N⋅ Log (N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 1463–1472. [Google Scholar] [CrossRef] [Green Version]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comp. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Anézo, C.; de Vries, A.H.; Höltje, H.-D.; Tieleman, D.P.; Marrink, S.-J. Methodological Issues in Lipid Bilayer Simulations. J. Chem. Phys. 2003, 107, 9424–9433. [Google Scholar] [CrossRef] [Green Version]
- Kellerman, M.Y.; Yoshinaga, M.Y.; Valentine, R.C.; Wörmer, L.; Valentine, D.L. Important roles for membrane lipids in haloarchaeal bioenergetics. Biochim. Biophys. Acta. Biomembr. 2016, 1858, 2940–2956. [Google Scholar] [CrossRef]












| Property | Ranking |
|---|---|
| clogP | MK-4 (8.86) > MK-3 (7.52) > MK-2 (5.67) > MK-1 (3.83) |
| Ability to disrupt a phospholipid monolayer based on increase in monolayer area between 30 and 35 mN/m | MK-4 > MK-2 > MK-1 ~ MK-3 |
| MK headgroup location relative to the midplane | MK-1 ~ MK-2 ~ MK-3 ~ MK-4 |
| Longest average distance from CT to naphthoquinone headgroup | MK-4 > MK-3 > MK-2 > MK-1 |
| Ability of CT to be within 0.6 nm of the naphthoquinone headgroup | MK-1 > MK-2 > MK-4 > MK-3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Cleave, C.; Koehn, J.T.; Pereira, C.S.; Haase, A.A.; Peters, B.J.; Croslow, S.W.; McLaughlin, K.G.; Werst, K.R.; Goach, A.L.; Crick, D.C.; et al. Interactions of Truncated Menaquinones in Lipid Monolayers and Bilayers. Int. J. Mol. Sci. 2021, 22, 9755. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189755
Van Cleave C, Koehn JT, Pereira CS, Haase AA, Peters BJ, Croslow SW, McLaughlin KG, Werst KR, Goach AL, Crick DC, et al. Interactions of Truncated Menaquinones in Lipid Monolayers and Bilayers. International Journal of Molecular Sciences. 2021; 22(18):9755. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189755
Chicago/Turabian StyleVan Cleave, Cameron, Jordan T. Koehn, Caroline Simões Pereira, Allison A. Haase, Benjamin J. Peters, Seth W. Croslow, Kyle G. McLaughlin, Katarina R. Werst, Audra L. Goach, Dean C. Crick, and et al. 2021. "Interactions of Truncated Menaquinones in Lipid Monolayers and Bilayers" International Journal of Molecular Sciences 22, no. 18: 9755. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189755