Possible Roles of Interleukin-4 and -13 and Their Receptors in Gastric and Colon Cancer
Abstract
:1. Introduction
2. Methods
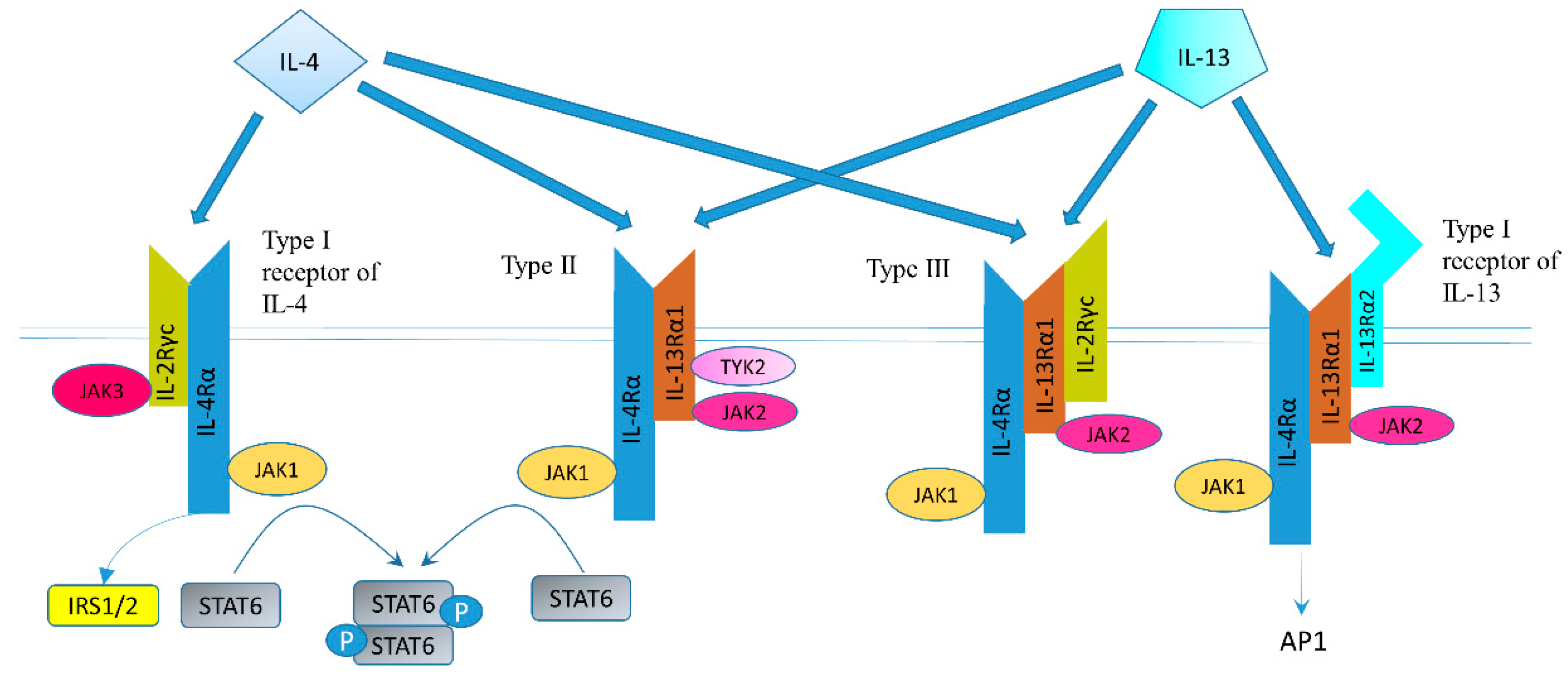
3. Summary of the IL-4/-13 Signaling Pathway
4. IL-13/IL-13R in Gastric Cancer
5. IL-4/IL-4R in Gastric Cancer
6. IL-13/IL-13R in Colon and Rectal Cancer
7. IL-4/IL-4R in Colon Cancer
8. SNPs in IL-4/13 and Their Receptors in Gastric and Colon Cancer
9. Discussion and Outlook
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GC | Gastric cancer |
| CRC | Colon and rectal cancer |
| IL-4 | Interleukin-4 |
| IL-13 | Interleukin-13 |
| SNPs | Single nucleotide polymorphisms |
| NKT | Natural killer T |
| IL-4R | Interleukin-4 receptors |
| IL-13R | Interleukin-13 receptors |
| Γc | IL-2Rγ-common chain |
| CHI3L1 | Chitinase-3-like protein 1 |
| STAT | Signal transducer and activator of transcription |
| CC | colon cancer |
| PI3K | Phosphatidylinositol 3-kinase |
| PTEN | phosphatase and tensin homolog |
| JAK | Janus kinase |
| FAM120A | Family with sequence similarity 120A |
| EMT | Epithelial–mesenchymal transition |
| 11βHSD2 | 11β-hydroxysteroid dehydrogenase type 2 |
| ADCC | Antibody-dependent cellular cytotoxicity |
| PBMCs | Peripheral blood mononuclear cells |
| GBM | Glioblastoma multiforme |
| PE | Pseudomonas exotoxin |
| TME | Tumor microenvironment |
| Th2 | T helper type 2 |
References
- Ilson, D.H. Advances in the treatment of gastric cancer. Curr. Opin. Gastroenterol. 2018, 34, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selim, J.H.; Shaheen, S.; Sheu, W.C.; Hsueh, C.T. Targeted and novel therapy in advanced gastric cancer. Exp. Hematol. Oncol. 2019, 8, 25. [Google Scholar] [CrossRef]
- Patel, T.H.; Cecchini, M. Targeted Therapies in Advanced Gastric Cancer. Curr. Treat. Options Oncol. 2020, 21, 70. [Google Scholar] [CrossRef]
- Tolba, M.F. Revolutionizing the landscape of colorectal cancer treatment: The potential role of immune checkpoint inhibitors. Int. J. Cancer 2020, 147, 2996–3006. [Google Scholar] [CrossRef]
- Vincent, A.; Ouelkdite-Oumouchal, A.; Souidi, M.; Leclerc, J.; Neve, B.; Van Seuningen, I. Colon cancer stemness as a reversible epigenetic state: Implications for anticancer therapies. World J. Stem Cells 2019, 11, 920–936. [Google Scholar] [CrossRef]
- Bess, S.N.; Greening, G.J.; Muldoon, T.J. Efficacy and clinical monitoring strategies for immune checkpoInt. inhibitors and targeted cytokine immunotherapy for locally advanced and metastatic colorectal cancer. Cytokine Growth Factor Rev. 2019, 49, 1–9. [Google Scholar] [CrossRef]
- Traub, B.; Sun, L.; Ma, Y.; Xu, P.; Lemke, J.; Paschke, S.; Henne-Bruns, D.; Knippschild, U.; Kornmann, M. Endogenously Expressed IL-4Ralpha Promotes the Malignant Phenotype of Human Pancreatic Cancer In Vitro and In Vivo. Int. J. Mol. Sci. 2017, 18, 716. [Google Scholar] [CrossRef]
- Bartolome, R.A.; Garcia-Palmero, I.; Torres, S.; Lopez-Lucendo, M.; Balyasnikova, I.V.; Casal, J.I. IL13 Receptor alpha2 Signaling Requires a Scaffold Protein, FAM120A, to Activate the FAK and PI3K Pathways in Colon Cancer Metastasis. Cancer Res. 2015, 75, 2434–2444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Antony, S.; Roy, K.; Juhasz, A.; Wu, Y.; Lu, J.; Meitzler, J.L.; Jiang, G.; Polley, E.; Doroshow, J.H. Interleukin-4 and interleukin-13 increase NADPH oxidase 1-related proliferation of human colon cancer cells. Oncotarget 2017, 8, 38113–38135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barderas, R.; Bartolome, R.A.; Fernandez-Acenero, M.J.; Torres, S.; Casal, J.I. High expression of IL-13 receptor alpha2 in colorectal cancer is associated with invasion, liver metastasis, and poor prognosis. Cancer Res. 2012, 72, 2780–2790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todaro, M.; Alea, M.P.; Di Stefano, A.B.; Cammareri, P.; Vermeulen, L.; Iovino, F.; Tripodo, C.; Russo, A.; Gulotta, G.; Medema, J.P.; et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 2007, 1, 389–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.; Liu, H.; Zhang, H.; He, H.; Li, H.; Shen, Z.; Qin, J.; Qin, X.; Xu, J.; Sun, Y. Interleukin-13 receptor alpha2 is associated with poor prognosis in patients with gastric cancer after gastrectomy. Oncotarget 2016, 7, 49281–49288. [Google Scholar] [CrossRef] [Green Version]
- Ostrand-Rosenberg, S. Immune surveillance: A balance between protumor and antitumor immunity. Curr. Opin. Genet. Dev. 2008, 18, 11–18. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.W.; Joyce, J.A. Alternative activation of tumor-associated macrophages by IL-4: Priming for protumoral functions. Cell Cycle 2010, 9, 4824–4835. [Google Scholar] [CrossRef] [Green Version]
- Sinha, P.; Clements, V.K.; Ostrand-Rosenberg, S. Interleukin-13-regulated M2 macrophages in combination with myeloid suppressor cells block immune surveillance against metastasis. Cancer Res. 2005, 65, 11743–11751. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.A.; Hural, J. Functions of IL-4 and Control of Its Expression. Crit. Rev. Immunol. 2017, 37, 181–212. [Google Scholar] [CrossRef]
- Minty, A.; Chalon, P.; Derocq, J.M.; Dumont, X.; Guillemot, J.C.; Kaghad, M.; Labit, C.; Leplatois, P.; Liauzun, P.; Miloux, B.; et al. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature 1993, 362, 248–250. [Google Scholar] [CrossRef]
- Gandhi, N.A.; Pirozzi, G.; Graham, N.M.H. Commonality of the IL-4/IL-13 pathway in atopic diseases. Expert Rev. Clin. Immunol. 2017, 13, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Junttila, I.S. Tuning the Cytokine Responses: An Update on Interleukin (IL)-4 and IL-13 Receptor Complexes. Front. Immunol. 2018, 9, 888. [Google Scholar] [CrossRef] [PubMed]
- Luzina, I.G.; Keegan, A.D.; Heller, N.M.; Rook, G.A.; Shea-Donohue, T.; Atamas, S.P. Regulation of inflammation by interleukin-4: A review of “alternatives”. J. Leukoc. Biol. 2012, 92, 753–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, C.K.; Geba, G.P.; Molfino, N. Investigational therapeutics targeting the IL-4/IL-13/STAT-6 pathway for the treatment of asthma. Eur. Respir. Rev. 2010, 19, 46–54. [Google Scholar] [CrossRef] [PubMed]
- LaPorte, S.L.; Juo, Z.S.; Vaclavikova, J.; Colf, L.A.; Qi, X.; Heller, N.M.; Keegan, A.D.; Garcia, K.C. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 2008, 132, 259–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ul-Haq, Z.; Naz, S.; Mesaik, M.A. Interleukin-4 receptor signaling and its binding mechanism: A therapeutic insight from inhibitors tool box. Cytokine Growth Factor Rev. 2016, 32, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Leland, P.; Joshi, B.H.; Puri, R.K. Targeting of IL-4 and IL-13 receptors for cancer therapy. Cytokine 2015, 75, 79–88. [Google Scholar] [CrossRef]
- Aman, M.J.; Tayebi, N.; Obiri, N.I.; Puri, R.K.; Modi, W.S.; Leonard, W.J. cDNA cloning and characterization of the human interleukin 13 receptor alpha chain. J. Biol. Chem. 1996, 271, 29265–29270. [Google Scholar] [CrossRef] [Green Version]
- Wills-Karp, M.; Finkelman, F.D. Untangling the complex web of IL-4- and IL-13-mediated signaling pathways. Sci. Signal. 2008, 1, pe55. [Google Scholar] [CrossRef] [Green Version]
- He, C.H.; Lee, C.G.; Dela Cruz, C.S.; Lee, C.M.; Zhou, Y.; Ahangari, F.; Ma, B.; Herzog, E.L.; Rosenberg, S.A.; Li, Y.; et al. Chitinase 3-like 1 regulates cellular and tissue responses via IL-13 receptor alpha2. Cell Rep. 2013, 4, 830–841. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.M.; He, C.H.; Nour, A.M.; Zhou, Y.; Ma, B.; Park, J.W.; Kim, K.H.; Dela Cruz, C.; Sharma, L.; Nasr, M.L.; et al. IL-13Ralpha2 uses TMEM219 in chitinase 3-like-1-induced signalling and effector responses. Nat. Commun. 2016, 7, 12752. [Google Scholar] [CrossRef] [PubMed]
- Geng, B.; Pan, J.; Zhao, T.; Ji, J.; Zhang, C.; Che, Y.; Yang, J.; Shi, H.; Li, J.; Zhou, H.; et al. Chitinase 3-like 1-CD44 interaction promotes metastasis and epithelial-to-mesenchymal transition through beta-catenin/Erk/Akt signaling in gastric cancer. J. Exp. Clin. Cancer Res. 2018, 37, 208. [Google Scholar] [CrossRef] [PubMed]
- Fichtner-Feigl, S.; Strober, W.; Kawakami, K.; Puri, R.K.; Kitani, A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat. Med. 2006, 12, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Wood, N.; Whitters, M.J.; Jacobson, B.A.; Witek, J.; Sypek, J.P.; Kasaian, M.; Eppihimer, M.J.; Unger, M.; Tanaka, T.; Goldman, S.J.; et al. Enhanced interleukin (IL)-13 responses in mice lacking IL-13 receptor alpha 2. J. Exp. Med. 2003, 197, 703–709. [Google Scholar] [CrossRef]
- Rahaman, S.O.; Sharma, P.; Harbor, P.C.; Aman, M.J.; Vogelbaum, M.A.; Haque, S.J. IL-13R(alpha)2, a decoy receptor for IL-13 acts as an inhibitor of IL-4-dependent signal transduction in glioblastoma cells. Cancer Res. 2002, 62, 1103–1109. [Google Scholar]
- Fujisawa, T.; Joshi, B.; Nakajima, A.; Puri, R.K. A novel role of interleukin-13 receptor alpha2 in pancreatic cancer invasion and metastasis. Cancer Res. 2009, 69, 8678–8685. [Google Scholar] [CrossRef] [Green Version]
- Murata, T.; Obiri, N.I.; Debinski, W.; Puri, R.K. Structure of IL-13 receptor: Analysis of subunit composition in cancer and immune cells. BioChem. Biophys Res. Commun. 1997, 238, 90–94. [Google Scholar] [CrossRef]
- Kornmann, M.; Kleeff, J.; Debinski, W.; Korc, M. Pancreatic cancer cells express interleukin-13 and -4 receptors, and their growth is inhibited by Pseudomonas exotoxin coupled to interleukin-13 and -4. Anticancer Res. 1999, 19, 125–131. [Google Scholar]
- Debinski, W.; Obiri, N.I.; Pastan, I.; Puri, R.K. A novel chimeric protein composed of interleukin 13 and Pseudomonas exotoxin is highly cytotoxic to human carcinoma cells expressing receptors for interleukin 13 and interleukin 4. J. Biol. Chem. 1995, 270, 16775–16780. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhang, S.; Wang, Q.; Zhang, X. Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J. Hematol. Oncol. 2017, 10, 36. [Google Scholar] [CrossRef] [Green Version]
- Gabitass, R.F.; Annels, N.E.; Stocken, D.D.; Pandha, H.A.; Middleton, G.W. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol. Immunother. 2011, 60, 1419–1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essner, R.; Huynh, Y.; Nguyen, T.; Rose, M.; Kojima, M.; Hoon, D.S. Functional interleukin-4 receptor and interleukin-2 receptor common gamma chain in human gastric carcinoma: A possible mechanism for cytokine-based therapy. J. Gastrointest. Surg. 2001, 5, 81–90. [Google Scholar] [CrossRef]
- Morisaki, T.; Uchiyama, A.; Yuzuki, D.; Essner, R.; Morton, D.L.; Hoon, D.S. Interleukin 4 regulates G1 cell cycle progression in gastric carcinoma cells. Cancer Res. 1994, 54, 1113–1118. [Google Scholar] [PubMed]
- Morisaki, T.; Yuzuki, D.H.; Lin, R.T.; Foshag, L.J.; Morton, D.L.; Hoon, D.S. Interleukin 4 receptor expression and growth inhibition of gastric carcinoma cells by interleukin 4. Cancer Res. 1992, 52, 6059–6065. [Google Scholar] [PubMed]
- Cardenas, D.M.; Sanchez, A.C.; Rosas, D.A.; Rivero, E.; Paparoni, M.D.; Cruz, M.A.; Suarez, Y.P.; Galvis, N.F. Preliminary analysis of single-nucleotide polymorphisms in IL-10, IL-4, and IL-4Ralpha genes and profile of circulating cytokines in patients with gastric Cancer. BMC Gastroenterol. 2018, 18, 184. [Google Scholar] [CrossRef] [Green Version]
- Diaz Orea, M.A.; Munoz Perez, V.; Gomez Conde, E.; Castellanos Sanchez, V.O.; Gonzalez Lopez, R.; Flores Alonso, J.C.; Cardenas, M.E.; Galicia, A.L.; Mendoza, A. Expression of Cytokines Interleukin-2, Interleukin-4, Interleukin-10 and Transforming Growth Factor beta in Gastric Adenocarcinoma Biopsies Obtained from Mexican Patients. Asian Pac. J. Cancer Prev. 2017, 18, 577–582. [Google Scholar]
- Formentini, A.; Braun, P.; Fricke, H.; Link, K.H.; Henne-Bruns, D.; Kornmann, M. Expression of interleukin-4 and interleukin-13 and their receptors in colorectal cancer. Int. J. Colorectal Dis. 2012, 27, 1369–1376. [Google Scholar] [CrossRef]
- Iwashita, J.; Sato, Y.; Sugaya, H.; Takahashi, N.; Sasaki, H.; Abe, T. mRNA of MUC2 is stimulated by IL-4, IL-13 or TNF-alpha through a mitogen-activated protein kinase pathway in human colon cancer cells. Immunol. Cell Biol. 2003, 81, 275–282. [Google Scholar] [CrossRef]
- Petiot, A.; Ogier-Denis, E.; Blommaart, E.F.; Meijer, A.J.; Codogno, P. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000, 275, 992–998. [Google Scholar] [CrossRef] [Green Version]
- Arico, S.; Petiot, A.; Bauvy, C.; Dubbelhuis, P.F.; Meijer, A.J.; Codogno, P.; Ogier-Denis, E. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J. Biol. Chem. 2001, 276, 35243–35246. [Google Scholar] [CrossRef] [Green Version]
- Murata, T.; Noguchi, P.D.; Puri, R.K. IL-13 induces phosphorylation and activation of JAK2 Janus kinase in human colon carcinoma cell lines: Similarities between IL-4 and IL-13 signaling. J. Immunol. 1996, 156, 2972–2978. [Google Scholar] [PubMed]
- Trejdosiewicz, L.K.; Morton, R.; Yang, Y.; Banks, R.E.; Selby, P.J.; Southgate, J. Interleukins 4 and 13 upregulate expression of cd44 in human colonic epithelial cell lines. Cytokine 1998, 10, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, K.; Walker, S.M.; Jessup, J.M.; Fidler, I.J. In vivo selection of highly metastatic cells from surgical specimens of different primary human colon carcinomas implanted into nude mice. Cancer Res. 1988, 48, 1943–1948. [Google Scholar]
- Xu, K.; Tao, W.; Su, Z. Propofol prevents IL-13-induced epithelial-mesenchymal transition in human colorectal cancer cells. Cell Biol. Int. 2018, 42, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Zhang, J.; Liu, H.; Wan, L.; Zhang, H.; Huang, Q.; Xu, E.; Lai, M. IL-13/STAT6 signaling plays a critical role in the epithelial-mesenchymal transition of colorectal cancer cells. Oncotarget 2016, 7, 61183–61198. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Cheng, Q.; Zhang, B.; Zhang, M. IL-13 induces the expression of 11betaHSD2 in IL-13Ralpha2 dependent manner and promotes the malignancy of colorectal cancer. Am. J. Transl. Res. 2016, 8, 1064–1072. [Google Scholar]
- Bartolome, R.A.; Martin-Regalado, A.; Jaen, M.; Zannikou, M.; Zhang, P.; de Los Rios, V.; Balyasnikova, I.V.; Casal, J.I. Protein Tyrosine Phosphatase-1B Inhibition Disrupts IL13Ralpha2-Promoted Invasion and Metastasis in Cancer Cells. Cancers 2020, 12, 500. [Google Scholar] [CrossRef] [Green Version]
- Cheadle, E.J.; Riyad, K.; Subar, D.; Rothwell, D.G.; Ashton, G.; Batha, H.; Sherlock, D.J.; Hawkins, R.E.; Gilham, D.E. Eotaxin-2 and colorectal cancer: A potential target for immune therapy. Clin. Cancer Res. 2007, 13, 5719–5728. [Google Scholar] [CrossRef] [Green Version]
- Matsui, S.; Okabayashi, K.; Tsuruta, M.; Shigeta, K.; Seishima, R.; Ishida, T.; Kondo, T.; Suzuki, Y.; Hasegawa, H.; Shimoda, M.; et al. Interleukin-13 and its signaling pathway is associated with obesity-related colorectal tumorigenesis. Cancer Sci. 2019, 110, 2156–2165. [Google Scholar] [CrossRef]
- Park, J.M.; Terabe, M.; van den Broeke, L.T.; Donaldson, D.D.; Berzofsky, J.A. Unmasking immunosurveillance against a syngeneic colon cancer by elimination of CD4+ NKT regulatory cells and IL-13. Int. J. Cancer 2005, 114, 80–87. [Google Scholar] [CrossRef]
- Bartolomé, R.A.; Jaén, M.; Casal, J.I. An IL13Rα2 peptide exhibits therapeutic activity against metastatic colorectal cancer. Br. J. Cancer 2018, 119, 940–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingram, N.; Northwood, E.L.; Perry, S.L.; Marston, G.; Snowden, H.; Taylor, J.C.; Scott, N.; Bishop, D.T.; Coletta, P.L.; Hull, M.A. Reduced type II interleukin-4 receptor signalling drives initiation, but not progression, of colorectal carcinogenesis: Evidence from transgenic mouse models and human case-control epidemiological observations. Carcinogenesis 2013, 34, 2341–2349. [Google Scholar] [CrossRef] [PubMed]
- Saigusa, S.; Tanaka, K.; Inoue, Y.; Toiyama, Y.; Okugawa, Y.; Iwata, T.; Mohri, Y.; Kusunoki, M. Low serum interleukin-13 levels correlate with poorer prognoses for colorectal cancer patients. Int. Surg. 2014, 99, 223–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.P.; Wang, Y.Y.; Pan, J.; Cen, R.; Cai, Y.K. Evaluation of specific fecal protein biochips for the diagnosis of colorectal cancer. World J. Gastroenterol. 2014, 20, 1332–1339. [Google Scholar] [CrossRef] [PubMed]
- Lahm, H.; Schnyder, B.; Wyniger, J.; Borbenyi, Z.; Yilmaz, A.; Car, B.D.; Fischer, J.R.; Givel, J.C.; Ryffel, B. Growth inhibition of human colorectal-carcinoma cells by interleukin-4 and expression of functional interleukin-4 receptors. Int. J. Cancer 1994, 59, 440–447. [Google Scholar] [CrossRef]
- Zhou, Y.; Ji, Y.; Wang, H.; Zhang, H.; Zhou, H. IL-33 Promotes the Development of Colorectal Cancer Through Inducing Tumor-Infiltrating ST2L(+) Regulatory T Cells in Mice. Technol. Cancer Res. Treat. 2018, 17, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaklamanis, L.; Gatter, K.C.; Mortensen, N.; Harris, A.L. Interleukin-4 receptor and epidermal growth factor receptor expression in colorectal cancer. Br. J. Cancer 1992, 66, 712–716. [Google Scholar] [CrossRef] [Green Version]
- Koller, F.L.; Hwang, D.G.; Dozier, E.A.; Fingleton, B. Epithelial interleukin-4 receptor expression promotes colon tumor growth. Carcinogenesis 2010, 31, 1010–1017. [Google Scholar] [CrossRef]
- Chang, T.L.; Peng, X.; Fu, X.Y. Interleukin-4 mediates cell growth inhibition through activation of Stat1. J. Biol. Chem. 2000, 275, 10212–10217. [Google Scholar] [CrossRef] [Green Version]
- Toi, M.; Bicknell, R.; Harris, A.L. Inhibition of colon and breast carcinoma cell growth by interleukin-4. Cancer Res. 1992, 52, 275–279. [Google Scholar]
- Topp, M.S.; Papadimitriou, C.A.; Eitelbach, F.; Koenigsmann, M.; Oelmann, E.; Koehler, B.; Oberberg, D.; Reufi, B.; Stein, H.; Thiel, E.; et al. Recombinant human interleukin 4 has antiproliferative activity on human tumor cell lines derived from epithelial and nonepithelial histologies. Cancer Res. 1995, 55, 2173–2176. [Google Scholar] [PubMed]
- Al-Tubuly, A.A.; Spijker, R.; Pignatelli, M.; Kirkland, S.C.; Ritter, M.A. Inhibition of growth and enhancement of differentiation of colorectal carcinoma cell lines by MAb MR6 and IL-4. Int. J. Cancer 1997, 71, 605–611. [Google Scholar] [CrossRef]
- Schnyder, B.; Lahm, H.; Pittet, M.; Schnyder-Candrian, S. Mono- or dual-phosphorylation of akt kinase is regulated by distinct receptors that involve the common insulin receptor substrate. J. Recept. Signal Transduct. Res. 2002, 22, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gong, C.; Mao, H.; Li, Z.; Fang, Z.; Chen, Q.; Lin, M.; Jiang, X.; Hu, Y.; Wang, W.; et al. E2F1/SP3/STAT6 axis is required for IL-4-induced epithelial-mesenchymal transition of colorectal cancer cells. Int. J. Oncol. 2018, 53, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Flieger, D.; Spengler, U.; Beier, I.; Sauerbruch, T.; Schmidt-Wolf, I. Combinations of the cytokines IL-12, IL-2 and IFN-alpha significantly augment whereas the cytokine IL-4 suppresses the cytokine-induced antibody-dependent cellular cytotoxicity of monoclonal antibodies 17-1A and BR55-2. Cytokine 2000, 12, 756–761. [Google Scholar] [CrossRef] [PubMed]
- Wersall, P.; Masucci, G.; Mellstedt, H. Interleukin-4 augments the cytotoxic capacity of lymphocytes and monocytes in antibody-dependent cellular cytotoxicity. Cancer Immunol. Immunother. 1991, 33, 45–49. [Google Scholar] [CrossRef]
- Nagler, A.; Lanier, L.L.; Phillips, J.H. The effects of IL-4 on human natural killer cells. A potent regulator of IL-2 activation and proliferation. J. Immunol. 1988, 141, 2349–2351. [Google Scholar]
- Qi, C.F.; Nieroda, C.; De Filippi, R.; Greiner, J.W.; Correale, P.; Schlom, J.; Tsang, K.Y. Macrophage colony-stimulating factor enhancement of antibody-dependent cellular cytotoxicity against human colon carcinoma cells. Immunol. Lett. 1995, 47, 15–24. [Google Scholar] [CrossRef]
- Flieger, D.; Spengler, U.; Beier, I.; Kleinschmidt, R.; Hoff, A.; Varvenne, M.; Sauerbruch, T.; Schmidt-Wolf, I. Enhancement of antibody dependent cellular cytotoxicity (ADCC) by combination of cytokines. Hybridoma 1999, 18, 63–68. [Google Scholar] [CrossRef]
- Voboril, R.; Weberova-Voborilova, J. Sensitization of colorectal cancer cells to irradiation by IL-4 and IL-10 is associated with inhibition of NF-kappaB. Neoplasma 2007, 54, 495–502. [Google Scholar]
- Flieger, D.; Hoff, A.S.; Sauerbruch, T.; Schmidt-Wolf, I.G. Influence of cytokines, monoclonal antibodies and chemotherapeutic drugs on epithelial cell adhesion molecule (EpCAM) and LewisY antigen expression. Clin. Exp. Immunol. 2001, 123, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Lahm, H.; Amstad, P.; Yilmaz, A.; Borbenyi, Z.; Wyniger, J.; Fischer, J.R.; Suardet, L.; Givel, J.C.; Odartchenko, N. Interleukin 4 down-regulates expression of c-kit and autocrine stem cell factor in human colorectal carcinoma cells. Cell Growth Differ. 1995, 6, 1111–1118. [Google Scholar] [PubMed]
- Wigmore, S.J.; Maingay, J.P.; Fearon, K.C.; Ross, J.A. Endogenous production of IL-8 by human colorectal cancer cells and its regulation by cytokines. Int. J. Oncol. 2001, 18, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Kanai, T.; Watanabe, M.; Hayashi, A.; Nakazawa, A.; Yajima, T.; Okazawa, A.; Yamazaki, M.; Ishii, H.; Hibi, T. Regulatory effect of interleukin-4 and interleukin-13 on colon cancer cell adhesion. Br. J. Cancer 2000, 82, 1717–1723. [Google Scholar]
- Uchiyama, A.; Essner, R.; Doi, F.; Nguyen, T.; Ramming, K.P.; Nakamura, T.; Morton, D.L.; Hoon, D.S. Interleukin 4 inhibits hepatocyte growth factor-induced invasion and migration of colon carcinomas. J. Cell BioChem. 1996, 62, 443–453. [Google Scholar] [CrossRef]
- Murata, T.; Noguchi, P.D.; Puri, R.K. Receptors for interleukin (IL)-4 do not associate with the common gamma chain, and IL-4 induces the phosphorylation of JAK2 tyrosine kinase in human colon carcinoma cells. J. Biol. Chem. 1995, 270, 30829–30836. [Google Scholar] [CrossRef] [Green Version]
- Herzberg, F.; Schoning, M.; Schirner, M.; Topp, M.; Thiel, E.; Kreuser, E.D. IL-4 and TNF-alpha induce changes in integrin expression and adhesive properties and decrease the lung-colonizing potential of HT-29 colon carcinoma cells. Clin. Exp. Metastasis 1996, 14, 165–175. [Google Scholar] [CrossRef]
- Gharib, A.F.; Shalaby, S.M.; Raafat, N.; Fawzy, W.M.S.; Abdel Hakim, N.H. Assessment of neutralizing interleukin-4 effect on CD133 gene expression in colon cancer cell line. Cytokine 2017, 97, 66–72. [Google Scholar] [CrossRef]
- Li, B.H.; Yang, X.Z.; Li, P.D.; Yuan, Q.; Liu, X.H.; Yuan, J.; Zhang, W.J. IL-4/Stat6 activities correlate with apoptosis and metastasis in colon cancer cells. BioChem. Biophys. Res. Commun. 2008, 369, 554–560. [Google Scholar] [CrossRef]
- Costamagna, D.; Duelen, R.; Penna, F.; Neumann, D.; Costelli, P.; Sampaolesi, M. Interleukin-4 administration improves muscle function, adult myogenesis, and lifespan of colon carcinoma-bearing mice. J. Cachexia Sarcopenia Muscle 2020. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.L.; Wang, N.; Wei, X.; Xie, G.F.; Li, J.J.; Liang, H.J. Interleukin-12 inhibits the survival of human colon cancer stem cells in vitro and their tumor initiating capacity in mice. Cancer Lett. 2012, 322, 92–97. [Google Scholar] [CrossRef]
- Kajiwara, A.; Doi, H.; Eguchi, J.; Ishii, S.; Hiraide-Sasagawa, A.; Sakaki, M.; Omori, R.; Hiroishi, K.; Imawari, M. Interleukin-4 and CpG oligonucleotide therapy suppresses the outgrowth of tumors by activating tumor-specific Th1-type immune responses. Oncol. Rep. 2012, 27, 1765–1771. [Google Scholar] [PubMed] [Green Version]
- Liu, Y.J.; Dou, X.Q.; Wang, F.; Zhang, J.; Wang, X.L.; Xu, G.L.; Xiang, S.S.; Gao, X.; Fu, J.; Song, H.F. IL-4Ralpha aptamer-liposome-CpG oligodeoxynucleotides suppress tumour growth by targeting the tumour microenvironment. J. Drug Target. 2017, 25, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Itsuki, Y.; Suzuki, S. Expression of interleukin-4 in colon 26 cells induces both eosinophil mediated local tumor killing and T-cell mediated systemic immunity in vivo. Nihon Ika Daigaku Zasshi 1996, 63, 275–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eguchi, J.; Hiroishi, K.; Ishii, S.; Baba, T.; Matsumura, T.; Hiraide, A.; Okada, H.; Imawari, M. Interleukin-4 gene transduced tumor cells promote a potent tumor-specific Th1-type response in cooperation with interferon-alpha transduction. Gene Ther. 2005, 12, 733–741. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Okamura, S.; Yamaji, T.; Iwasaki, M.; Tsugane, S.; Shetty, V.; Koizumi, T. Plasma cytokine levels and the presence of colorectal cancer. PLoS ONE 2019, 14, e0213602. [Google Scholar] [CrossRef] [Green Version]
- Berghella, A.M.; Pellegrini, P.; Del Beato, T.; Maccarone, D.; Adorno, D.; Casciani, C.U. Prognostic significance of immunological evaluation in colorectal cancer. Cancer BioTher. Radiopharm. 1996, 11, 355–361. [Google Scholar] [CrossRef]
- Berghella, A.M.; Contasta, I.; Pellegrini, P.; Del Beato, T.; Adorno, D. Peripheral blood immunological parameters for use as markers of pre-invasive to invasive colorectal cancer. Cancer BioTher. Radiopharm. 2002, 17, 43–50. [Google Scholar] [CrossRef]
- Zaloudik, J.; Lauerova, L.; Janakova, L.; Talac, R.; Simickova, M.; Nekulova, M.; Mikulikova, I.; Kovarik, J.; Sheard, M. Significance of pre-treatment immunological parameters in colorectal cancer patients with unresectable metastases to the liver. Hepatogastroenterology 1999, 46, 220–227. [Google Scholar]
- Pellegrini, P.; Berghella, A.M.; Del Beato, T.; Maccarone, D.; Cencioni, S.; Adorno, D.; Casciani, C.U. The sCEA molecule suppressive role in NK and TH1 cell functions in colorectal cancer. Cancer BioTher. Radiopharm. 1997, 12, 257–264. [Google Scholar] [CrossRef]
- Kim, Y.W.; Kim, S.K.; Kim, C.S.; Kim, I.Y.; Cho, M.Y.; Kim, N.K. Association of serum and intratumoral cytokine profiles with tumor stage and neutrophil lymphocyte ratio in colorectal cancer. Anticancer Res. 2014, 34, 3481–3487. [Google Scholar]
- Di Stefano, A.B.; Iovino, F.; Lombardo, Y.; Eterno, V.; Hoger, T.; Dieli, F.; Stassi, G.; Todaro, M. Survivin is regulated by interleukin-4 in colon cancer stem cells. J. Cell Physiol. 2010, 225, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Todaro, M.; Lombardo, Y.; Francipane, M.G.; Alea, M.P.; Cammareri, P.; Iovino, F.; Di Stefano, A.B.; Di Bernardo, C.; Agrusa, A.; Condorelli, G.; et al. Apoptosis resistance in epithelial tumors is mediated by tumor-cell-derived interleukin-4. Cell Death Differ. 2008, 15, 762–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.T.; Sohn, I.; Do, I.G.; Jang, J.; Kim, S.H.; Jung, I.H.; Park, J.O.; Park, Y.S.; Talasaz, A.; Lee, J.; et al. Transcriptome analysis of CD133-positive stem cells and prognostic value of survivin in colorectal cancer. Cancer Genom. Proteom. 2014, 11, 259–266. [Google Scholar] [CrossRef]
- Volonte, A.; Di Tomaso, T.; Spinelli, M.; Todaro, M.; Sanvito, F.; Albarello, L.; Bissolati, M.; Ghirardelli, L.; Orsenigo, E.; Ferrone, S.; et al. Cancer-initiating cells from colorectal cancer patients escape from T cell-mediated immunosurveillance in vitro through membrane-bound IL-4. J. Immunol. 2014, 192, 523–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barth, R.J., Jr.; Camp, B.J.; Martuscello, T.A.; Dain, B.J.; Memoli, V.A. The cytokine microenvironment of human colon carcinoma. Lymphocyte expression of tumor necrosis factor-alpha and interleukin-4 predicts improved survival. Cancer 1996, 78, 1168–1178. [Google Scholar] [CrossRef]
- Correale, P.; Botta, C.; Staropoli, N.; Nardone, V.; Pastina, P.; Ulivieri, C.; Gandolfo, C.; Baldari, T.C.; Lazzi, S.; Ciliberto, D.; et al. Systemic inflammatory status predict the outcome of k-RAS WT metastatic colorectal cancer patients receiving the thymidylate synthase poly-epitope-peptide anticancer vaccine. Oncotarget 2018, 9, 20539–20554. [Google Scholar] [CrossRef]
- Evans, C.F.; Galustian, C.; Bodman-Smith, M.; Dalgleish, A.G.; Kumar, D. The effect of colorectal cancer upon host peripheral immune cell function. Colorectal Dis. Off. J. Assoc. Coloproctol. Great Br. Irel. 2010, 12, 561–569. [Google Scholar] [CrossRef]
- Purasiri, P.; Murray, A.; Richardson, S.; Heys, S.D.; Horrobin, D.; Eremin, O. Modulation of cytokine production in vivo by dietary essential fatty acids in patients with colorectal cancer. Clin. Sci. 1994, 87, 711–717. [Google Scholar] [CrossRef]
- Risch, N.J. Searching for genetic determinants in the new millennium. Nature 2000, 405, 847–856. [Google Scholar] [CrossRef]
- Botstein, D.; Risch, N. Discovering genotypes underlying human phenotypes: Past successes for mendelian disease, future approaches for complex disease. Nat. Genet. 2003, 33, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Cascorbi, I.; Henning, S.; Brockmoller, J.; Gephart, J.; Meisel, C.; Muller, J.M.; Loddenkemper, R.; Roots, I. Substantially reduced risk of cancer of the aerodigestive tract in subjects with variant--463A of the myeloperoxidase gene. Cancer Res. 2000, 60, 644–649. [Google Scholar] [PubMed]
- Piedrafita, F.J.; Molander, R.B.; Vansant, G.; Orlova, E.A.; Pfahl, M.; Reynolds, W.F. An Alu element in the myeloperoxidase promoter contains a composite SP1-thyroid hormone-retinoic acid response element. J. Biol. Chem. 1996, 271, 14412–14420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, L.J.; Ratner, E.; Leng, S.; Zhai, R.; Nallur, S.; Babar, I.; Muller, R.U.; Straka, E.; Su, L.; Burki, E.A.; et al. A SNP in a let-7 microRNA complementary site in the KRAS 3’ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008, 68, 8535–8540. [Google Scholar] [CrossRef] [Green Version]
- Erichsen, H.C.; Chanock, S.J. SNPs in cancer research and treatment. Br. J. Cancer 2004, 90, 747–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Omar, E.M.; Rabkin, C.S.; Gammon, M.D.; Vaughan, T.L.; Risch, H.A.; Schoenberg, J.B.; Stanford, J.L.; Mayne, S.T.; Goedert, J.; Blot, W.J.; et al. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology 2003, 124, 1193–1201. [Google Scholar] [CrossRef]
- Wu, M.S.; Wu, C.Y.; Chen, C.J.; Lin, M.T.; Shun, C.T.; Lin, J.T. Interleukin-10 genotypes associate with the risk of gastric carcinoma in Taiwanese Chinese. Int. J. Cancer 2003, 104, 617–623. [Google Scholar] [CrossRef]
- Lai, K.C.; Chen, W.C.; Jeng, L.B.; Li, S.Y.; Chou, M.C.; Tsai, F.J. Association of genetic polymorphisms of MK, IL-4, p16, p21, p53 genes and human gastric cancer in Taiwan. Eur. J. Surg. Oncol. 2005, 31, 1135–1140. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, M.A.; Lanas, A.; Quintero, E.; Nicolas, D.; Parra-Blanco, A.; Strunk, M.; Benito, R.; Angel Simon, M.; Santolaria, S.; Sopena, F.; et al. Gastric cancer susceptibility is not linked to pro-and anti-inflammatory cytokine gene polymorphisms in whites: A Nationwide Multicenter Study in Spain. Am. J. Gastroenterol. 2007, 102, 1878–1892. [Google Scholar] [CrossRef]
- Crusius, J.B.; Canzian, F.; Capella, G.; Pena, A.S.; Pera, G.; Sala, N.; Agudo, A.; Rico, F.; Del Giudice, G.; Palli, D.; et al. Cytokine gene polymorphisms and the risk of adenocarcinoma of the stomach in the European prospective investigation into cancer and nutrition (EPIC-EURGAST). Ann. Oncol. 2008, 19, 1894–1902. [Google Scholar] [CrossRef]
- Zambon, C.F.; Basso, D.; Marchet, A.; Fasolo, M.; Stranges, A.; Schiavon, S.; Navaglia, F.; Greco, E.; Fogar, P.; Falda, A.; et al. IL-4 -588C>T polymorphism and IL-4 receptor alpha [Ex5+14A>G.; Ex11+828A>G] haplotype concur in selecting H. pylori cagA subtype infections. Clin. Chim. Acta 2008, 389, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Ishikawa, T.; Kato, H.; Yoshida, N.; Naito, Y.; Kokura, S.; Yagi, N.; Takagi, T.; Handa, O.; Kitawaki, J.; et al. Synergistic effect of HLA class II loci and cytokine gene polymorphisms on the risk of gastric cancer in Japanese patients with Helicobacter pylori infection. Int. J. Cancer 2009, 125, 2595–2602. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.P.; Park, S.K.; Cho, L.Y.; Gwack, J.; Yang, J.J.; Shin, A.; Kim, C.S.; Kim, Y.; Kang, D.; Chang, S.H.; et al. Soybean product intake modifies the association between interleukin-10 genetic polymorphisms and gastric cancer risk. J. Nutr. 2009, 139, 1008–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Lu, Y.; Ding, Y.B.; Ke, Q.; Hu, Z.B.; Yan, Z.G.; Xue, Y.; Zhou, Y.; Hua, Z.L.; Shu, Y.Q.; et al. Promoter polymorphisms of IL2, IL4, and risk of gastric cancer in a high-risk Chinese population. Mol. Carcinog. 2009, 48, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.F.; Wen, Y.; Loh, M.; Wen, Y.Y.; Yang, S.J.; Zhao, Z.M.; Tian, Z.; Huang, H.; Lan, H.; Chen, F.; et al. Interleukin-4 and -8 gene polymorphisms and risk of gastric cancer in a population in Southwestern China. Asian Pac. J. Cancer Prev. 2014, 15, 2951–2957. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.; Wang, X.; Wei, J.; Wang, L.; Shi, Y.; Zheng, L.; Tang, W.; Ding, G.; Liu, C.; Liu, R.; et al. Interleukin 12B rs3212227 T > G polymorphism was associated with an increased risk of gastric cardiac adenocarcinoma in a Chinese population. Dis. Esophagus 2015, 28, 291–298. [Google Scholar] [CrossRef]
- Martinez-Campos, C.; Torres-Poveda, K.; Camorlinga-Ponce, M.; Flores-Luna, L.; Maldonado-Bernal, C.; Madrid-Marina, V.; Torres, J. Polymorphisms in IL-10 and TGF-beta gene promoter are associated with lower risk to gastric cancer in a Mexican population. BMC Cancer 2019, 19, 453. [Google Scholar] [CrossRef]
- He, B.; Pan, B.; Pan, Y.; Sun, H.; Xu, T.; Qin, J.; Xu, X.; Wang, S. IL-4/IL-4R and IL-6/IL-6R genetic variations and gastric cancer risk in the Chinese population. Am. J. Transl. Res. 2019, 11, 3698–3706. [Google Scholar]
- Yun, Y.; Dong, W.; Chen, C.; Zhang, H.; Shi, N.; He, M.; Chen, X. Roles of IL-4 genetic polymorphisms and haplotypes in the risk of gastric cancer and their interaction with environmental factors. Int. J. Clin. Exp. Pathol. 2017, 10, 8936–8943. [Google Scholar]
- Burada, F.; Angelescu, C.; Mitrut, P.; Ciurea, T.; Cruce, M.; Saftoiu, A.; Ioana, M. Interleukin-4 receptor -3223T→C polymorphism is associated with increased gastric adenocarcinoma risk. Can. J. Gastroenterol. 2012, 26, 532–536. [Google Scholar] [CrossRef] [Green Version]
- Xia, H.Z.; Du, W.D.; Wu, Q.; Chen, G.; Zhou, Y.; Tang, X.F.; Tang, H.Y.; Liu, Y.; Yang, F.; Ruan, J.; et al. E-selectin rs5361 and FCGR2A rs1801274 variants were associated with increased risk of gastric cancer in a Chinese population. Mol. Carcinog. 2012, 51, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Bhayal, A.C.; Krishnaveni, D.; Rao, K.P.; Kumar, A.R.; Jyothy, A.; Nallari, P.; Venkateshwari, A. Significant Association of Interleukin4 Intron 3 VNTR Polymorphism with Susceptibility to Gastric Cancer in a South Indian Population from Telangana. PLoS ONE 2015, 10, e0138442. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, A.M.; Balseiro, S.C.; Silva, M.R.; Alarcao, A.; d’Aguiar, M.J.; Ferreira, T.; Carvalho, L. Association Between IL-4 and IL-6 Expression Variants and Gastric Cancer Among Portuguese Population. GE Port. J. Gastroenterol. 2015, 22, 143–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seno, H.; Satoh, K.; Tsuji, S.; Shiratsuchi, T.; Harada, Y.; Hamajima, N.; Sugano, K.; Kawano, S.; Chiba, T. Novel interleukin-4 and interleukin-1 receptor antagonist gene variations associated with non-cardia gastric cancer in Japan: Comprehensive analysis of 207 polymorphisms of 11 cytokine genes. J. Gastroenterol. Hepatol. 2007, 22, 729–737. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Wang, X.; Gao, F.; Yu, L.; Chen, X. Association between four SNPs in IL-4 and the risk of gastric cancer in a Chinese population. Int. J. Mol. Epidemiol. Genet. 2017, 8, 45–52. [Google Scholar]
- Pavithra, D.; Gautam, M.; Rama, R.; Swaminathan, R.; Gopal, G.; Ramakrishnan, A.S.; Rajkumar, T. TGFbeta C-509T, TGFbeta T869C, XRCC1 Arg194Trp, IKBalpha C642T, IL4 C-590T Genetic polymorphisms combined with socio-economic, lifestyle, diet factors and gastric cancer risk: A case control study in South Indian population. Cancer Epidemiol. 2018, 53, 21–26. [Google Scholar] [CrossRef]
- Cavalcante, G.C.; Amador, M.A.; Ribeiro Dos Santos, A.M.; Carvalho, D.C.; Andrade, R.B.; Pereira, E.E.; Fernandes, M.R.; Costa, D.F.; Santos, N.P.; Assumpcao, P.P.; et al. Analysis of 12 variants in the development of gastric and colorectal cancers. World J. Gastroenterol. 2017, 23, 8533–8543. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Park, Y.; Lee, J.; Choi, I.J.; Kim, Y.W.; Ryu, K.W.; Sung, J.; Kim, J. Effects of Soy Product Intake and Interleukin Genetic Polymorphisms on Early Gastric Cancer Risk in Korea: A Case-Control Study. Cancer Res. Treat. 2017, 49, 1044–1056. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.M.; Li, Z.X.; Tang, F.B.; Zhang, Y.; Zhou, T.; Zhang, L.; Ma, J.L.; You, W.C.; Pan, K.F. Association of genetic polymorphisms of interleukins with gastric cancer and precancerous gastric lesions in a high-risk Chinese population. Tumour Biol. 2016, 37, 2233–2242. [Google Scholar] [CrossRef]
- Schmidt, H.M.; Ha, D.M.; Taylor, E.F.; Kovach, Z.; Goh, K.L.; Fock, K.M.; Barrett, J.H.; Forman, D.; Mitchell, H. Variation in human genetic polymorphisms, their association with Helicobacter pylori acquisition and gastric cancer in a multi-ethnic country. J. Gastroenterol. Hepatol. 2011, 26, 1725–1732. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, M.A.; Nicolas-Perez, D.; Lanas, A.; Bujanda, L.; Carrera, P.; Benito, R.; Strunk, M.; Sopena, F.; Santolaria, S.; Piazuelo, E.; et al. Prognostic role of host cyclooxygenase and cytokine genotypes in a Caucasian cohort of patients with gastric adenocarcinoma. PLoS ONE 2012, 7, e46179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talebkhan, Y.; Doozbakhshan, M.; Saberi, S.; Esmaeili, M.; Karami, N.; Mohajerani, N.; Abdirad, A.; Eshagh Hosseini, M.; Nahvijou, A.; Mohagheghi, M.A.; et al. Serum Antibodies against Helicobacter pylori Neutrophil Activating Protein in Carriers of IL-4 C-590T Genetic Polymorphism Amplify the Risk of Gastritis and Gastric Cancer. Iran. Biomed. J. 2017, 21, 321–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walczak, A.; Przybylowska, K.; Trzcinski, R.; Sygut, A.; Dziki, L.; Dziki, A.; Majsterek, I. Association of -1112 c/t promoter region polymorphism of the interleukin 13 gene with occurrence of colorectal cancer. Pol. J. Surg. 2011, 83, 27–31. [Google Scholar] [CrossRef] [Green Version]
- Ibrahimi, M.; Moossavi, M.; Mojarad, E.N.; Musavi, M.; Mohammadoo-Khorasani, M.; Shahsavari, Z. Positive correlation between interleukin-1 receptor antagonist gene 86bp VNTR polymorphism and colorectal cancer susceptibility: A case-control study. Immunol. Res. 2019, 67, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Landi, S.; Bottari, F.; Gemignani, F.; Gioia-Patricola, L.; Guino, E.; Osorio, A.; de Oca, J.; Capella, G.; Canzian, F.; Moreno, V.; et al. Interleukin-4 and interleukin-4 receptor polymorphisms and colorectal cancer risk. Eur. J. Cancer 2007, 43, 762–768. [Google Scholar] [CrossRef]
- Lee, Y.S.; Choi, H.B.; Lee, I.K.; Kim, T.G.; Oh, S.T. Association between interleukin-4R and TGF-beta1 gene polymorphisms and the risk of colorectal cancer in a Korean population. Colorectal Dis. Off. J. Assoc. Coloproctol. Great Br. Irel. 2010, 12, 1208–1212. [Google Scholar]
- Cozar, J.M.; Romero, J.M.; Aptsiauri, N.; Vazquez, F.; Vilchez, J.R.; Tallada, M.; Garrido, F.; Ruiz-Cabello, F. High incidence of CTLA-4 AA (CT60) polymorphism in renal cell cancer. Hum. Immunol. 2007, 68, 698–704. [Google Scholar] [CrossRef]
- Suchy, J.; Klujszo-Grabowska, E.; Kladny, J.; Cybulski, C.; Wokolorczyk, D.; Szymanska-Pasternak, J.; Kurzawski, G.; Scott, R.J.; Lubinski, J. Inflammatory response gene polymorphisms and their relationship with colorectal cancer risk. BMC Cancer 2008, 8, 112. [Google Scholar] [CrossRef] [Green Version]
- Wilkening, S.; Tavelin, B.; Canzian, F.; Enquist, K.; Palmqvist, R.; Altieri, A.; Hallmans, G.; Hemminki, K.; Lenner, P.; Forsti, A. Interleukin promoter polymorphisms and prognosis in colorectal cancer. Carcinogenesis 2008, 29, 1202–1206. [Google Scholar] [CrossRef]
- Shamoun, L.; Skarstedt, M.; Andersson, R.E.; Wagsater, D.; Dimberg, J. Association study on IL-4, IL-4Ralpha and IL-13 genetic polymorphisms in Swedish patients with colorectal cancer. Clin. Chim. Acta 2018, 487, 101–106. [Google Scholar] [CrossRef]
- Yu, Y.; Zhou, J.; Gong, C.; Long, Z.; Tian, J.; Zhu, L.; Li, J.; Yu, H.; Wang, F.; Zhao, Y. Dietary factors and microRNA-binding site polymorphisms in the IL13 gene: Risk and prognosis analysis of colorectal cancer. Oncotarget 2017, 8, 47379–47388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Zheng, S.; Zhang, S.; Jin, W.; Liu, H.; Jin, M.; Chen, Z.; Ding, Z.; Wang, L.; Chen, K. Polymorphisms of inflammation-related genes and colorectal cancer risk: A population-based case-control study in China. Int. J. Immunogenet. 2014, 41, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Marques, D.; Ferreira-Costa, L.R.; Ferreira-Costa, L.L.; Correa, R.D.S.; Borges, A.M.P.; Ito, F.R.; Ramos, C.C.O.; Bortolin, R.H.; Luchessi, A.D.; Ribeiro-Dos-Santos, A.; et al. Association of insertion-deletions polymorphisms with colorectal cancer risk and clinical features. World J. Gastroenterol. 2017, 23, 6854–6867. [Google Scholar] [CrossRef] [PubMed]
- Walczak, A.; Przybylowska, K.; Dziki, L.; Sygut, A.; Chojnacki, C.; Chojnacki, J.; Dziki, A.; Majsterek, I. The lL-8 and IL-13 gene polymorphisms in inflammatory bowel disease and colorectal cancer. DNA Cell Biol. 2012, 31, 1431–1438. [Google Scholar] [CrossRef] [Green Version]
- Zamora-Ros, R.; Shivappa, N.; Steck, S.E.; Canzian, F.; Landi, S.; Alonso, M.H.; Hebert, J.R.; Moreno, V. Dietary inflammatory index and inflammatory gene interactions in relation to colorectal cancer risk in the Bellvitge colorectal cancer case-control study. Genes Nutr. 2015, 10, 447. [Google Scholar] [CrossRef] [Green Version]
- Wen, X.; Xin, X.; Li, J.; Qiao, L.; Liu, F.; Guo, Y.; Qu, Z.; Wang, R.; Li, X. The correlation between IL-4 polymorphisms and colorectal cancer risk in a population in Northwest China. Eur. J. Cancer Prev. 2020, 29, 95–99. [Google Scholar] [CrossRef]
- Burada, F.; Dumitrescu, T.; Nicoli, R.; Ciurea, M.E.; Rogoveanu, I.; Ioana, M. Cytokine promoter polymorphisms and risk of colorectal cancer. Clin. Lab. 2013, 59, 773–779. [Google Scholar] [CrossRef]
- Yannopoulos, A.; Nikiteas, N.; Chatzitheofylaktou, A.; Tsigris, C. The (-590 C/T) polymorphism in the interleukin-4 gene is associated with increased risk for early stages of corolectal adenocarcinoma. In Vivo 2007, 21, 1031–1035. [Google Scholar]
- Talseth, B.A.; Meldrum, C.; Suchy, J.; Kurzawski, G.; Lubinski, J.; Scott, R.J. Lack of association between genetic polymorphisms in cytokine genes and disease expression in patients with hereditary non-polyposis colorectal cancer. Scand J. Gastroenterol. 2007, 42, 628–632. [Google Scholar] [CrossRef]
- Sainz, J.; Rudolph, A.; Hoffmeister, M.; Frank, B.; Brenner, H.; Chang-Claude, J.; Hemminki, K.; Forsti, A. Effect of type 2 diabetes predisposing genetic variants on colorectal cancer risk. J. Clin. Endocrinol. Metab. 2012, 97, E845–E851. [Google Scholar] [CrossRef] [Green Version]
- Xiao, L.; Yu, X.; Zhang, R.; Chang, H.; Xi, S.; Xiao, W.; Zeng, Z.; Zhang, H.; Xu, R.; Gao, Y. Can an IL13 -1112 C/T (rs1800925) polymorphism predict responsiveness to neoadjuvant chemoradiotherapy and survival of Chinese Han patients with locally advanced rectal cancer? Oncotarget 2016, 7, 34149–34157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho-Pun-Cheung, A.; Assenat, E.; Bascoul-Mollevi, C.; Bibeau, F.; Boissiere-Michot, F.; Thezenas, S.; Cellier, D.; Azria, D.; Rouanet, P.; Senesse, P.; et al. A large-scale candidate gene approach identifies SNPs in SOD2 and IL13 as predictive markers of response to preoperative chemoradiation in rectal cancer. Pharm. J. 2011, 11, 437–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Wang, B.; Chai, Y.; Chen, J. Estimation of associations between 10 common gene polymorphisms and gastric cancer: Evidence from a meta-analysis. J. Clin. Pathol. 2020, 73, 318–321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xie, D.; Zhou, H.; Fan, R.; Zhang, L.; Li, C.; Jin, S.; Meng, Q.; Lu, J. The -590C/T polymorphism in the IL-4 gene and the risk of cancer: A meta-analysis. Tumour Biol. 2013, 34, 2261–2268. [Google Scholar] [CrossRef]
- Cho, Y.A.; Kim, J. Association of IL4, IL13, and IL4R polymorphisms with gastrointestinal cancer risk: A meta-analysis. J. Epidemiol. 2017, 27, 215–220. [Google Scholar] [CrossRef]
- Sun, Z.; Cui, Y.; Jin, X.; Pei, J. Association between IL-4 -590C>T polymorphism and gastric cancer risk. Tumour Biol. 2014, 35, 1517–1521. [Google Scholar] [CrossRef]
- Wang, T.; Tian, L.; Gao, M.; Song, H.; Wei, Y.; Xue, Y. Interleukin (IL)-4 -590C>T polymorphism is not associated with the susceptibility of gastric cancer: An updated meta-analysis. Ann. Med. Surg. 2016, 9, 1–5. [Google Scholar] [CrossRef]
- Zhang, C.; Huang, J.Y.; He, Z.Q.; Weng, H. Genetic association between interluekin-4 rs2243250 polymorphism and gastric cancer susceptibility: Evidence based on a meta-analysis. Onco Targets Ther. 2016, 9, 2403–2408. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Xie, X.; Shi, X.; Li, S. Associations of common IL-4 gene polymorphisms with cancer risk: A meta-analysis. Mol. Med. Rep. 2017, 16, 1927–1945. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xu, Y.; Wang, Y.; Yao, Y.; Yang, J. Associations between interleukin gene polymorphisms and the risk of gastric cancer: A meta-analysis. Clin. Exp. Pharmacol. Physiol. 2018, 45, 1236–1244. [Google Scholar] [CrossRef]
- Loh, M.; Koh, K.X.; Yeo, B.H.; Song, C.M.; Chia, K.S.; Zhu, F.; Yeoh, K.G.; Hill, J.; Iacopetta, B.; Soong, R. Meta-analysis of genetic polymorphisms and gastric cancer risk: Variability in associations according to race. Eur. J. Cancer 2009, 45, 2562–2568. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Yamaoka, Y.; Furuta, T. Influence of interleukin polymorphisms on development of gastric cancer and peptic ulcer. World J. Gastroenterol. 2010, 16, 1188–1200. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Hu, J.; Liu, B.; Tao, Y.; Zhou, X.; Yuan, X. Lack of association between interleukin-4 -524C>T polymorphism and colorectal cancer susceptibility. Tumour Biol. 2014, 35, 3657–3662. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, Q.; Xu, X.; Ren, S.; Wang, L. Association between IL-4 -589C>T polymorphism and colorectal cancer risk. Tumour Biol. 2014, 35, 2675–2679. [Google Scholar] [CrossRef] [PubMed]
- Hallett, M.A.; Venmar, K.T.; Fingleton, B. Cytokine stimulation of epithelial cancer cells: The similar and divergent functions of IL-4 and IL-13. Cancer Res. 2012, 72, 6338–6343. [Google Scholar] [CrossRef] [Green Version]
- Prokopchuk, O.; Liu, Y.; Henne-Bruns, D.; Kornmann, M. Interleukin-4 enhances proliferation of human pancreatic cancer cells: Evidence for autocrine and paracrine actions. Br. J. Cancer 2005, 92, 921–928. [Google Scholar] [CrossRef] [Green Version]
- Ito, S.E.; Shirota, H.; Kasahara, Y.; Saijo, K.; Ishioka, C. IL-4 blockade alters the tumor microenvironment and augments the response to cancer immunotherapy in a mouse model. Cancer Immunol. Immunother. 2017, 66, 1485–1496. [Google Scholar] [CrossRef]
- Rubin, J.T.; Lotze, M.T. Acute gastric mucosal injury associated with the systemic administration of interleukin-4. Surgery 1992, 111, 274–280. [Google Scholar]
- Braddock, M.; Hanania, N.A.; Sharafkhaneh, A.; Colice, G.; Carlsson, M. Potential Risks Related to Modulating Interleukin-13 and Interleukin-4 Signalling: A Systematic Review. Drug Saf. 2018, 41, 489–509. [Google Scholar] [CrossRef] [Green Version]
- Debinski, W.; Puri, R.K.; Kreitman, R.J.; Pastan, I. A wide range of human cancers express interleukin 4 (IL4) receptors that can be targeted with chimeric toxin composed of IL4 and Pseudomonas exotoxin. J. Biol. Chem. 1993, 268, 14065–14070. [Google Scholar] [CrossRef]
- Shimamura, T.; Royal, R.E.; Kioi, M.; Nakajima, A.; Husain, S.R.; Puri, R.K. Interleukin-4 cytotoxin therapy synergizes with gemcitabine in a mouse model of pancreatic ductal adenocarcinoma. Cancer Res. 2007, 67, 9903–9912. [Google Scholar] [CrossRef] [Green Version]
- Ishige, K.; Shoda, J.; Kawamoto, T.; Matsuda, S.; Ueda, T.; Hyodo, I.; Ohkohchi, N.; Puri, R.K.; Kawakami, K. Potent in vitro and in vivo antitumor activity of interleukin-4-conjugated Pseudomonas exotoxin against human biliary tract carcinoma. Int. J. Cancer 2008, 123, 2915–2922. [Google Scholar] [CrossRef]
- Debinski, W.; Obiri, N.I.; Powers, S.K.; Pastan, I.; Puri, R.K. Human glioma cells overexpress receptors for interleukin 13 and are extremely sensitive to a novel chimeric protein composed of interleukin 13 and pseudomonas exotoxin. Clin. Cancer Res. 1995, 1, 1253–1258. [Google Scholar] [PubMed]
- NCT00014677. Available online: https://clinicaltrials.gov/ct2/show/NCT00014677 (accessed on 1 August 2020).
- Kunwar, S.; Chang, S.; Westphal, M.; Vogelbaum, M.; Sampson, J.; Barnett, G.; Shaffrey, M.; Ram, Z.; Piepmeier, J.; Prados, M.; et al. Phase III randomized trial of CED of IL13-PE38QQR vs Gliadel wafers for recurrent glioblastoma. Neuro-Oncology 2010, 12, 871–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Horibe, T.; Kohno, M.; Haramoto, M.; Ohara, K.; Puri, R.K.; Kawakami, K. Targeting interleukin-4 receptor alpha with hybrid peptide for effective cancer therapy. Mol. Cancer Ther. 2012, 11, 235–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.Y.; Wong, T.T.; Teng, M.C.; Liu, R.S.; Lu, M.; Liang, H.F.; Wei, M.C. Focused ultrasound and interleukin-4 receptor-targeted liposomal doxorubicin for enhanced targeted drug delivery and antitumor effect in glioblastoma multiforme. J. Control. Release 2012, 160, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Ulasov, I.V.; Tyler, M.A.; Han, Y.; Glasgow, J.N.; Lesniak, M.S. Novel recombinant adenoviral vector that targets the interleukin-13 receptor alpha2 chain permits effective gene transfer to malignant glioma. Hum. Gene Ther. 2007, 18, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Balyasnikova, I.V.; Wainwright, D.A.; Solomaha, E.; Lee, G.; Han, Y.; Thaci, B.; Lesniak, M.S. Characterization and immunotherapeutic implications for a novel antibody targeting interleukin (IL)-13 receptor alpha2. J. Biol. Chem. 2012, 287, 30215–30227. [Google Scholar] [CrossRef] [Green Version]
- Pollack, I.F.; Jakacki, R.I.; Butterfield, L.H.; Hamilton, R.L.; Panigrahy, A.; Potter, D.M.; Connelly, A.K.; Dibridge, S.A.; Whiteside, T.L.; Okada, H. Antigen-specific immune responses and clinical outcome after vaccination with glioma-associated antigen peptides and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose in children with newly diagnosed malignant brainstem and nonbrainstem gliomas. J. Clin. Oncol. 2014, 32, 2050–2058. [Google Scholar]
- Pollack, I.F.; Jakacki, R.I.; Butterfield, L.H.; Hamilton, R.L.; Panigrahy, A.; Normolle, D.P.; Connelly, A.K.; Dibridge, S.; Mason, G.; Whiteside, T.L.; et al. Immune responses and outcome after vaccination with glioma-associated antigen peptides and poly-ICLC in a pilot study for pediatric recurrent low-grade gliomas. Neuro-Oncology 2016, 18, 1157–1168. [Google Scholar] [CrossRef]
- NCT01280552. Available online: http://clinicaltrials.gov/show/NCT01280552 (accessed on 1 August 2020).
- Kong, S.; Sengupta, S.; Tyler, B.; Bais, A.J.; Ma, Q.; Doucette, S.; Zhou, J.; Sahin, A.; Carter, B.S.; Brem, H.; et al. Suppression of human glioma xenografts with second-generation IL13R-specific chimeric antigen receptor-modified T cells. Clin. Cancer Res. 2012, 18, 5949–5960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terabe, M.; Matsui, S.; Noben-Trauth, N.; Chen, H.; Watson, C.; Donaldson, D.D.; Carbone, D.P.; Paul, W.E.; Berzofsky, J.A. NKT cell-mediated repression of tumor immunosurveillance by IL-13 and the IL-4R-STAT6 pathway. Nat. Immunol. 2000, 1, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [Green Version]
- Pitt, J.M.; Marabelle, A.; Eggermont, A.; Soria, J.C.; Kroemer, G.; Zitvogel, L. Targeting the tumor microenvironment: Removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 2016, 27, 1482–1492. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coussens, L.M.; Zitvogel, L.; Palucka, A.K. Neutralizing tumor-promoting chronic inflammation: A magic bullet? Science 2013, 339, 286–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santoni, M.; Massari, F.; Amantini, C.; Nabissi, M.; Maines, F.; Burattini, L.; Berardi, R.; Santoni, G.; Montironi, R.; Tortora, G.; et al. Emerging role of tumor-associated macrophages as therapeutic targets in patients with metastatic renal cell carcinoma. Cancer Immunol. Immunother. 2013, 62, 1757–1768. [Google Scholar] [CrossRef]
- Zhao, G.; Liu, L.; Peek, R.M., Jr.; Hao, X.; Polk, D.B.; Li, H.; Yan, F. Activation of Epidermal Growth Factor Receptor in Macrophages Mediates Feedback Inhibition of M2 Polarization and Gastrointestinal Tumor Cell Growth. J. Biol. Chem. 2016, 291, 20462–20472. [Google Scholar] [CrossRef] [Green Version]
| Cell Lines | Medium | Source | Concentration | Time | Type of Assay | Proliferation | Reference |
|---|---|---|---|---|---|---|---|
| HT-29, DLD-1 | McCoy’s 5A + 10% FBS | R&D systems (Minneapolis, MN, USA) | 50 ng/mL | 1–4 days | Cell counting | ↑ | [12] |
| HT-29, HCT 116 | DMEM + 10% FCS | Sigma (St Louis, MO, USA) | 0–50 ng/mL | 24 h | MTT assay | ↑ | [68] |
| HT-29, WiDr | McCoy’s 5A + 10% FBS; DMEM+ 10% FBS | Schering-Plow Research Institute (Kenilworth, NJ, USA) | 0–50 ng/mL for HT-29, 0–100 ng/mL for WiDr | 3 days | [3H]thymidine incorporation assay | ↓ | [69] |
| HT-29, WiDr | EMEM with amino acids, 25 mM HEPES, and 0.5% FBS | Schering Corp. (Kenilworth, NJ, USA) | NA | 3 days | [3H]thymidine uptake studies | ↓ | [86] |
| Colo205 | RPMI-1640 + 10% FCS, 2 mM glutamine, PS and 5 × 10–5 M 2-ME | Pharmingen (San Diego, CA, USA) | 1–100 U/mL | 1–6 days | Trypan blue viability cell counting | NS | [84] |
| HT-29 | DMEM/F-12 + 2 mM L-glutamine (serum-free) | Biermann | 100 U/mL | 48 h | [3H]thymidine incorporation assay | NS | [87] |
| SW620 | RPMI 1640 + 10% FBS and antibiotics penicillin/streptomycin | Sigma (St. Louis, MO, USA) | 1–100 ng/mL | 6 h | Colorimetric method using the CellTiter 96 AQueous One Solution Assay | NS | [80] |
| HT-29 | DMEM + 1% FBS | Immunex Corp. (Seattle, WA, USA) | 1–20 nM (about 14–280 ng/mL) | 1–5 days | Cell counting, MTT assay | ↓ | [70] |
| SW1222, HT-29 | RPMI 1640 + 10% FCS + sodium bicarbonate 2 g/L, sodium pyruvate 2 mM, PS and L-glutamine 1 mM | Genzyme (West Malling, UK) | 1–100 U/mL | 48 h | Liquid scintillation counting using [3H]-TdR | ↓ | [72] |
| HT-29, WiDr, SW1116, Co-115, LS411N, LS513 and LS1034 cells | 1:1 mixture of DMEM and Ham’s F-12 + HEPES (10 mM), L-glutamine (2 mM), PS +1%FBS | Genzyme (Cambridge, MA, USA) | multiple concentrations including 100 U/mL (1 ng/mL) | 6 days | MTT assay and incorporation of tritiated thymidine. | In HT29, WiDr, LS411N, LS513, LS1034 cells, ↓; in CO-115 and SW1116, NS. | [65] |
| LS513 | The same as the above line | Schering Plough (Dardilly, France) | 0–10 nM (about 0–140 ng/mL) | 6 days | Liquid scintillation counting using [3H]-TdR | ↓ | [73] |
| HTB 38 | NA | Immunex Corp. (Seattle, WA, USA) | 0.01–10 ng/mL | 10 days | Human tumor cloning assay | ↓ | [71] |
| First Author (Year) | Number of Studies/Articles | SNPs | Result |
|---|---|---|---|
| Zongjing Xie (2019) [163] | 18 | polymorphisms in IL-4 | No significant association was found between polymorphisms in IL-4 and GC in combined analyses. |
| Jie Zhang (2013) [164] | 8 studies about GC, 3 studies about CRC | IL-4 -590C>T (rs2243250) | No significant association was found in GC and CRC. |
| Young Ae Cho (2017) [165] | 27 | IL-4: rs2243250, rs2070874; IL-13: rs1800925, rs20541; IL-4R: rs1805010, rs1801275. | The IL-4 rs2070874 T allele was associated with an increased risk of gastrointestinal cancer. The IL-4R rs1801275 heterozygote was associated with a reduced risk of gastrointestinal cancer. |
| Sun Z (2014) [166] | 7 | IL-4 -590C>T (rs2243250) | IL-4 -590C>T polymorphism was associated with a lower GC risk under dominant model and allelic model in Caucasians. |
| Tie Wang (2016) [167] | 9 | IL-4 -590C>T (rs2243250) | IL-4 -590C>T polymorphism was not associated with the susceptibility of GC. |
| Zhang C (2016) [168] | 11 | IL-4 -590C>T (rs2243250) | IL-4 rs2243250 polymorphism was not associated with GC susceptibility. |
| Jia Y (2017) [169] | 7 studies about GC, 4 studies about CRC for rs2243250; 2 studies about GC for rs2070874; 2 studies about GC for rs79071878. | IL-4: rs2243250, rs2070874, rs79071878 | rs2243250 polymorphism was found to be associated with an increased risk of GC. |
| Liu Y (2018) [170] | 3 studies about GC for rs2227284; 2 studies about GC for rs2243248; 16 studies about GC for rs2243250; | IL-4 -33T>C (rs2227284); IL-4 -1098T>G (rs2243248); IL-4 -590C>T (rs2243250) | IL-4 rs2243250 polymorphisms was associated with elevated GC risk in Asians. |
| Loh M (2009) [171] | 203 | 225 polymorphisms across 95 genes, including IL-4 -590C>T | IL-4 -590C>T displayed conflicting effects between Asian and Caucasian populations in GC. |
| Mitsushige Sugimoto (2010) [172] | 5 | IL-4 -590C>T | The risk of gastric non-cardia cancer development was significantly associated with IL-4-590 T allele carrier status. |
| Huanlei Wu (2014) [173] | 5 | IL-4 -524C>T | IL-4 -524C>T polymorphism was not associated with an increased CRC susceptibility. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Traub, B.; Shi, J.; Kornmann, M. Possible Roles of Interleukin-4 and -13 and Their Receptors in Gastric and Colon Cancer. Int. J. Mol. Sci. 2021, 22, 727. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22020727
Song X, Traub B, Shi J, Kornmann M. Possible Roles of Interleukin-4 and -13 and Their Receptors in Gastric and Colon Cancer. International Journal of Molecular Sciences. 2021; 22(2):727. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22020727
Chicago/Turabian StyleSong, Xujun, Benno Traub, Jingwei Shi, and Marko Kornmann. 2021. "Possible Roles of Interleukin-4 and -13 and Their Receptors in Gastric and Colon Cancer" International Journal of Molecular Sciences 22, no. 2: 727. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22020727





